Abstract
Polarizing effects of productive dendritic cell (DC)–T-cell interactions on DC cytoskeleton have been known in some detail, but the effects on DC membrane have been studied to a lesser extent. We found that T-cell incubation led to DC elongation and segregation of characteristic DC veils to the broader pole of the cell. On the opposite DC pole, we observed a novel membrane feature in the form of bundled microvilli. Each villus was approximately 100 nm in diameter and 600 to 1200 nm long. Microvilli exhibited high density of antigen-presenting molecules and costimulatory molecules and provided the physical basis for the multifocal immune synapse we observed during human DC and T-cell interactions. T cells preferentially bound to this site in clusters often contained both CD4+ and CD8+ T cells.
Introduction
Dendritic cells (DCs) are sentinels of immunity as they respond to environmental cues resulting in antigen uptake and processing and stimulation of antigen-specific T cells.1,2 In addition, DCs activate the cells of innate immunity, eg, natural killer (NK) cells, NK T cells, and neutrophils.3 Langerhans cells, the DCs predominantly located in the skin, were originally discovered because of their unique morphology of large neuron-like dendrites. These cells were recognized for their myeloid origin, role in lymphocyte activation,4-8 and characteristic translucent membrane veils.5,9 Similar to Langerhans cells in situ, DCs matured from monocytes in vitro display analogous membrane veils.5,9
T cells are activated through cell-cell contact and by soluble molecules. Early response to cell-cell contact includes the formation of a large macromolecular membrane assembly: the immunologic synapse10,11 and the consequent cell polarization.12,13 The initial idea that immunologic synapses mediate T-cell activation has been challenged by the finding that T-cell receptor (TCR)–mediated signaling precedes or is independent of synapse formation.14-17 Consequently, the full function of the synapse has been modified to include directed cytokine secretion, enhanced secondary signals, or internalization of TCRs.18-20 Most studies of the synapse have focused on those formed between T cells and B cells21-25 ; consequently, the synapse between T cells and DCs has been studied to a much lesser extent. In contrast to the unifocal T-cell–B-cell synapse, the structure of the DC–T-cell interaction site, studied mostly in murine cells, appears “immature” or “multifocal,” containing numerous segregated microdomains.26 Microdomain segregation could be important for the physical separation of ligands and receptors in the membrane plane; such separation appears essential in preventing the cis-acting inhibition, such as that observed within the Notch family ligands and receptors during DC–T-cell interaction.27 In addition, recent evidence suggests that mechanical TCR entrapment into segregated microdomains of the synapse is required for optimal T-cell activation.28 Even though changes in membrane structure have been implicated in the formation of multifocal synapses,29 no available model of membrane organization explains how segregation develops and is maintained. The multifocal character of the synapse, the requirement for active membrane segregation, the formation of the DC–T-cell synapse even in the absence of antigen,30 and the unique DC role in immunity prompted us to investigate the changes in membrane structure in the course of interactions of human DCs with human T cells.
Methods
Cell isolation and culture
Cells were obtained from the blood of normal healthy donors at the Mayo Clinic Division of Transfusion Medicine according to institutional guidelines and informed consent in accordance with the Declaration of Helsinki. Peripheral blood mononuclear cells were isolated from buffy coats or from apheresis leukoreduction system chambers31 by buoyant density separation using Lymphoprep separation medium (ICN, Aurora, OH) according to the manufacturer's instructions. Cells were counted, assayed for viability by trypan-blue exclusion, and isolated by CD14-specific immunoadsorption (CD14-specfic microbeads; Miltenyi Biotec, Auburn, CA). All cytokines were from R&D Systems (Minneapolis, MN) except granulocyte-macrophage colony-stimulating factor (GM-CSF) that was from Berlex Laboratories (Berkeley, CA). DCs were cultured according to a commonly used DC generation protocol32 with minor modifications. Briefly, CD14+ cells were plated at 2 × 106 per mL X-VIVO 15 medium (Cambrex, Walkersville, MD) containing 1.0% human AB serum (Sigma-Aldrich, St Louis, MO), GM-CSF (800 IU/mL), interleukin-4 (IL-4; 1000 IU/mL), penicillin (100 U/mL), and streptomycin (100 mg/mL) for 5 days. Nonadherent cells were collected, counted, and resuspended at 106 cells/mL in the fresh maturation medium (as stated earlier in this paragraph, with the addition of 1100 IU/mL tumor necrosis factor-α [TNF-α], and 1.0 mg/mL prostaglandin E2; Sigma-Aldrich). After 2 days, the nonadherent mature DCs were collected, counted, and used in experiments. These cells expressed CD54, human leukocyte antigen (HLA) class I and class II molecules, and more than 85% of cells expressed CD80 and CD83.33,34
T cells were isolated by negative immunoadsorption (Pan T Cell Kit; Miltenyi Biotec), pooled from 7 or more normal donors and cryopreserved until use. Thawed T cells were washed, counted, and assessed for viability by trypan blue exclusion and propidium iodide accumulation. Typically, more than 95% of thawed cells were viable and expressed CD3 and TCR.
For assays of influenza-specific T cells, we isolated CD14+ cells (for differentiation into DCs), CD4+ cells, and CD8+ cells from individual buffy coats by positive immunoadsorption (Miltenyi Biotec). All cells were frozen until used. Thawed DCs (106/mL) were “pulsed” with an influenza vaccine (600 ng/mL; Fluvirin, NDC 66521-106-10; Evans Vaccines, Liverpool, United Kingdom) for 2 hours under tissue culture conditions, washed, and irradiated (3000 cGy) before incubation with T cells.
Influenza-specific CD8+ T cells were expanded by 3 one-week incubations at ratios of one vaccine-loaded DC per 5 T cells. To the second and third incubations, we added IL-2 (25 U/mL) and IL-7 (10 ng/mL). Influenza-specific CD4+ T cells were expanded similarly except that the DC to T-cell ratio was 1:10 and IL-6 (1000 U/mL) and IL-12 (10 ng/mL) were included for the first week followed by the change of medium and addition of IL-2 (20 U/mL) and IL-7 (2.0 ng/mL). Antigen specificity of expanded cells was assayed by quantifying interferon-γ (IFN-γ) release by ELISpot assay (eBioscience, San Diego, CA; data not shown).
Sample preparation for microscopy
For confocal microscopy, live cells were stained with the near-red–emitting fluorescent membrane dye dioctadecyl tetramethylindocarbocyanine perchlorate (DiI), far-red-emitting dioctadecyl tetramethyldindocarbocyanine perchlorate (DiD; DiD fluorescence has been pseudocolored blue for easier visualization), or green-emitting cytoplasmic dye 5-chloromethylfluorescein diacetate (CMFDA; all Invitrogen, Carlsbad, CA). T cells (106/mL) in phenol red–free X-VIVO 15 medium were incubated with DiI or DiD at the final dye concentration of 2.0 μM at 37°C for 10 minutes and washed twice with phenol red–free X-VIVO 15 for 5 minutes. CMFDA was added to the medium to the final concentration of 5.0 μM. HLA-DR molecules, CD54 molecules, and CD80 molecules were stained by fluorescently labeled monoclonal antibodies phycoerythrin (PE)–Cy5–anti–HLA-DR (batch 41821; BD Biosciences PharMingen, San Diego, CA), fluorescein isothiocyanate (FITC)–anti-CD54 (clone 8.4A6; Biosource, Camarillo, CA), and PE–anti-CD80 (batch 71524; BD Biosciences PharMingen).
For analysis of shape and T-cell binding, DCs were plated on poly-L-lysine–coated chambered slides (Lab-Tek; EMS, Hatfield, PA). CD4+ T cells were prestained with DiI and CD8+ T cells with DiD. T cells were gently laid over DCs and incubated in 5% CO2 at 37°C for 1 hour. CMFDA was added to the cells, incubated for 20 minutes, and washed out by gentle decanting and reintroduction of the medium. The cells were fixed, washed, and mounted with Prolong Gold antifade reagent (Molecular Probes, Eugene, OR). Before staining for actin and vasodilator-stimulated phosphoprotein (VASP), DCs were incubated with allogeneic T cells at the 1:3 ratio for 2 hours and fixed with 2% paraformaldehyde for 30 minutes. Subsequently, the cells were washed twice with phosphate-buffered saline (PBS; pH 7.4) and refrigerated overnight. Actin-binding phalloidin labeled with Alexa Fluor 488 (Molecular Probes) or mouse antihuman VASP immunoglobulin G1 (IgG1; BD Biosciences, San Jose, CA) was added according to the manufacturer's instructions, incubated for 40 minutes, and washed. VASP was visualized by Alexa Fluor 488 coupled to goat antimouse serum (Invitrogen). Nuclei were counterstained with Hoechst 33258 (Invitrogen).
For scanning electron microscopy (SEM), the cells were prepared as described previously.35 Briefly, they were plated onto poly-L-lysine–coated slides and fixed by incremental addition of Trump fixative (1.0% glutaraldehyde, 4.0% formaldehyde in 0.1 M phosphate buffer, pH 7.2). The samples were washed with water and dried by successive immersions into increasing concentrations of ethanol (60%-100%) followed by critical-point drying with liquid CO2 in a pressure- and temperature-controlled tank (Ted Pella, Redding, CA). Finally, the cells were sputter-coated with gold/palladium for 15 seconds.35
For transmission electron microscopy (TEM), the cells were fixed in Trump fixative and washed 3 times for 30 minutes in 0.1 M phosphate buffer, pH 7.2. Then, they were embedded in 2.0% agar and incubated in a 1.0% solution of phosphate-buffered osmium tetroxide. After three 30-minute washes in distilled water, the cells were stained en bloc with 2.0% uranyl acetate for 30 minutes at 60°C and washed with 3 changes of distilled water. The bloc was dehydrated by incubating in successively increasing concentrations of ethanol and 100% propylene oxide, embedded in resin, sectioned into 0.10-μm- to 0.12-μm-thick sections, and mounted on nickel grids.
Microscopy
Confocal and differential interference contrast (DIC) images were obtained by an LSM 510 Confocal Laser Scanning microscope (Zeiss, Oberkochen, Germany) using a C-Apochromat 40×/1.20 W Corr objective (unless noted otherwise) with excitation at 488 nm and 568 nm from an ArKr laser and at 633 nm from a HeNe laser. Live cells were incubated in phenol red–free X-VIVO 15 medium (Cambrex, Walkersville, MD) supplemented with 1.0% human AB serum (Sigma-Aldrich, St Louis, MO) in an atmosphere of 5.0% CO2 at 37°C. We recorded the images at the rate of one scan per minute using the inherent Zeiss LSM software and reconstructed video clips at 15 frames per second. SEM images were acquired by a Hitachi S-4700 FE-SEM (Hitachi, Schaumburg, IL) at the accelerating voltage of 5 kV, emission current of 9 to 11 μA, and a working distance of 9 to 12 mm. For TEM images we used a JEOL 1200 EXII electron microscope (JEOL, Tokyo, Japan) operating at 60 kV.
Image processing
We processed images by Adobe Photoshop software (Adobe Systems, San Jose, CA). First, we maximized the tonal range of each RGB image by setting the darkest pixels to black (intensity level, 0) and the lightest pixels to white (intensity level, 255). Background noise was determined as the average signal level in areas outside cells and was subtracted from the overall output. Fluorescence of monoclonal antibody specific for CD83 (a molecule rather evenly distributed on the DC surface) conjugated to the fluorophore used in the particular experiment was used to distinguish specific fluorescence from autofluorescence. The images were not processed further. Time-resolved images were collected into video format with Metamorph software (Molecular Devices, Sunnyvale, CA).
Analysis of cell shape
We used a C-Apochromat 40×/1.20 W Corr objective with the gain and brightness on the color channel set to highlight the cell body and eliminate the fine coloring in the veils. This allowed us to exclude from the image the DC veils and record only cell “bodies.” Then, we applied Metamorph software to analyze the changes in shape of CMFDA-stained DCs induced by binding of nonactivated allogeneic DiI-stained CD4+ T cells and DiD-stained CD8+ T cells. We established color ranges and background intensity thresholds in free DCs, CD4+ T cells, and CD8+ T cells and established size ranges for these cells. We identified each cell perimeter as the largest continuous isoluminous pixel loop and calculated the number of pixels contained within the loop as a measure of the cell's orthogonal projection (“footprint”). Similarly, we quantified the length, width, and location of the cell center. For each DC, we determined the area (A) and perimeter (P) and calculated its Shape Factor (SF = 4πA/P2)36,37 ; the numerical expression of roundness (SF = 1 for a perfect circle; SF = 0 for a straight line). Although the cell orientation can affect the appearance of polarity, the effect of orientation is probably the same in control cells and is thus not expected to influence the magnitude of observed changes.
Analysis of cell-cell binding
We quantified the interactions of influenza vaccine “pulsed” DCs with CD4+ T cells and CD8+ T cells expanded in the presence of the same vaccine. To minimize the possibility that T cells bind underneath the DCs, we added T cells to the previously plated DCs. Cells were incubated on slides, fixed, and optically dissected (by confocal microscopy) into sections less than 3 μm thick. The image was focused at the portion of each DC closest to the objective to detect T cells bound on its sides and upper surface. We identified individual cell boundaries as described in “Analysis of cell shape” and calculated the mean radius of each cell from several measurements taken at different angles around the cell center. Finally, we measured the center-to-center distances for each cell pair (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). To avoid the difficulties of assigning cell boundaries in aggregates of numerous overlapping cells, we analyzed only aggregates of one DC with one or 2 T cells. If the distance between 2 cells was equal to or less than the sum of the mean radii of the measured cells, we considered them bound to each other. From the data obtained from 3 independent experiments, we determined the frequencies of binding of single CD4+ T cells, single CD8+ T cells, and all T cells to single DCs, calculated the expected number of 2 T cells to one DC-binding event, and compared these values with the observed number 2 T cells to one DC event.
Statistical analysis
Statistical significance of differences between groups of data were tested by the 2-sided Student t test for paired samples or χ2 analysis using Prism, version 5.0 software (GraphPad Software, San Diego, CA). The significance level was set at P equals .05.
Results
DC membrane forms veils and bundles of microvilli
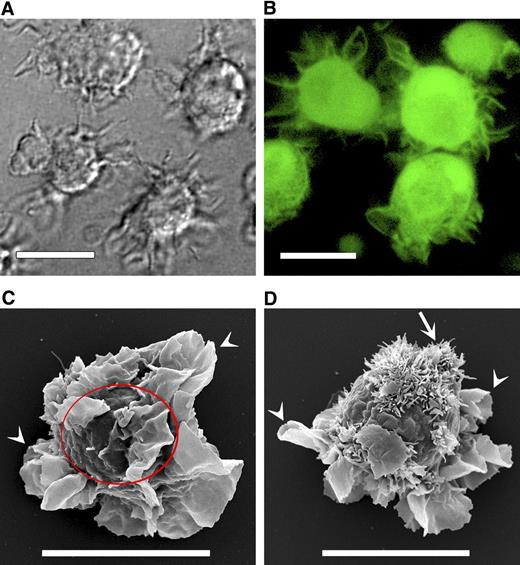
Typical monocyte-derived DCs are characterized by thin flexible membrane extensions known as veils.5,9 The DIC image of a typical human in vitro–matured myeloid DC reveals the veils (Figure 1A) replete with cytoplasm (Figure 1B). The resolution of DIC microscopy limits the appearance of these structures to the long thin extensions or blurred flattened protrusions of the membrane. To overcome these limitations, we used confocal microscopy and SEM for a more detailed characterization of the DC membrane. High-resolution SEM images (Figure 1C,D) show more membrane detail and allow measurements of particular membrane features. For example, from the measurements of 76 veils on 14 typical mature DCs, we calculated the average veil thickness of 130 plus or minus 30 nm and an outer edge to cell surface length of 4.0 plus or minus 1.4 μm (Figure 1C arrowheads). The length measured in fixed cells is comparable with the length assessed in live cells by light microscopy (Figure 1A,B).
DC cell membrane veils and microvilli. (A) Differential interference contrast micrograph of live human mature DCs showing membrane veils. (B) Confocal micrograph of live DCs labeled with the cytoplasmic dye CMFDA to demonstrate cytoplasmic content of the veils (objective: C-Apochromat 63×/1.2 W Corr excitation: 488 nm; emission band pass filter: 505-550 nm). (C,D) Scanning electron micrographs of fixed DCs. Arrowheads indicate veils; arrow, microvilli. The line in panel C circumscribes the DC body (excluding veils). Bars represent 10 μm.
DC cell membrane veils and microvilli. (A) Differential interference contrast micrograph of live human mature DCs showing membrane veils. (B) Confocal micrograph of live DCs labeled with the cytoplasmic dye CMFDA to demonstrate cytoplasmic content of the veils (objective: C-Apochromat 63×/1.2 W Corr excitation: 488 nm; emission band pass filter: 505-550 nm). (C,D) Scanning electron micrographs of fixed DCs. Arrowheads indicate veils; arrow, microvilli. The line in panel C circumscribes the DC body (excluding veils). Bars represent 10 μm.
Figure 1 reveals a distinctive and novel DC feature, the bundle of small villi-like protrusions (Figure 1D arrow). We examined 300 individual DCs and identified cells with the villi unobstructed and clearly visible. The villi were between 0.6 μm and 1.2 μm in length (L) and approximately 0.1 μm in diameter (DV) with a surface area of 0.2 to 0.4 μm2 (πDV(L)); the calculated size of the “footprint” of each villus on the cell surface was 0.008 μm2 (π(DV/2)2). The villi were typically bundled and separated from each other by approximately 0.1 μm (DE). In 300 DCs studied in 10 independent SEM experiments, we observed veiled DCs without the microvilli but never identified a DC exhibiting the microvilli but not veils. Usually, a single bundle was found on any DC, and it was mostly positioned at the narrow end of the egg-shaped DC.
T-cell presence increases DC polarity
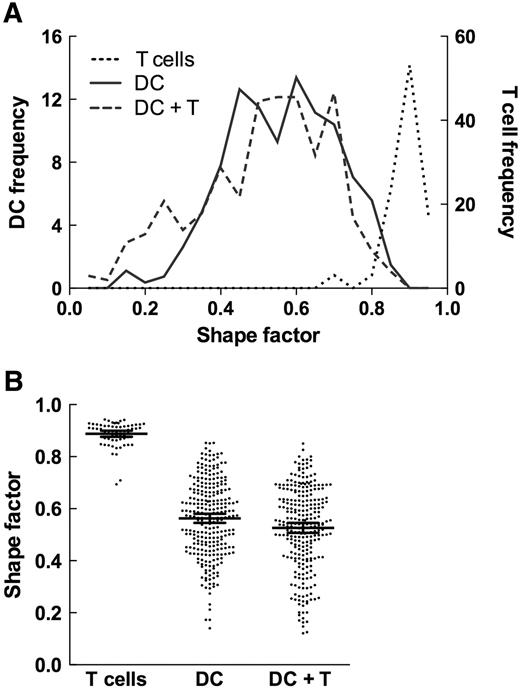
To determine whether T cells affected the shape of live DCs similarly to the changes observed in fixed DCs (Figure 1), we recorded confocal micrographs of live DCs before and after a 20-minute incubation with live allogeneic T cells; this incubation length was selected to allow the completion of any morphologic changes induced by T cells. We analyzed only DCs without bound T cells to determine whether the mere presence of T cells in culture induced changes in DC shape consistent with the SEM observations. We measured the size and shape of the orthogonal projection of each T cell–free DC “body” (defined as cell mass devoid of membrane protrusions > 1.0 μm) and calculated the SF value for each projection; Figure 2). Isolated T cells were characterized by SF = 0.889 plus or minus 0.047 (n = 64), the value identical to other nonactivated hematopoietic cells.37 In the absence of T cells, DCs were not as round as T cells (SF = 0.563 ± 0.15; n = 269, P = .001 relative to T cells), but incubation with allogeneic T cells reduced the SF value to 0.512 plus or minus 0.17 (n = 379, P < .001 relative to DCs incubated without T cells). Although the redistribution of membrane ruffles and veils in the presence of T cells could have affected the values of the shape factor, the high-resolution SEM micrographs support the notion that DC bodies became elongated.
DCs elongate in the presence of T cells. (A) Frequency of T cells, DCs, and T cell–free DCs incubated in the presence of T cells plotted as a function of their SF values. Orthogonal projections of T cell were roughly circular (SF = 0.89 ± 0.05; n = 64), whereas those of DCs were elongated (SF = 0.56 ± 0.17; n = 269; P < .001 compared with T cells). In the presence of T cells, DCs become more elongated (SF = 0.51 ± 0.17; n = 379; P = 0.001 relative to DCs alone). (B) SF values determined for individual cells are plotted for each cell type; mean values plus or minus 95% confidence intervals are indicated.
DCs elongate in the presence of T cells. (A) Frequency of T cells, DCs, and T cell–free DCs incubated in the presence of T cells plotted as a function of their SF values. Orthogonal projections of T cell were roughly circular (SF = 0.89 ± 0.05; n = 64), whereas those of DCs were elongated (SF = 0.56 ± 0.17; n = 269; P < .001 compared with T cells). In the presence of T cells, DCs become more elongated (SF = 0.51 ± 0.17; n = 379; P = 0.001 relative to DCs alone). (B) SF values determined for individual cells are plotted for each cell type; mean values plus or minus 95% confidence intervals are indicated.
DCs capture T cells by veils but retain them at the microvilli
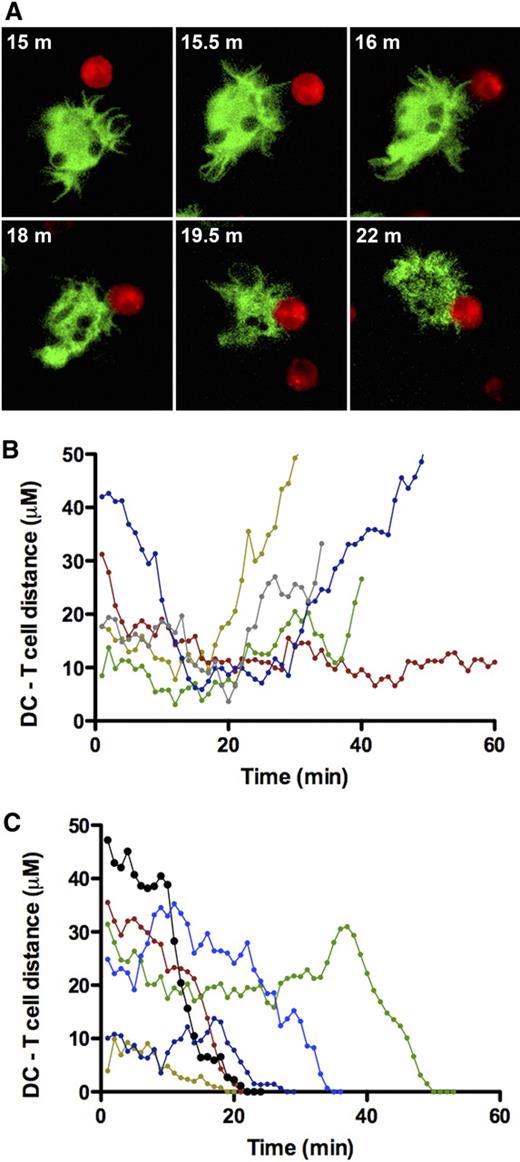
To investigate the role of the veils and microvilli in DC contacts with other cells, we monitored the interactions of DCs with other DCs and with T cells (Figure 3A). In the absence of T cells, the veils made only transient contact with other DCs (Video S1). Similarly, the veils mediated initial contacts with T cells that lasted up to 30 minutes before the T cell either left the DC (Figure 3B) or was attracted closer to the DC body (Figure 3C; Video S2).
DC–T-cell encounters measured in real time. (A) Representative sequence of a time-lapse video recording of a T cell (red) binding to a DC (green), obtained by confocal microscopy with excitation of Dil (red) at 568 nm and CMFDA (green) at 488 nm; m indicates minute. (B,C) Time dependence of the separation of centers of cells in individually monitored DC–T-cell pairs; the sequence in panel A is represented by the pertinent segment of the black line in panel C. (B) DC–T-cell encounters that resulted in separation of cells. (C) DC–T-cell encounters that ended in the formation of a stable DC–T-cell complex.
DC–T-cell encounters measured in real time. (A) Representative sequence of a time-lapse video recording of a T cell (red) binding to a DC (green), obtained by confocal microscopy with excitation of Dil (red) at 568 nm and CMFDA (green) at 488 nm; m indicates minute. (B,C) Time dependence of the separation of centers of cells in individually monitored DC–T-cell pairs; the sequence in panel A is represented by the pertinent segment of the black line in panel C. (B) DC–T-cell encounters that resulted in separation of cells. (C) DC–T-cell encounters that ended in the formation of a stable DC–T-cell complex.
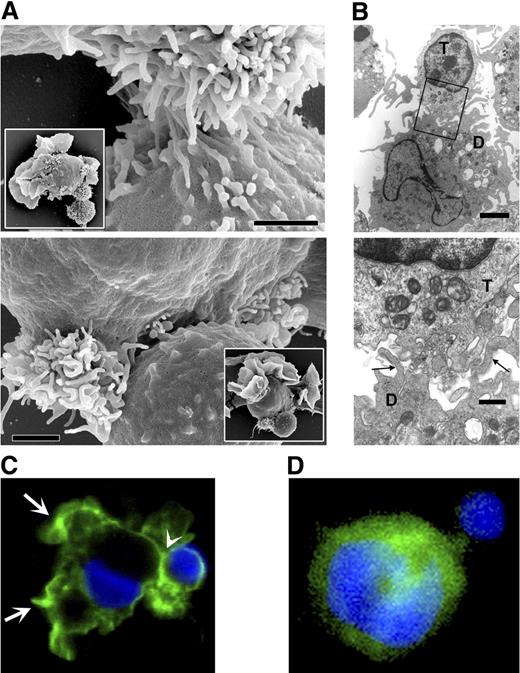
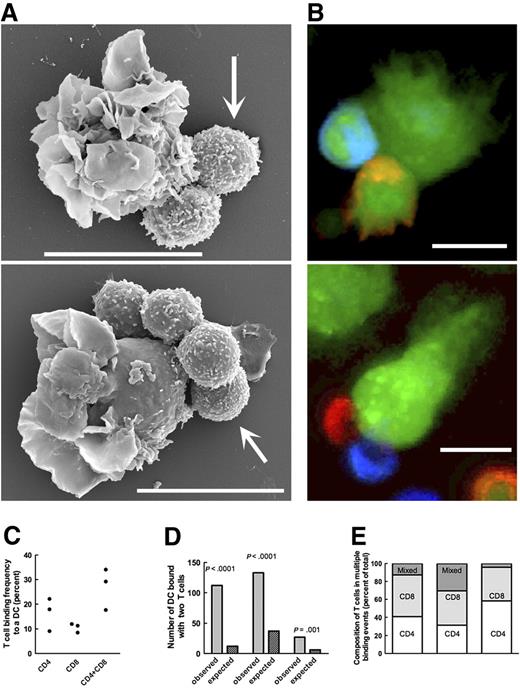
To characterize the site of T-cell retention, we incubated DCs with T cells at a 1:3 ratio for 4 hours before fixation. We analyzed SEM images of 200 DC–T-cell complexes. In 47 complexes, the binding region of both cells was visible (ie, the contact plane was sufficiently aligned with the direction of viewing); in 39 of these cases (82%), the cells interacted exclusively by the microvilli (Figure 4A), whereas other images were inconclusive (microvilli could not be seen). Inspection of TEM images confirmed the presence of villous structures in the space between the interacting DCs and T cells (Figure 4B).
T cells interact with DCs at the microvilli site. (A) Scanning electron micrographs recorded after a 4-hour coincubation of T cells and DCs. (Large panels) T cells bound to bundled DC microvilli. The bundle in the top panel is compact; in the bottom panel it is split, yet the T cell interacts with the microvilli. Bars represent 1 μm. Insets show entire cells demonstrating the disposition of veils and microvilli on DCs. (B) Transmission electron micrograph showing DC protrusions (→) consistent with the sections of microviilli at the DC–T-cell contact. Bars represent 2 μm (top panel) and 1 μm (bottom panel). (C) Distribution of intracellular actin (green, Alexa 488–conjugated phalloidin; see “Sample preparation for microscopy”) in a DC–T-cell pair. Actin density was higher at the perimeter of veils (arrows) and at the T cell–binding site (arrowhead). (D) Distribution of vasodilator-stimulated phosphoprotein (VASP; green, Alexa 488–conjugated to secondary antibody; “Sample preparation for microscopy”) in a DC interacting with a T cell. In panels C and D, nuclei were counterstained with Hoechst 33258 (blue). The T cell can be recognized by the round nucleus, smaller diameter, and lack of membrane extensions. Panels C and D each are composed of 2 stacked adjacent 1.0-μm-thick optical sections.
T cells interact with DCs at the microvilli site. (A) Scanning electron micrographs recorded after a 4-hour coincubation of T cells and DCs. (Large panels) T cells bound to bundled DC microvilli. The bundle in the top panel is compact; in the bottom panel it is split, yet the T cell interacts with the microvilli. Bars represent 1 μm. Insets show entire cells demonstrating the disposition of veils and microvilli on DCs. (B) Transmission electron micrograph showing DC protrusions (→) consistent with the sections of microviilli at the DC–T-cell contact. Bars represent 2 μm (top panel) and 1 μm (bottom panel). (C) Distribution of intracellular actin (green, Alexa 488–conjugated phalloidin; see “Sample preparation for microscopy”) in a DC–T-cell pair. Actin density was higher at the perimeter of veils (arrows) and at the T cell–binding site (arrowhead). (D) Distribution of vasodilator-stimulated phosphoprotein (VASP; green, Alexa 488–conjugated to secondary antibody; “Sample preparation for microscopy”) in a DC interacting with a T cell. In panels C and D, nuclei were counterstained with Hoechst 33258 (blue). The T cell can be recognized by the round nucleus, smaller diameter, and lack of membrane extensions. Panels C and D each are composed of 2 stacked adjacent 1.0-μm-thick optical sections.
The role of actin in DC–T-cell interactions has been extensively documented,38,39 but its role in the maintenance of DC shape is not equally understood. Therefore, we sought to determine the distribution of cytoskeletal actin and of the actin-modulating VASP on DC membranes. VASP is of interest because it participates in anchoring TCRs to actin cytoskeleton,40 maintains cytoskeletal actin at the leading edges of cell movement and at the sites of cell-cell focal adhesion interactions,41,42 and accumulates at the tips of filopodia during their formation.43 As expected,38,39 we found actin concentrated at the edges of DC veils and distally at the site of T-cell binding (Figure 4C). We also found VASP expressed at the DC membrane but concentrated proximally to the T-cell binding site (Figure 4D). Such distribution of the 2 proteins is compatible with VASP association with actin and its accumulation at sites of the cell-cell interaction.
T cells cluster at DC microvilli
Frequently, a single DC bound multiple T cells. When 2 or more T cells were bound to a DC, T cells almost always clustered at the single microvilli bundle (Figure 5A). To determine the mode of T-cell binding into the cluster (ie, whether T cells bind to the same high-avidity DC site), we recorded time-lapse videos of DCs incubated with a 3-fold excess of T cells. We observed that the second T cell bound to a DC at sites distant from the first T cell, but that later the 2 T cells coalesced at a single site (Video S3).
Multiple T cells cluster on DCs. (A) Typical examples of DC–T-cell clusters. Bars represent 10 μm. (B) Confocal images of DC–T-cell clusters used to determine the binding frequency of CD4+ (red, Dil) and CD8+ (blue, DiD) T cells to DCs. DCs were pulsed with an influenza vaccine and coincubated with T cells previously in vitro expanded in the presence of the same vaccine. DCs, CD4+ cells, and CD8+ cells were incubated at the ratio of 1:3:3 for 4 hours when all cells were counterstained with CMFDA (green). Optical sections were 1.5 μm thick. Bars represent 10 μm. (C) Frequencies of single CD4+ cells bound to a DC, of single CD8+ cells bound to a DC, or of a CD4+ cell and a CD8+ cell bound to a single DC. Each dot represents a single experiment. In 3 independent experiments, we analyzed the total of 1070 DCs. The frequency of a CD4+ binding on a DC did not differ from CD8+ binding frequency (P = .2). (D) Numbers of DCs that bound 2 T cells (light columns) compared with the numbers predicted on the assumption of independent T-cell binding (dark columns). Each column pair represents an independent experiment. (E) Relative distribution of DCs in complex with 2 T cells: CD4+/CD4+, CD4+/CD8+, and CD8+/CD8+ observed in 3 independent experiments.
Multiple T cells cluster on DCs. (A) Typical examples of DC–T-cell clusters. Bars represent 10 μm. (B) Confocal images of DC–T-cell clusters used to determine the binding frequency of CD4+ (red, Dil) and CD8+ (blue, DiD) T cells to DCs. DCs were pulsed with an influenza vaccine and coincubated with T cells previously in vitro expanded in the presence of the same vaccine. DCs, CD4+ cells, and CD8+ cells were incubated at the ratio of 1:3:3 for 4 hours when all cells were counterstained with CMFDA (green). Optical sections were 1.5 μm thick. Bars represent 10 μm. (C) Frequencies of single CD4+ cells bound to a DC, of single CD8+ cells bound to a DC, or of a CD4+ cell and a CD8+ cell bound to a single DC. Each dot represents a single experiment. In 3 independent experiments, we analyzed the total of 1070 DCs. The frequency of a CD4+ binding on a DC did not differ from CD8+ binding frequency (P = .2). (D) Numbers of DCs that bound 2 T cells (light columns) compared with the numbers predicted on the assumption of independent T-cell binding (dark columns). Each column pair represents an independent experiment. (E) Relative distribution of DCs in complex with 2 T cells: CD4+/CD4+, CD4+/CD8+, and CD8+/CD8+ observed in 3 independent experiments.
The observation of 2 or more T cells residing on a single DC simultaneously in close proximity to each other raises the possibility that DCs can facilitate contemporaneous interactions with CD4+ and CD8+ T cells, the immunologic equivalent of the “3-body problem” for which a “temporal bridge” solution has been proposed.44,45 Based on the propensity for multiple T-cell binding to DCs, we hypothesized that the binding is cooperative, that is, that the binding of the first T cell to the DC increases the avidity for binding of subsequent T cells to a single DC-binding site. To test this hypothesis, we combined DCs with the separately in vitro–expanded and individually fluorescently labeled CD4+ T cells and CD8+ at the ratio 1:3:3. After 4 hours of incubation, cells were fixed and examined by confocal microscopy (Figure 5B). If T-cell binding to DCs is random (ie, not cooperative), the probability of a single DC binding 2 T cells (p2) is the product of probabilities of a DC binding one T cell (p1), that is, p2 = p12. If the binding is cooperative, p2 > p12, that is, more DCs will bind 2 (or more) T cells than predicted by this calculation.
We found that 27.0% plus or minus 8.5% of DCs bound a single T cell (determined in a total of 1070 DCs in 3 independent experiments; Figure 5C). Based on this finding, we expect that 7.3% (range, 3.4%–12.6%) of DCs would bind 2 T cells. Significantly, counting only 2 T cells bound at a single DC site, we found that 21.6% plus or minus 7.0% of DCs bound 2 T cells (P < .001 for each experiment analyzed separately by χ2 analysis; Figure 5D). Clearly, T-cell binding to DCs is cooperative. When a single T cell bound to a DC, there was no difference between binding of CD4+ cells (16.4% ± 6.6%) and CD8+ cells (10.6% ± 1.9%; P = .22). When CD4+ cells and CD8+ cells were bound to a DC simultaneously, they were found in all possible combinations (CD4+/CD4+, CD4+/CD8+, CD8+/CD8+; Figure 5E). When DCs and T were incubated together for more then 4 hours (the time frame of experiments in this work), T-cell binding continued and eventually resulted in large cell aggregates (data not shown).
The synapse between DCs and T cells contains microdomains
When a T cell interacts with a B cell, antigen-presenting molecules and costimulatory molecules aggregate and organize into the planar immunologic synapse.11,25 The end-stage of synapse formation is characterized by the arrangement of TCRs and signaling molecules (eg, CD4, CD8, CD28) within the “central supramolecular activation cluster” (cSMAC). This cluster is surrounded by the peripheral SMAC (pSMAC) replete with talin, leukocyte function-associated antigen 1, and other molecules.23 Because T cells interact with DCs by way of microvilli (Figures 4, 5), we hypothesized that the spatial organization of antigen-presenting and costimulatory molecules differs from the organization of planar immunologic synapses. Hence, we incubated DCs with allogeneic T cells, fixed the samples, labeled with CMFDA and fluorescent antibodies specific for HLA-DR and CD80, and observed the organization of these molecules on DC surface by confocal microscopy. When DCs bound T cells, we found that HLA-DR and CD80 molecules concentrated at the interaction site (Figure S2).
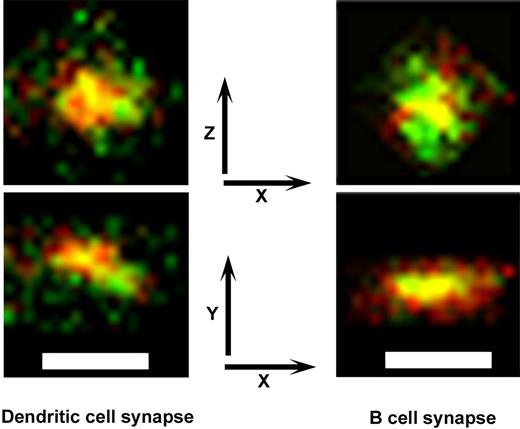
To compare the organization of membrane molecules at synapses that T cells form with DCs or B cells, we analyzed the 3-dimensional distribution of the TCR and CD28 molecules at the DC–T-cell interaction site and compared it with the distribution at the B-cell–T-cell site under conditions leading to the formation of stable pSMACs.25 We optically sectioned the cell-cell interaction sites and reconstructed their images in 3 dimensions; Figure 6 shows the distribution of the TCR and CD28 molecules in the plane of the cell-cell interaction surface (x-z) and the plane perpendicular to that surface (x-y) in the B-cell–T-cell synapse and DC–T-cell synapse. As expected, the B-cell–T-cell synapse was typical in the concentration of T-cell membrane molecules TCR and CD28 within the cSMAC.11,25 However, within the DC–T-cell interaction site, TCR and CD28 molecules were distributed as punctuate aggregates within a larger and deeper space than in the B-cell–T-cell synapse. Thus, the DC–T-cell punctuate synapse appears consistent with the presence of microdomains within the binding site.
Spatial distribution of T-cell molecules in the complex with a DC or a B cell. Antigen-loaded DCs or B cells were incubated with autologous CD4+ T cells (expanded in the presence of an influenza vaccine) for 4 hours; TCR and CD28 were stained (green and red, respectively) and the cells fixed. By confocal optical sectioning (using a C-Apochromat 63×/1.20 W Corr lens) and 3-D reconstruction with Metamorph software we show the plane of contact between 2 cells. The x-y plane is coincidental with the plane of maximum fluorescence; the z-axis is parallel with the line connecting the centers of the T and the DC or B cell. Images are reconstructed in the x-y and x-z planes to compare the size, depth, and distribution of molecules in the respective synapses. Bars represent 1 μm.
Spatial distribution of T-cell molecules in the complex with a DC or a B cell. Antigen-loaded DCs or B cells were incubated with autologous CD4+ T cells (expanded in the presence of an influenza vaccine) for 4 hours; TCR and CD28 were stained (green and red, respectively) and the cells fixed. By confocal optical sectioning (using a C-Apochromat 63×/1.20 W Corr lens) and 3-D reconstruction with Metamorph software we show the plane of contact between 2 cells. The x-y plane is coincidental with the plane of maximum fluorescence; the z-axis is parallel with the line connecting the centers of the T and the DC or B cell. Images are reconstructed in the x-y and x-z planes to compare the size, depth, and distribution of molecules in the respective synapses. Bars represent 1 μm.
HLA-DR and CD54 form small, interspersed aggregates in the DC–T-cell synapse
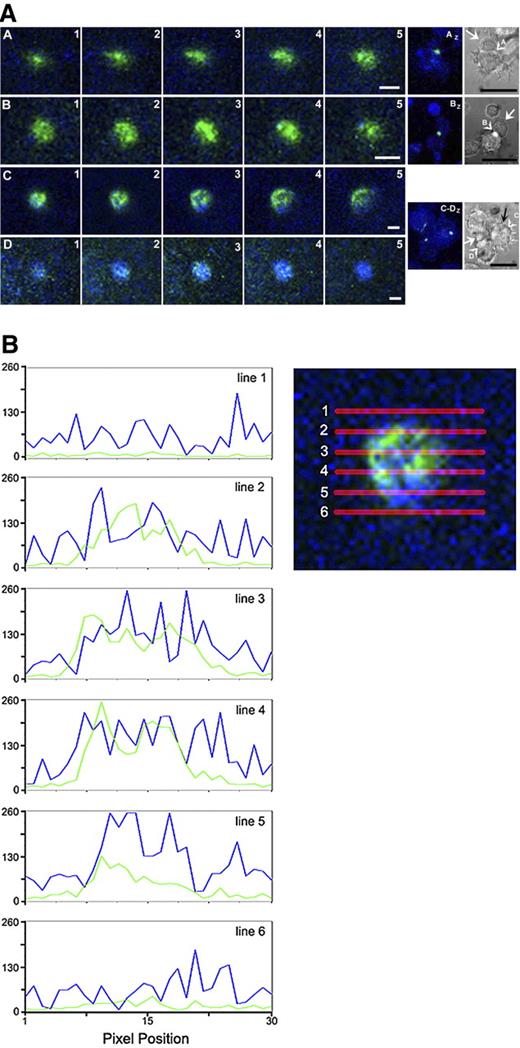
The observation of T-cell membrane microdomains at the synapse with a DC prompted us to explore the distribution of characteristic molecules on the interacting DCs. We “pulsed” the DCs with an influenza vaccine and coincubated with autologous CD4+ cells previously expanded in the presence of the same influenza vaccine. After a 40-minute incubation, the cells were fixed and their HLA-DR and CD54 (ICAM-1) molecules were immunofluorescently labeled; in general, these molecules are concentrated in the cSMAC and pSMAC of the mature immune planar synapse, respectively.26,46 Figure 7A displays the DC–T-cell interaction sites (DIC micrographs right) that we optically dissected in 0.5-μm-thick sections starting from the plane of the slide. Optical sections (rows A-D, panels 1-5) show the distribution of HLA-DR and CD54 molecules at different depths of the cell-cell interaction site. Panels AZ, BZ and CZ, DZ show optical sections 1 to 5 superimposed on the respective DIC image to demonstrate the precise location of optically sectioned sites. Figure 7A shows that, under the conditions otherwise favoring the formation of the planar concentric immune synapse, the molecules were not segregated into a cSMAC (HLA-DR) and a pSMAC (CD54). Rather, HLA-DR and CD54 molecules appeared as aggregates scattered across the synapse; aggregates could be less than 0.2 μm in diameter.
Spatial distribution of DC molecules in the synapse with T cells. (A) DCs were pulsed with an influenza vaccine, combined with autologous, influenza-expanded CD4+ T cells, incubated for 40 minutes, fixed, and labeled. Rows A through D show the contact area (indicated by arrowhead in DIC images at far right) of distinct T cells (indicated by arrows) where they are bound to a DC. T cells C and D are bound to the same DC. Panels 1 to 5: Confocal microscopy of 0.5-μm-thick sections taken in 0.25-μm steps with a C-Apochromat 63×/1.20 W Corr objective; image 1 was closest to the slide. CD54, green; HLA-DR, blue. Panels AZ to DZ are overlays of sections 1 to 5 and the pertinent DIC image. Arrows point to T cells (the black arrow points to a cell underneath the DC). Arrowheads show the contact site analyzed in respective panels 1 to 5. Image resolution was increased 5-fold by linear maximization of the tonal range for each color and by background reduction by equal increase of input levels of shadows for all color channels. Bars in fluorescence images represent 1 μm; bars in DIC images represent 10 μm. (B) Line-by-line fluorescence intensity measured and plotted for each fluorophore in an optical slice through frame C3, panel A (as diagrammed to the right of plots). Intensity distribution along the lines demonstrates the unorganized distribution of synapse molecules in the x-y plane.
Spatial distribution of DC molecules in the synapse with T cells. (A) DCs were pulsed with an influenza vaccine, combined with autologous, influenza-expanded CD4+ T cells, incubated for 40 minutes, fixed, and labeled. Rows A through D show the contact area (indicated by arrowhead in DIC images at far right) of distinct T cells (indicated by arrows) where they are bound to a DC. T cells C and D are bound to the same DC. Panels 1 to 5: Confocal microscopy of 0.5-μm-thick sections taken in 0.25-μm steps with a C-Apochromat 63×/1.20 W Corr objective; image 1 was closest to the slide. CD54, green; HLA-DR, blue. Panels AZ to DZ are overlays of sections 1 to 5 and the pertinent DIC image. Arrows point to T cells (the black arrow points to a cell underneath the DC). Arrowheads show the contact site analyzed in respective panels 1 to 5. Image resolution was increased 5-fold by linear maximization of the tonal range for each color and by background reduction by equal increase of input levels of shadows for all color channels. Bars in fluorescence images represent 1 μm; bars in DIC images represent 10 μm. (B) Line-by-line fluorescence intensity measured and plotted for each fluorophore in an optical slice through frame C3, panel A (as diagrammed to the right of plots). Intensity distribution along the lines demonstrates the unorganized distribution of synapse molecules in the x-y plane.
For a more objective analysis of the distribution of HLA-DR and CD54 at the DC–T-cell interaction site, we quantified the associated fluorescence intensity across the plane perpendicular to the cell-cell interaction surface of the section C3 in Figure 7A. Light intensity profiles of each optical section (Figure 7B) indicate no apparent pattern in the arrangement of molecules within the DC–T-cell interaction site. This finding is in line with evidence that the DC immune synapse is apparently less ordered than the concentrically structured immune synapse observed at the interface of a T cell interacting with a B cell.
Discussion
This study demonstrates that DC veils “capture” T cells, that T cells migrate along the veils, and that some T cells ultimately bind at the newly described DC microvilli. A previous study of DC morphology and polarization did not detect these structures,26 possibly because it analyzed murine cells and probably because it used TEM, a method less discernible of 3-dimensional features of membrane ultrastructure.
Here, we routinely observed polarized DCs with microvilli located opposite to the veils. Our data cannot resolve the mechanistic aspects of microvilli formation, but it is intriguing that microvilli enhances cooperative T cell binding. We hypothesize not only that DC membrane polarization accompanies DC cytoskeleton polarization in the course of the physical contact with T cells,38,39,47-50 but also that polarization and/or significant changes (such as membrane ruffling) in the DC membrane can take place before this contact. These changes are probably mediated by chemokines implicated in induction of cell polarization and in routing the CD8+ cell migration to CD4+ cells bound to DCs.51 The appearance of bundled microvilli increases the surface area available for membrane-membrane contact and concentrates antigen-presenting, costimulatory, and adhesion molecules to densities required for productive T-cell stimulation. Consequently, the immunologic synapse between a DC and a T cell can involve a larger contact area than that of the synapse between a B cell and a T cell.
Molecules participating in antigen presentation and T-cell costimulation aggregate at the site of DC–T-cell contact, but they do not assume the concentric arrangement typical for the immunologic synapse found at the B-cell–T-cell contact. This finding extends a previous observation in murine cells26 to human cells. Whereas SMACs have been reported at the interface of T cells and a DC-like cell line, DCs apparently can stimulate productive T-cell responses in the absence of ordered SMACs.14,15,26,47,48,52,53 Thus, the density of pertinent antigen-presenting and costimulatory molecules in the microvilli could replace the requirement of membrane reorganization that culminates in SMAC formation. Further studies to correlate microvilli interactions and T-cell activation are required to fully test this hypothesis.
In addition to the absence of SMACs at the DC–T-cell interface, DCs differ from B cells in their sensitivity to inhibitors of cytoskeletal organization. The effect of such inhibitors on the ability of B cells to activate T cells is limited,54 but their effect on the same ability of DCs is substantial38,47-50 despite the observation that multifocal DC synapses are formed nonetheless.26 The complete role of cytoskeletal organization in membrane restructuring, microvilli formation, and multifocal synapse assembly and function has yet to be determined.
It has been suggested recently that multifocal clustering of T-cell ligands within the immune synapse enhances T-cell activation.26,28,55 In this case, mechanical restriction of the movement of TCR-containing microclusters leads to prolonged TCR-mediated signaling. In murine cells, DC–T-cell interactions result in a “mature” synapse only rarely; the common outcome is the formation of discrete areas of ligand concentration separated by membrane undulations.26 Our data extend these observations to human cells and provide the first potential mechanism of multifocal concentration. DC microvilli potentially restrict the movement of surface molecules and segregate T-cell ligands into discrete microclusters extending the DC–T-cell interactions in space and time.
We found that the concentration of microvilli in a single DC area enhances T-cell clustering. Clustering is particularly intriguing in the light of current models of cytotoxic T lymphocyte (CTL) stimulation. The prevailing model of T-cell help requires mutual interaction of an antigen-presenting cell, a T helper cell, and a CTL. The low probability of simultaneous random encounters among DCs, CD4+ helper cells, and CD8+ CTL precursors of the same specificity has necessitated the postulate of different models of CTL activation. For example, chemokine gradients direct CD8+ cells to lymph nodes during immunization.51 Likewise, in certain circumstances, DCs can activate naive T cells independently of helper T cells.56 Otherwise, DC can transfer information from previously encountered T-helper cells to CTLs.44,45 Furthermore, CD4+ T cells could acquire molecules from a DC and directly stimulate CD8+ cells.57 Although simultaneous binding of a DC, CD4+ cell, and CD8+ cell may not be highly probable in vivo, our data show that DCs can aggregate T cells at the same binding site and thus increase the likelihood of such 3-cell interaction. Multiple-cell interactions could underlie the T-cell cooperation observed when a polarized CD4+ T cells skews polarization of naive T cells within DC clusters58 or when DCs mediate information transfer between memory T cells and naive T cells44 or between helper T cells and cytotoxic T cells.45
Our data provide evidence for distinct membrane structures participating in the recently defined kinetic steps in T-cell activation by DCs.59,60 The tips of DC veils are enriched in β-integrin,61 which can enhance “probing” of T cells reminiscent of transient T-cell interactions with DC veils followed by long-term (hours) interactions and T-cell clustering.59,60 We show that the bundles of DC microvilli serve as the structural basis for stable DC–T-cell interactions and T-cell clustering on DCs; the surface area of the DC committed to microvilli is limited and is probably related to the DC cytoskeleton polarization during T-cell activation.47,48 Our in vitro data suggest a model in which T cells induce a DC restructuring before binding and that, on binding, the T cell of proper TCR specificity translocates to the high-avidity binding area. Thus, DC veils can enhance the efficiency of probing for epitope specificity, whereas high avidity binding by the microvilli facilitates stable interactions leading to T-cell clustering and subsequent activation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Ranjeny Thomas (University of Queensland, Brisbane, Australia) for helpful discussions.
This work was supported by a generous grant from Mrs Adelyn L. Luther (Singer Island, FL), the Commonwealth Cancer Foundation for Research (Richmond, VA), a Glen and Florence Voyles Foundation Scholarship (Terre Haute, IN; A.B.D.), and the Mayo Clinic Cancer Center.
Authorship
Contribution: P.J.F. and A.B.D. designed the research; P.J.F. and P.A.B. performed experiments; P.J.F., S.V.-P., and A.B.D. analyzed results; and P.J.F., A.B.D., S.V-P., and F.G.P. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Allan B. Dietz, Division of Transfusion Medicine, Mayo Clinic, Rochester, MN 55905; e-mail: dietz.allan@mayo.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal