Abstract
We compared the transcriptomes of marrow-derived mesenchymal stem cells (MSCs) with differentiated adipocytes, osteocytes, and chondrocytes derived from these MSCs. Using global gene-expression profiling arrays to detect RNA transcripts, we have identified markers that are specific for MSCs and their differentiated progeny. Further, we have also identified pathways that MSCs use to differentiate into adipogenic, chondrogenic, and osteogenic lineages. We identified activin-mediated transforming growth factor (TGF)–β signaling, platelet-derived growth factor (PDGF) signaling and fibroblast growth factor (FGF) signaling as the key pathways involved in MSC differentiation. The differentiation of MSCs into these lineages is affected when these pathways are perturbed by inhibitors of cell surface receptor function. Since growth and differentiation are tightly linked processes, we also examined the importance of these 3 pathways in MSC growth. These 3 pathways were necessary and sufficient for MSC growth. Inhibiting any of these pathways slowed MSC growth, whereas a combination of TGF-β, PDGF, and β-FGF was sufficient to grow MSCs in a serum-free medium up to 5 passages. Thus, this study illustrates it is possible to predict signaling pathways active in cellular differentiation and growth using microarray data and experimentally verify these predictions.
Introduction
Mesenchymal stem cells (MSCs) have the capacity to differentiate into cells of connective tissue lineages, including bone, fat, cartilage, and muscle.1,2 In addition, some reports also show their potential to differentiate into ectodermal and endodermal lineages.3,4 MSCs also play a role in providing the stromal support system for hematopoietic stem cells (HSCs) in the marrow.5
MSCs can be obtained in relatively large numbers from a variety of connective tissue sources like adipose6 and dermal tissue,7 synovial fluid,8,9 decidous teeth,10-12 cord blood,13,14 amniotic fluid,15-17 and placenta.18-20 However, the largest body of data available is on postpubertal stromal MSCs,21,22 and these are often used as the prototypical MSC population. Adult bone marrow–derived MSCs have been shown to be easy to cultivate and expand,1 and they maintain pluripotency after prolonged culture, enabling the use of cultured MSCs in autologous and allogeneic transplants.23 MSCs have been used in cardiac repair,24 enhancing marrow engraftment,25 reducing graft-versus-host disease,26,27 and in generating connective tissue elements that may be abnormal.28-32
Despite the therapeutic potential of adult MSC populations, much remains unknown about their growth regulation and markers for isolation. Gene-expression analysis is a valuable tool for answering many of these questions. Early MSC gene-expression studies investigating this issue33-37 have looked at single snapshots of gene expression and therefore do not give any information on the changes occurring in the MSC transcriptome as cells are passaged in culture. In this study we have analyzed the MSC transcriptome to identify transcripts expressed selectively in undifferentiated MSCs, as well their differentiated progeny. From this analysis, we identified 3 pathways important in MSC differentiation into fat, cartilage, and bone and modulating these pathways in vitro also modulates differentiation of MSCs into these 3 lineages. We also experimentally validated these predictions. Further, we show that these 3 pathways are also important in MSC proliferation. This study also shows that it is possible to predict active signaling pathways in differentiating cells using microarray data, and these predictions can be experimentally tested using loss-of-function and gain-of-function studies.
Methods
Cell culture
Human bone marrow–derived MSCs were purchased frozen from Lonza (Gaithersburg, MD) at passage 2. These cells were thawed, cultured for an additional passage (passage 3), and pooled before being used in the experiments. This pool, designated as M1 (Table 1), consisted of cells from 3 donors. The use of pooled samples minimizes donor-specific variation in the microarray data. MSCs were seeded at 3000 cells/cm2 and expanded on tissue culture treated–plastic dishes (Corning Life Sciences, Acton, MA). The MSC culture medium comprised of Dulbecco modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) or MesenPRO medium, 1× GlutaMAX, and 0.5 μg/mL gentamycin (all from Invitrogen). Cells were fed every alternate day and passaged using TryPLE, a recombinant protease (Invitrogen) when 80% confluence was reached. These samples were grown less than a month in culture (passage 3).
Samples used in this study
| Sample ID . | No. of days differentiated . | Lineage . |
|---|---|---|
| M1 | — | Undifferentiated |
| O07 | 7 | Osteogenic |
| A07 | 7 | Adipogenic |
| C07 | 7 | Chondrogenic |
| O14 | 14 | Osteogenic |
| A14 | 14 | Adipogenic |
| C14 | 14 | Chondrogenic |
| Sample ID . | No. of days differentiated . | Lineage . |
|---|---|---|
| M1 | — | Undifferentiated |
| O07 | 7 | Osteogenic |
| A07 | 7 | Adipogenic |
| C07 | 7 | Chondrogenic |
| O14 | 14 | Osteogenic |
| A14 | 14 | Adipogenic |
| C14 | 14 | Chondrogenic |
MSCs from 3 donors at passage 3 and their progeny were used in this study. Samples on the humanRef-8v1 chip were organized such that replicate samples were on different chips. Each chip contained samples of all undifferentiated MSCs and their differentiated progeny, but samples from day 7 and day 14 were on separate chips. In total 6 replicates were hybridized for each sample. Each replicate was hybridized on a separate chip. Having each replicate on a different chip averages factors affecting the microarray signal such as hybridization conditions, scanning conditions, etcetera, and reduce variation from these sources in the final dataset. This design also serves as a good internal control for assessing the reproducibility of the arrays. Undifferentiated MSCs cultured for 7 and 14 days, respectively, were used to compare their transcriptome with their differentiated progeny at these time points.
— indicates not applicable.
Excess cells at passage3 were frozen at a controlled rate in a −70°C freezer using a Mr Frosty (Nalgene Scientific, Rochester, NY) cryobox. The cell stocks were transferred to liquid nitrogen storage after 24 hours. For experiments in serum-free medium, these pooled MSCs were thawed, expanded for 4 days in DMEM supplemented with 10% FBS, harvested upon near confluence (passage 4), and seeded into StemPro serum-free medium (MSC SFM Invitrogen) with or without growth factor supplementation. The StemPro MSC SFM was supplemented with β-FGF, TGF-β1, BB-PDGF, and insulin. These cells were further cultured for 5 passages (passage 5 to passage 9), and MSCs cultured in serum-free medium were seeded in differentiation media.
MSC differentiation
MSCs from cultures mentioned above were expanded in culture and seeded in T125 flasks for differentiation to either adipocytes, osteocytes, or chondrocytes at different passages. Cells were also seeded in parallel on chamber slides for differentiation into ostegenic and adipogenic lineages and were stained with Von Kossa and Oil red O, respectively.
MSCs were seeded at a density of 20 000 cells/cm2 in the presence of adipogeneic differentiation media containing 0.5 mM isobutyl-methylxanthine (Aldrich, Milwaukee, WI), 10 μM bovine insulin, 1 μM dexamathasone, and 200 μM indomethacin (all from Sigma-Aldrich, St Louis, MO).
Osteogeneic differentiation (seeding density 5000 cells/cm2) media comprised of MSC growth media with 10 mM glycerol-2-phosphate, 50 μM L-ascorbic acid, and 100 nM dexamathasone (all from Sigma-Aldrich).
Chondrogenic differentiation of MSCs was induced through aggregate culture. Aliquots of 2.5 × 105 cells were centrifuged for 160g for 5 minutes in 15 mL polypropylene conical tubes to form pellets, which were cultured in medium containing high glucose DMEM supplemented with 4 mM proline, 50 μg/mL ascorbic acid, 1% ITS-Premix (BD Biosciences, San Jose, CA), 1 mM sodium pyruvate, and 0.1 μM dexamethasone for 15 days in the presence of 10 ng/mL of TGF-β3 (R&D Systems, Minneapolis, MN). Media was changed every 3 days. Table 1 shows the samples used for this study.
For differentiation of MSCs cultured in StemPro serum-free medium, passage 9 MSCs were seeded in 12-well plates in the following differentiation media: (1) Adipogenic differentiation: cells were seeded at 1.24 × 105 cells/cm2 in low glucose DMEM supplemented with 10% MSC qualified FBS, 2 mM GlutaMAX, 5 μg/mL gentamicin, 58 μg/mL recombinant human insulin (these 5 from Invitrogen), 1 μM dexamethasone, 0.5 mM isobutyl-methylxanthine, and 200 μM indomethacin (these 3 from Sigma-Aldrich). (2) Osteogenic differentiation: cells were seeded at 3.08 × 104 cells/cm2 in low glucose DMEM supplemented with 10% MSC qualified FBS (Invitrogen), 2 mM GlutaMAX, 5 μg/mL gentamicin (these 3 from Invitrogen), 10 mM glycerol-2-phosphate, 50 μM L-ascorbic acid (both from Sigma-Aldrich), 10 ng/mL recombinant human BMP-2 (BioSource International, Camarillo, CA), and 100 nM dexamethasone (Sigma-Aldrich). (3) Chondrogenic differentiation: cells were seeded as 10 μL drops into the middle of each well of a 12-well plate (8.0 × 104 cells/drop). After a sufficient attachment period (2 hours), cells were fed differentiation medium containing low glucose DMEM supplemented with 10% MSC-qualified FBS, 2 mM GlutaMAX, 5 μg/mL gentamicin, 6.25 μg/mL recombinant human insulin (these 5 from Invitrogen), 10 ng/mL recombinant human TGFβ1 (Biosource), and 50 nM L-ascorbic acid (Sigma-Aldrich).
For each lineage described above, control cultures were seeded under identical conditions in low glucose DMEM supplemented with 10% MSC-qualified FBS, 2 mM GlutaMAX, and 5 μg/mL gentamicin (all from Invitrogen). It is important to note that the differentiation conditions used in these experiments are slightly different from those used in differentiation of MSCs cultured for microarray experiments and for the experiments involving pathway inhibitors. However, this did not affect the ability of the MSCs cultured in serum-free medium to differentiate into the 3 lineages.
RNA extraction and microarray
Marrow-derived MSC samples at passage 3 were profiled using the Illumina Bead Array system. Cells (95 × 106) from undifferentiated MSCs as well as their differentiated progeny were dissolved in Trizol and stored at −80°C for RNA extraction. RNA extraction from Trizol samples was carried out using the RNAeasy kit (Qiagen, Valencia, CA). Sample amplification was performed using 500 ng of total RNA with the Illumina TotalPrep RNA amplification kit (Ambion, Austin, TX) following the manufacturers' instructions. The biotin-labeled RNA was hybridized to a human Ref-8v1 bead chip (Illumina, San Diego, CA), which is able to generate whole genome expression profile for up to 8 samples in parallel.
Hybridized arrays were scanned with an Illumina Bead array Reader confocal scanner and data uploaded to Beadstudio for background subtraction and then exported to Genespring. Normalization was performed using median values at the chip and gene level. Good-quality signals were obtained by filtering for detection score of at least 0.99 in all replicates. Detection values of at least 0.99 characterize the chance that the target sequence was distinguishable from the negative controls and are analogous to present signals in the Affymetrix platform. This step of the preprocessing aims to scale all datasets from different chips by removing systematic errors and allow comparison across all samples.
To evaluate the quality of replicate samples, an unsupervised hierarchical clustering with Pearson correlation was applied to all datasets. To assess the quality of the data, the distribution of all chips were analyzed, and the correlation coefficient was calculated for each pair of replicate samples in Matlab (Mathworks, Natick, MA).38
The array data are deposited in ArrayExpress with accession number E-TABM-318.
MSC marker analysis
To identify marker genes that are statistically significant, we used results from the post hoc test. Statistically significant genes were identified using 1-way ANOVA with Student-Newman-Keuls post-hoc test. The false discovery rate (FDR) was kept to below 0.05 (Benjamini and Hochberg method). Only genes with significant differential expression observed exclusively in one lineage pass through this analysis. Expression of a few of these markers was verified by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR). The primers used are listed in Table S9 (available on the Blood website; see the Supplemental Materials link at the top of the online article). We compared the relative expression of a marker in a specific lineage to its combined expression in all other lineages using the comparative Ct method. This was achieved by pooling RNA from the other lineages before the cDNA conversion and amplification. For example, to compare relative expression of Finkel-Biskis-Jinkins (FBJ) murine osteosarcoma viral oncogene homolog B, (FOSB), RNA from M1 was compared with pooled RNA from A14, C14, and O14. Similarly, putative lymphocyte G0/G1 switch gene (GOS2) expression in A14 was compared with pooled RNA from M1, C14, and O14.
Signaling pathways analysis
We performed a Welch t test between differentiated progeny of MSC (adipocytes, chondrocytes, or osteocytes) and undifferentiated MSCs to identify genes that are differentially regulated between each pair samples. Each comparison yields a list of genes up/down-regulated in each lineage compared with undifferentiated MSCs. Contrary to the markers analysis, genes may be significantly up/down-regulated in one or more differentiated lineage. We also filtered for genes with differential expression of more than 1.5-fold compared with undifferentiated MSC samples. Thus, we produced 6 lists (one for each sample) of genes that were uploaded to Ingenuity Pathways Analysis (IPA) and then mapped to each canonical pathway in the database. A list of systematic IDs was obtained for each differentiated lineage and mapped to Entrez Gene IDs.
Mapped IDs were used as input data into IPA version 5.0 (Ingenuity Systems, www.ingenuity.com). Statistics of input data can be found in Table 7. IPA reported enriched pathways and a P value related to the probability of observing the enrichment using the right-tailed Fisher exact test. The P value is calculated by comparing the number of genes in the differentiated samples that participate in a given pathway, relative to the total number of occurrences of these genes in all pathway annotations stored in the Ingenuity Pathways KnowledgeBase (IPKB). Significant pathways (P < .05) were assigned a significance score by taking the negative log of the P value. Therefore, a P value of less than .05 corresponds to a significance score of more than 1.3. This approach enabled us to represent gene-expression data with a signaling pathway as the basic unit (instead of genes) and identify differentially expressed pathways in MSCs versus differentiated cells.
Perturbation experiments to assess signaling pathways
We verified the hypothesis that PDGF, FGF, and TGF-β signaling are important in MSC differentiation using perturbation studies. We inhibited PDGF signaling using AG-370 (Sigma-Aldrich), which is a member of the tyrphostin family of tyrosine kinase inhibitors and selectively inhibits PDGF receptor kinase. We investigated the effect of blocking FGFR on MSC survival and differentiation, using SU5402 (Calbiochem, La Jolla, CA), which inhibits the tyrosine kinase activity of the basic fibroblast growth factor receptor 1 (FGFR1). We also investigated the role of blocking activin receptor by SB431542 (Sigma-Aldrich), which is a potent and selective inhibitor of the transforming growth factor-β (TGF-β) type I receptor activin receptor-like kinase ALK5.
These experiments were carried out on marrow-derived MSCs isolated from donors at the National University Hospital, Singapore. Informed consent was obtained from donors in accordance with the Declaration of Helsinki, and guidelines of Institutional Review Board of the National University Hospital, Singapore, were followed. Aliquots were frozen at passage 2, thawed, and used for these experiments. MSCs cultured from a single donor (P3) were seeded in 6-well plates (10 000 cells/cm2) and incubated in the presence of inhibitors. Cells were cultured in MesenPRO medium or differentiation media to differentiate these cells into osteogenic and adipogenic lineages. Chondrogenic cultures were set up as pellet cultures in 15-mL conical tubes. Viability of these cells was checked by trypan blue dye exclusion test. Cells in differentiation medium were also stained with oil Red O (for adipocytes), von Kossa (for osteocytes), and alcian blue39 (for chondrocytes) on day 14.
We also confirmed the inhibition of phosphorylation of FGFR-1 by Western blotting using a phospho-FGFR-Y766 antibody (Abgent, San Diego, CA). This is a rabbit polyclonal antibody that recognizes epitopes surrounding the Y766 amino acid residue of human FGFR. SU5402 also binds the FGFR1 in this region.
Results
Preliminary assessment of cell and data quality
In the present report, we filtered for good quality signals using detection values of 0.99 or greater. This filter ensures that only good-quality signals are included in the analysis. Using this cutoff, cells expressed approximately 8000 transcripts on average (Tables S1, S2). Intensity values are expressed in arbitrary units, and in our samples, signals typically range from 0 to 60 000 units. The distribution of genes in this range is as shown in Table S2. All samples were found to have similar distribution of gene expression.
To assess the quality of sample, we used the Genespring (Agilent Technologies, Palo Alto, CA) software package to perform the clustering (Figure S1). All replicate samples cluster closer together compared with samples that are nonreplicates (data not shown).
Marker genes analysis
We used analysis of variance (ANOVA) to identify markers that were differentially expressed in MSCs compared with their differentiated progeny. From this analysis, we identified 4199 target IDs that are significantly different on both day 7 and day 14 in MSCs. We then used the post-hoc test to further explore the genes that show differences in expression levels between specific lineages. We were interested in genes that are up/down-regulated specifically in one cell type only. The results from this test were used to identify both markers of undifferentiated MSCs and cells of differentiated lineages derived from these MSC.
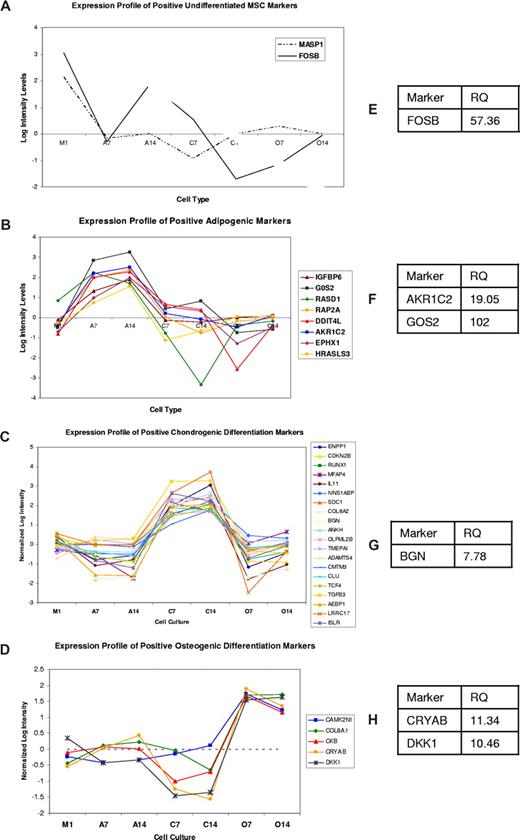
Figure 1 shows the normalized log intensity values of differentially expressed genes as analyzed by ANOVA and post-hoc test. Table 2 shows the log intensity values of expression of these genes in the different samples included in this study. MASP1 and FOSB are differentially expressed and have high transcript levels in undifferentiated MSCs (Table 2). Verification of expression of FOSB qRT-PCR (Figure 1E) shows 57-fold higher transcripts for this gene in M1 compared with combined expression in A14, C14, and O14 cells. These are defined as the putative positive markers because of their differential expression in MSCs and mean signal intensity (MSI) greater than 1000. The complete list of marker genes in MSCs (both positive and negative) is shown in Table S3.
Differentially expressed genes selected using ANOVA yield good classifiers for undifferentiated MSCs and their lineages. The x-axis represents the different samples and the y-axis represent log intensity values. Assuming that high transcript levels correlate with strong protein expression, we filtered the post hoc analysis results for genes showing strong intensity for putative positive markers of MSCs. The filtering criterion was mean signal intensity (MSI)—the average signal intensity for a particular condition—of more than 1000 units. Based on analysis of genes with known expression levels, Illumina reported the intensity of 1000 units to correspond to a concentration of 10 pM of RNA. Similarly for differentiation markers, we identified genes with statistical significance between cells of that lineage and other lineages from the post hoc test. All putative markers show a strong up-regulation compared with the expression in other lineages. (A-D) Putative markers for undifferentiated MSCs and MSC-derived adipocytes, chondrocytes, and osteocytes as predicted by the microarray data. (E-H) Quantitative RT-PCR (qRT-PCR) validation of some of these markers. Relative quantification was calculated using the comparative Ct method. Expression of a gene in one lineage was compared with the expression of the same gene in pooled RNA from other lineages. This approach enabled a better comparison of qRT-PCR data with the array data and also indicated the specificity of these markers in a particular lineage. The tables show relative quantification (RQ) values of expression of these genes in the respective lineage compared with their expression in all other lineages.
Differentially expressed genes selected using ANOVA yield good classifiers for undifferentiated MSCs and their lineages. The x-axis represents the different samples and the y-axis represent log intensity values. Assuming that high transcript levels correlate with strong protein expression, we filtered the post hoc analysis results for genes showing strong intensity for putative positive markers of MSCs. The filtering criterion was mean signal intensity (MSI)—the average signal intensity for a particular condition—of more than 1000 units. Based on analysis of genes with known expression levels, Illumina reported the intensity of 1000 units to correspond to a concentration of 10 pM of RNA. Similarly for differentiation markers, we identified genes with statistical significance between cells of that lineage and other lineages from the post hoc test. All putative markers show a strong up-regulation compared with the expression in other lineages. (A-D) Putative markers for undifferentiated MSCs and MSC-derived adipocytes, chondrocytes, and osteocytes as predicted by the microarray data. (E-H) Quantitative RT-PCR (qRT-PCR) validation of some of these markers. Relative quantification was calculated using the comparative Ct method. Expression of a gene in one lineage was compared with the expression of the same gene in pooled RNA from other lineages. This approach enabled a better comparison of qRT-PCR data with the array data and also indicated the specificity of these markers in a particular lineage. The tables show relative quantification (RQ) values of expression of these genes in the respective lineage compared with their expression in all other lineages.
Putative markers for undifferentiated MSCs and their differentiated progeny identified from ANOVA and post hoc analysis
| Probe ID . | Gene symbol . | Description . | Entrez Gene ID . | Sample ID . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M1 . | A07 . | A14 . | C07 . | C14 . | O07 . | O14 . | ||||
| MSC markers | ||||||||||
| GI_21264358-I | MASP1 | Homo sapiens mannan-binding lectin serine protease 1 (C4/C2 activating component of Ra-reactive factor) (MASP1), transcript variant 2, mRNA | 5648 | 2.1247 | −0.1619 | 0.0000 | −0.9388 | 0.0000 | 0.2726 | 0.0000 |
| GI_5803016-S | FOSB | H sapiens FBJ murine osteosarcoma viral oncogene homolog B (FOSB), mRNA | 2354 | 3.0524 | −0.3024 | 1.8793 | 0.5454 | −1.7003 | −1.1363 | −0.0643 |
| Adipogenic markers | ||||||||||
| GI_12669906-S | IGFBP6 | H sapiens acyl-CoA synthetase long-chain family member 1 (ACSL1), mRNA | 2180 | −0.1096 | 1.3134 | 1.8922 | −0.1455 | −0.2354 | 0.0038 | 0.0985 |
| GI_20070269-S | G0S2 | H sapiens putative lymphocyte G0/G1 switch gene (G0S2), mRNA | 50486 | −0.4908 | 2.8306 | 3.2601 | 0.4378 | 0.8131 | −0.7489 | −0.5906 |
| GI_22027484-S | >RASD1 | H sapiens RAS, dexamethasone-induced 1 (RASD1), mRNA | 51655 | 0.8445 | 2.2295 | 1.7099 | −0.7705 | −3.3403 | −0.3931 | −0.1897 |
| GI_31377551-S | RAP2A | H sapiens RAP2A, member of RAS oncogene family (RAP2A), mRNA | 5911 | −0.2610 | 1.9745 | 2.3584 | 0.0316 | −0.7797 | −0.1838 | −0.0538 |
| GI_34222182-S | DDIT4L | H sapiens DNA-damage-inducible transcript 4-like (DDIT4L), mRNA | 115265 | −0.8135 | 1.9974 | 2.2798 | 0.6702 | 0.3898 | −2.5613 | −0.3804 |
| GI_45446741-S | AKR1C2 | H sapiens aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) (AKR1C2), transcript variant 1, mRNA | 1646 | −0.3467 | 2.1801 | 2.4928 | 0.1962 | −0.0946 | −0.4847 | 0.0891 |
| Chondrogenic markers | ||||||||||
| GI_13324676-S | ENPP1 | H sapiens ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1), mRNA | 5167 | 0.2188 | −0.7887 | −0.5631 | 1.9896 | 3.0516 | −1.1652 | −0.4323 |
| GI_17981693-A | CDKN2B | H sapiens cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) (CDKN2B), transcript variant 1, mRNA | 1030 | 0.3149 | −0.0605 | −0.0050 | 1.8528 | 1.8740 | −0.8466 | −0.9401 |
| GI_19923197-S | RUNX1 | H sapiens runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene; RUNX1), mRNA | 861 | 0.0426 | 0.0033 | −0.0123 | 1.6058 | 2.0494 | −0.7632 | −0.3739 |
| GI_23111004-S | MFAP4 | H sapiens microfibrillar-associated protein 4 (MFAP4), mRNA | 4239 | −0.2909 | −0.5414 | −1.7318 | 2.1228 | 1.7113 | 0.0428 | 0.6477 |
| GI_24430217-S | IL11 | H sapiens interleukin 11 (IL11), mRNA | 3589 | 0.4145 | −1.0965 | −0.7898 | 2.0906 | 2.4231 | −1.7893 | −1.0528 |
| GI_24475846-S | IVNS1ABP | H sapiens influenza virus NS1A binding protein (IVNS1ABP), mRNA | 10625 | −0.2214 | −0.4830 | −0.4939 | 1.4905 | 2.2335 | 0.4409 | 0.3169 |
| GI_29568085-S | SDC1 | H sapiens syndecan 1 (SDC1), mRNA | 6382 | 0.6379 | −1.7792 | −1.7169 | 2.0289 | 2.5978 | −1.7140 | −1.2265 |
| GI_32964829-S | COL8A2 | H sapiens collagen, type VIII, alpha 2 (COL8A2), mRNA | 1296 | −0.4979 | −0.1827 | −0.1771 | 2.3983 | 1.1285 | −0.1501 | 0.5740 |
| GI_34304351-S | BGN | H sapiens biglycan (BGN), mRNA | 633 | 0.1863 | −0.4741 | −0.9904 | 1.8441 | 2.0613 | −0.5917 | −0.1769 |
| GI_34452701-S | ANKH | H sapiens ankylosis, progressive homolog (mouse; ANKH), mRNA | 56172 | −0.1405 | −0.3468 | −0.3980 | 1.9032 | 2.6121 | 0.1362 | 0.2076 |
| GI_37548655-S | OLFML2B | H sapiens olfactomedin-like 2B (OLFML2B) mRNA | 25903 | −0.1088 | −0.0092 | −0.1600 | 1.5639 | 1.6795 | −0.5550 | 0.1048 |
| GI_40317614-A | TMEPAI | H sapiens transmembrane, prostate androgen induced RNA (TMEPAI), transcript variant 1, mRNA | 56937 | −0.4838 | 0.2304 | 0.0321 | 2.3013 | 2.3872 | −0.1680 | −0.0132 |
| GI_41327755-S | ADAMTS4 | H sapiens a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 4 (ADAMTS4), mRNA | 9507 | −0.7378 | 0.3704 | 0.2196 | 2.1222 | 2.4524 | −0.2427 | −0.1231 |
| GI_32130529-A | CMTM3 | H sapiens CKLF-like MARVEL transmembrane domain containing 3 (CMTM3), mRNA | 123920 | −0.0054 | 0.0109 | −0.1266 | 1.0540 | 1.7720 | −0.3590 | 0.0973 |
| GI_42716296-S | CLU | H sapiens clusterin (complement lysis inhibitor, SP-40,40, sulfated glycoprotein 2, testosterone-repressed prostate message 2, apolipoprotein J; CLU), transcript variant 1, mRNA | 1191 | −0.0165 | −0.4015 | −0.6002 | 1.8815 | 1.7451 | −0.1837 | −0.0081 |
| GI_4507398-S | TCF4 | H sapiens transcription factor 4 (TCF4), mRNA | 6925 | 0.4581 | −0.6464 | −0.8820 | 1.4544 | 1.8483 | −0.2636 | −0.0804 |
| GI_4507464-S | TGFB3 | H sapiens transforming growth factor, beta 3 (TGFB3), mRNA | 7043 | −0.0634 | 0.1799 | 0.3213 | 3.2386 | 3.2736 | −0.3335 | −0.4560 |
| GI_4755145-S | AEBP1 | H sapiens AE binding protein 1 (AEBP1), mRNA | 165 | 0.2491 | −1.5791 | −1.6139 | 1.9207 | 2.0746 | −0.3012 | 0.0292 |
| GI_5031970-S | LRRC17 | H sapiens leucine rich repeat containing 17 (LRRC17), mRNA | 10234 | 0.5369 | −0.0269 | 0.0042 | 2.6441 | 3.7125 | −2.4749 | −0.3653 |
| GI_41582237-A | ISLR | H sapiens immunoglobulin superfamily containing leucine-rich repeat (ISLR), transcript variant 1, mRNA | 3671 | 0.1956 | −0.8433 | −1.2155 | 2.6252 | 2.2217 | −0.7244 | −0.0703 |
| Osteogenic markers | ||||||||||
| GI_31324542-S | CAMK2N1 | H sapiens calcium/calmodulin-dependent protein kinase II (CaMKIINalpha), mRNA | 55450 | −0.22461 | −0.4192 | −0.34172 | −0.14237 | 0.118001 | 1.729828 | 1.220218 |
| GI_32895369-I | COL8A1 | H sapiens collagen, type VIII, alpha 1 (COL8A1), transcript variant 1, mRNA | 1295 | −0.44275 | 0.120977 | 0.2237 | −0.03571 | −0.64901 | 1.696772 | 1.708998 |
| GI_34335231-S | CKB | H sapiens creatine kinase, brain (CKB), mRNA | 1152 | −0.10611 | 0.06397 | 0.009828 | −1.00722 | −0.69708 | 1.666986 | 1.155259 |
| GI_4503056-S | CRYAB | H sapiens crystallin, alpha B (CRYAB), mRNA | 1410 | −0.54266 | 0.029788 | 0.42692 | −1.25125 | −1.56576 | 1.890013 | 1.347231 |
| GI_7110718-S | DKK1 | H sapiens dickkopf homolog 1 (Xenopus laevis; DKK1), mRNA | 22943 | 0.341599 | −0.41979 | −0.34115 | −1.46302 | −1.35283 | 1.528545 | 1.629448 |
| Probe ID . | Gene symbol . | Description . | Entrez Gene ID . | Sample ID . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M1 . | A07 . | A14 . | C07 . | C14 . | O07 . | O14 . | ||||
| MSC markers | ||||||||||
| GI_21264358-I | MASP1 | Homo sapiens mannan-binding lectin serine protease 1 (C4/C2 activating component of Ra-reactive factor) (MASP1), transcript variant 2, mRNA | 5648 | 2.1247 | −0.1619 | 0.0000 | −0.9388 | 0.0000 | 0.2726 | 0.0000 |
| GI_5803016-S | FOSB | H sapiens FBJ murine osteosarcoma viral oncogene homolog B (FOSB), mRNA | 2354 | 3.0524 | −0.3024 | 1.8793 | 0.5454 | −1.7003 | −1.1363 | −0.0643 |
| Adipogenic markers | ||||||||||
| GI_12669906-S | IGFBP6 | H sapiens acyl-CoA synthetase long-chain family member 1 (ACSL1), mRNA | 2180 | −0.1096 | 1.3134 | 1.8922 | −0.1455 | −0.2354 | 0.0038 | 0.0985 |
| GI_20070269-S | G0S2 | H sapiens putative lymphocyte G0/G1 switch gene (G0S2), mRNA | 50486 | −0.4908 | 2.8306 | 3.2601 | 0.4378 | 0.8131 | −0.7489 | −0.5906 |
| GI_22027484-S | >RASD1 | H sapiens RAS, dexamethasone-induced 1 (RASD1), mRNA | 51655 | 0.8445 | 2.2295 | 1.7099 | −0.7705 | −3.3403 | −0.3931 | −0.1897 |
| GI_31377551-S | RAP2A | H sapiens RAP2A, member of RAS oncogene family (RAP2A), mRNA | 5911 | −0.2610 | 1.9745 | 2.3584 | 0.0316 | −0.7797 | −0.1838 | −0.0538 |
| GI_34222182-S | DDIT4L | H sapiens DNA-damage-inducible transcript 4-like (DDIT4L), mRNA | 115265 | −0.8135 | 1.9974 | 2.2798 | 0.6702 | 0.3898 | −2.5613 | −0.3804 |
| GI_45446741-S | AKR1C2 | H sapiens aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) (AKR1C2), transcript variant 1, mRNA | 1646 | −0.3467 | 2.1801 | 2.4928 | 0.1962 | −0.0946 | −0.4847 | 0.0891 |
| Chondrogenic markers | ||||||||||
| GI_13324676-S | ENPP1 | H sapiens ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1), mRNA | 5167 | 0.2188 | −0.7887 | −0.5631 | 1.9896 | 3.0516 | −1.1652 | −0.4323 |
| GI_17981693-A | CDKN2B | H sapiens cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) (CDKN2B), transcript variant 1, mRNA | 1030 | 0.3149 | −0.0605 | −0.0050 | 1.8528 | 1.8740 | −0.8466 | −0.9401 |
| GI_19923197-S | RUNX1 | H sapiens runt-related transcription factor 1 (acute myeloid leukemia 1; aml1 oncogene; RUNX1), mRNA | 861 | 0.0426 | 0.0033 | −0.0123 | 1.6058 | 2.0494 | −0.7632 | −0.3739 |
| GI_23111004-S | MFAP4 | H sapiens microfibrillar-associated protein 4 (MFAP4), mRNA | 4239 | −0.2909 | −0.5414 | −1.7318 | 2.1228 | 1.7113 | 0.0428 | 0.6477 |
| GI_24430217-S | IL11 | H sapiens interleukin 11 (IL11), mRNA | 3589 | 0.4145 | −1.0965 | −0.7898 | 2.0906 | 2.4231 | −1.7893 | −1.0528 |
| GI_24475846-S | IVNS1ABP | H sapiens influenza virus NS1A binding protein (IVNS1ABP), mRNA | 10625 | −0.2214 | −0.4830 | −0.4939 | 1.4905 | 2.2335 | 0.4409 | 0.3169 |
| GI_29568085-S | SDC1 | H sapiens syndecan 1 (SDC1), mRNA | 6382 | 0.6379 | −1.7792 | −1.7169 | 2.0289 | 2.5978 | −1.7140 | −1.2265 |
| GI_32964829-S | COL8A2 | H sapiens collagen, type VIII, alpha 2 (COL8A2), mRNA | 1296 | −0.4979 | −0.1827 | −0.1771 | 2.3983 | 1.1285 | −0.1501 | 0.5740 |
| GI_34304351-S | BGN | H sapiens biglycan (BGN), mRNA | 633 | 0.1863 | −0.4741 | −0.9904 | 1.8441 | 2.0613 | −0.5917 | −0.1769 |
| GI_34452701-S | ANKH | H sapiens ankylosis, progressive homolog (mouse; ANKH), mRNA | 56172 | −0.1405 | −0.3468 | −0.3980 | 1.9032 | 2.6121 | 0.1362 | 0.2076 |
| GI_37548655-S | OLFML2B | H sapiens olfactomedin-like 2B (OLFML2B) mRNA | 25903 | −0.1088 | −0.0092 | −0.1600 | 1.5639 | 1.6795 | −0.5550 | 0.1048 |
| GI_40317614-A | TMEPAI | H sapiens transmembrane, prostate androgen induced RNA (TMEPAI), transcript variant 1, mRNA | 56937 | −0.4838 | 0.2304 | 0.0321 | 2.3013 | 2.3872 | −0.1680 | −0.0132 |
| GI_41327755-S | ADAMTS4 | H sapiens a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 4 (ADAMTS4), mRNA | 9507 | −0.7378 | 0.3704 | 0.2196 | 2.1222 | 2.4524 | −0.2427 | −0.1231 |
| GI_32130529-A | CMTM3 | H sapiens CKLF-like MARVEL transmembrane domain containing 3 (CMTM3), mRNA | 123920 | −0.0054 | 0.0109 | −0.1266 | 1.0540 | 1.7720 | −0.3590 | 0.0973 |
| GI_42716296-S | CLU | H sapiens clusterin (complement lysis inhibitor, SP-40,40, sulfated glycoprotein 2, testosterone-repressed prostate message 2, apolipoprotein J; CLU), transcript variant 1, mRNA | 1191 | −0.0165 | −0.4015 | −0.6002 | 1.8815 | 1.7451 | −0.1837 | −0.0081 |
| GI_4507398-S | TCF4 | H sapiens transcription factor 4 (TCF4), mRNA | 6925 | 0.4581 | −0.6464 | −0.8820 | 1.4544 | 1.8483 | −0.2636 | −0.0804 |
| GI_4507464-S | TGFB3 | H sapiens transforming growth factor, beta 3 (TGFB3), mRNA | 7043 | −0.0634 | 0.1799 | 0.3213 | 3.2386 | 3.2736 | −0.3335 | −0.4560 |
| GI_4755145-S | AEBP1 | H sapiens AE binding protein 1 (AEBP1), mRNA | 165 | 0.2491 | −1.5791 | −1.6139 | 1.9207 | 2.0746 | −0.3012 | 0.0292 |
| GI_5031970-S | LRRC17 | H sapiens leucine rich repeat containing 17 (LRRC17), mRNA | 10234 | 0.5369 | −0.0269 | 0.0042 | 2.6441 | 3.7125 | −2.4749 | −0.3653 |
| GI_41582237-A | ISLR | H sapiens immunoglobulin superfamily containing leucine-rich repeat (ISLR), transcript variant 1, mRNA | 3671 | 0.1956 | −0.8433 | −1.2155 | 2.6252 | 2.2217 | −0.7244 | −0.0703 |
| Osteogenic markers | ||||||||||
| GI_31324542-S | CAMK2N1 | H sapiens calcium/calmodulin-dependent protein kinase II (CaMKIINalpha), mRNA | 55450 | −0.22461 | −0.4192 | −0.34172 | −0.14237 | 0.118001 | 1.729828 | 1.220218 |
| GI_32895369-I | COL8A1 | H sapiens collagen, type VIII, alpha 1 (COL8A1), transcript variant 1, mRNA | 1295 | −0.44275 | 0.120977 | 0.2237 | −0.03571 | −0.64901 | 1.696772 | 1.708998 |
| GI_34335231-S | CKB | H sapiens creatine kinase, brain (CKB), mRNA | 1152 | −0.10611 | 0.06397 | 0.009828 | −1.00722 | −0.69708 | 1.666986 | 1.155259 |
| GI_4503056-S | CRYAB | H sapiens crystallin, alpha B (CRYAB), mRNA | 1410 | −0.54266 | 0.029788 | 0.42692 | −1.25125 | −1.56576 | 1.890013 | 1.347231 |
| GI_7110718-S | DKK1 | H sapiens dickkopf homolog 1 (Xenopus laevis; DKK1), mRNA | 22943 | 0.341599 | −0.41979 | −0.34115 | −1.46302 | −1.35283 | 1.528545 | 1.629448 |
Genes with lineage-specific expression as shown in Figure 1. These putative markers were identified from ANOVA and post hoc analysis. All genes shown in this table have MSI more than 1000. Data are the normalized log intensity values for this analysis since it normalizes the signal in multiple replicates and accounts for variations in hybridization, scanning, and so on. This enables a direct comparison of expression across multiple samples carried out on different arrays, which is not always possible with MSI values.
Results for adipogenic marker analysis shows 25 putative markers were identified at either day 7 or day 14, and 9 of these were identified on both days. Of these, IGFBP6, GOS2, RASD1, RAP2A, DDIT4L, and AKR1C2 are strongly expressed at the transcriptome level on day 7 and day 14 adipocytes differentiated from MSCs. The analysis for chondrogenic markers identified the largest number of putative markers. Sixty-three genes met the criteria for this analysis, and 20 of these were identified at day 7 and day 14. Of these, ENPP1, CDKN2B, IL11, BGN, ANKH, and TMEPAI are expressed strongly. Osteogenic marker analysis produced 20 putative markers, where 7 are common to both lists. The genes CAMK2N1, COL8A1, CKB, CRYAB, and DKK1 are expressed at high levels in osteogenic cells. Good markers for osteogenic differentiation of MSCs have been a challenge, since the osteogenic and chondrogenic lineage share many common pathways. Positive differentiation markers are listed in Table 2, while the complete lists of differentiation markers (negative and positive) are shown in Tables S4 to S6.
TGF-β, PDGF, and FGF signaling is essential for MSC differentiation
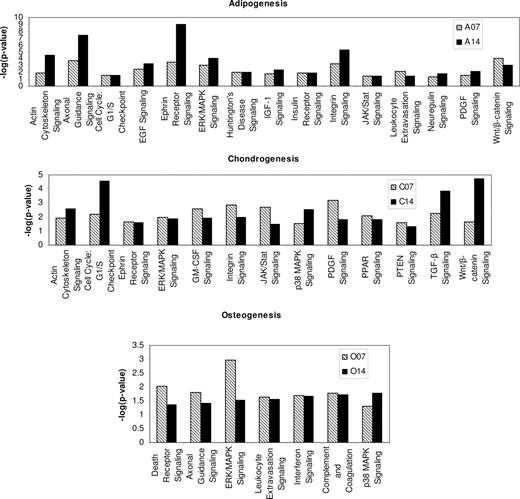
The progression from undifferentiated MSC to differentiated cells involves the activation or suppression of certain signaling pathways. These pathways may be specific to one lineage or may be important for the differentiation process in general. To predict signaling pathways active in differentiating MSCs, we examined the statistical significance values for known signaling pathways in IPA, based on the probability of gene enrichment. Table 3 identifies signaling pathways that are statistically significant in MSC differentiation, based on gene expression. These 9 pathways were predicted to be important in MSC differentiation in general, since these pathways have significant enrichment in at least 1 sample in all 3 lineages compared with undifferentiated MSCs. ERK-MAPK signaling was active in all samples of differentiated cells, whereas PDGF and axonal guidance signaling was active in 5 of 6 samples of differentiated cells. Figure 2 highlights the pathways that are predicted to be significant for commitment to a specific lineage based on differential gene expression of these cells compared with undifferentiated MSCs. Our microarray data analysis results predicted 15 signaling pathways activated during adipogenesis, 13 signaling pathways activated during chondrogenesis, and 7 signaling pathways activated during osteogenesis. Of the pathways important in MSC differentiation (Table 3), ERK-MAPK signaling was significant in all 3 lineages, PDGF signaling was significant in adipogenesis and chondrogenesis, TGF-β was significant in chondrogenesis, whereas axonal guidance signaling was significant in adipogenesis and osteogenesis. In addition to these pathways the death receptor signaling, p38 MAP kinase were also differentially expressed in differentiated cells (Figure S3). It is important to note the absence of FGF signaling, although FGF has been shown to be important in MSC differentiation.40 Of the pathways predicted to be important in MSC differentiation as well as lineage commitment, we chose PDGF and TGF-β for further investigation since these were predicted to be important in MSC differentiation in general as well as in lineage commitment. We hypothesized that since FGF signals through the ERK-MAPK pathway (predicted to be important in MSC differentiation as well as lineage commitment), FGF signaling is important in MSC differentiation even though FGFR was not strongly expressed by differentiating MSCs. Although we did not test this hypothesis, it has been reported that FGF treatment of mesenchymal cells of the chick embryo wing bud elevates endogenous ERK phosphorylation in micromass cultures.41
Ingenuity pathway analysis predicts differentially expressed canonical signaling pathways across the differentiated lineages of MSCs
| Pathways . | Sample ID . | |||||
|---|---|---|---|---|---|---|
| A07 . | A14 . | C07 . | C14 . | O07 . | O14 . | |
| Axonal guidance signaling | 3.67 | 7.4 | — | 2.16 | 1.81 | 1.42 |
| Complement and coagulation cascades | 2.28 | — | 1.7 | — | 1.77 | 1.73 |
| EGF signaling | 2.51 | 3.27 | 1.98 | — | 2.47 | — |
| ERK/MAPK signaling | 3.02 | 4.02 | 1.98 | 1.89 | 2.98 | 1.54 |
| Integrin signaling | 3.22 | 5.31 | 2.86 | 1.96 | 1.47 | — |
| Interferon signaling | — | 1.31 | 3.25 | — | 1.70 | 1.67 |
| Leukocyte extravasation signaling | 2.19 | 1.47 | — | 1.93 | 1.63 | 1.56 |
| PDGF signaling | 1.58 | 2.12 | 3.17 | 1.82 | 1.65 | — |
| TGF-β signaling | — | 2.00 | 2.28 | 3.82 | 2.29 | — |
| Pathways . | Sample ID . | |||||
|---|---|---|---|---|---|---|
| A07 . | A14 . | C07 . | C14 . | O07 . | O14 . | |
| Axonal guidance signaling | 3.67 | 7.4 | — | 2.16 | 1.81 | 1.42 |
| Complement and coagulation cascades | 2.28 | — | 1.7 | — | 1.77 | 1.73 |
| EGF signaling | 2.51 | 3.27 | 1.98 | — | 2.47 | — |
| ERK/MAPK signaling | 3.02 | 4.02 | 1.98 | 1.89 | 2.98 | 1.54 |
| Integrin signaling | 3.22 | 5.31 | 2.86 | 1.96 | 1.47 | — |
| Interferon signaling | — | 1.31 | 3.25 | — | 1.70 | 1.67 |
| Leukocyte extravasation signaling | 2.19 | 1.47 | — | 1.93 | 1.63 | 1.56 |
| PDGF signaling | 1.58 | 2.12 | 3.17 | 1.82 | 1.65 | — |
| TGF-β signaling | — | 2.00 | 2.28 | 3.82 | 2.29 | — |
Data are −log(P values). Pathways with −log(P value) greater than 1.3 (P < .05) in at least one sample (day 7 or day 14) were considered to be statistically differentially expressed in MSC differentiation. These pathways are predicted to be important in MSC differentiation to all 3 lineages. Since FGF signaling pathway was not significantly enriched in any sample, it is not represented in this table.
— indicates not available.
Ingenuity pathway analysis predictions. Pathways important for adipogenesis, chondrogenesis, and osteogenesis are based on gene expression data. Only pathways that are significantly enriched on both day 7 and day 14 are shown. The x-axis represents the negative log of the P value (also called the significance score) calculated from the Fisher exact test. Pathways with significance score of more than 1.3 are considered significant. Contrary to the marker analysis, statistically significant genes in all 3 lineages may overlap between cells cultured for 7 and 14 days, as well as MSCs differentiating into all 3 lineages (Figure S2). For example, in our study 138 genes are associated with the MSC differentiation process but are not necessarily lineage-specific genes.
Ingenuity pathway analysis predictions. Pathways important for adipogenesis, chondrogenesis, and osteogenesis are based on gene expression data. Only pathways that are significantly enriched on both day 7 and day 14 are shown. The x-axis represents the negative log of the P value (also called the significance score) calculated from the Fisher exact test. Pathways with significance score of more than 1.3 are considered significant. Contrary to the marker analysis, statistically significant genes in all 3 lineages may overlap between cells cultured for 7 and 14 days, as well as MSCs differentiating into all 3 lineages (Figure S2). For example, in our study 138 genes are associated with the MSC differentiation process but are not necessarily lineage-specific genes.
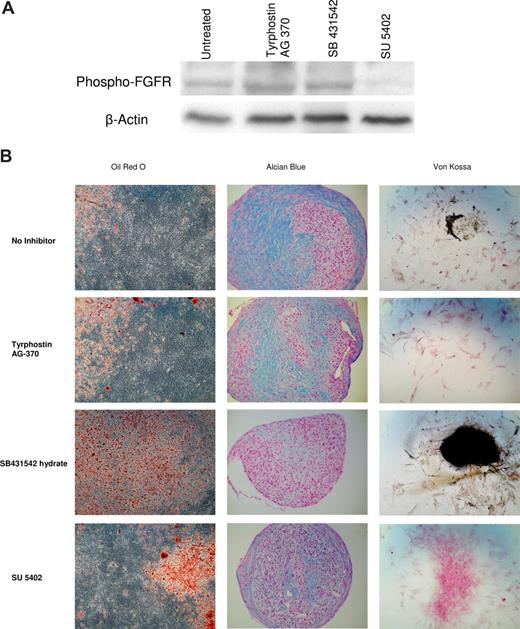
To test our predictions, we perturbed the TGF-β, PDGF, and FGF pathways in differentiating MSCs using specific receptor inhibitors and studied their effect on cell differentiation. Figure 3 shows the effect of these inhibitors on MSC differentiation. Inhibition of PDGF signaling resulted in fewer osteocytes and also the absence of mineralized bone nodules. Inhibition of ALK-5–mediated TGF-β signaling resulted in enhanced adipogenic and osteogenic differentiation, but a complete lack of chondrogenic differentiation. Inhibition of FGFR signaling resulted in complete lack of osteogenic differentiation and a reduced chondrogenic differentiation compared with no inhibitor control.
Activin-mediated TGF-β, PDGF, and FGF signaling are important for MSC differentiation. Marrow-derived MSCs were cultured in the presence of inhibitors of these pathways. (A) Western blot of MSCs cultured in the presence of SU5402 probed with phospho-FGFR-Y766 antibody. Untreated MSCs as well as MSCs cultured in the presence of tyrphostin AG 370 and SB431542 show the presence of the phosphoyrlated FGFR, whereas cells cultured in the presence of SU5402 do not show phospho-FGFR. SU5402 is reported to be a weak inhibitor of tyrosine phosphorylation of the PDGF receptor and does not inhibit phosphorylation of the TGF-β receptor. The Western blot in this figure confirms this. (B) MSCs were cultured in differentiation medium for 14 days and stained with Oil Red O, Alcian Blue, and Von Kossa for staining adipocytes, chondrocytes, and osteocytes, respectively. Inhibition of any of these 3 pathways leads to an altered differentiation potential of MSCs, proving that these pathways are essential for MSC differentiation. Oil Red O and Von Kossa micrographs aquired with a Zeiss Axiovert 40 C microscope (Carl Zeiss, Singapore) fitted with a 20×/0.3 NA objective. Alcian Blue micrographs were acquired with a 10×/0.25 NA objective on the same microscope. All images were captured on a 7.1 megapixel Canon Powershot A620 camera (Canon, Singapore) and the contrast was adjusted using the auto-contrast feature in Picasa (Google, Mountain View, CA).
Activin-mediated TGF-β, PDGF, and FGF signaling are important for MSC differentiation. Marrow-derived MSCs were cultured in the presence of inhibitors of these pathways. (A) Western blot of MSCs cultured in the presence of SU5402 probed with phospho-FGFR-Y766 antibody. Untreated MSCs as well as MSCs cultured in the presence of tyrphostin AG 370 and SB431542 show the presence of the phosphoyrlated FGFR, whereas cells cultured in the presence of SU5402 do not show phospho-FGFR. SU5402 is reported to be a weak inhibitor of tyrosine phosphorylation of the PDGF receptor and does not inhibit phosphorylation of the TGF-β receptor. The Western blot in this figure confirms this. (B) MSCs were cultured in differentiation medium for 14 days and stained with Oil Red O, Alcian Blue, and Von Kossa for staining adipocytes, chondrocytes, and osteocytes, respectively. Inhibition of any of these 3 pathways leads to an altered differentiation potential of MSCs, proving that these pathways are essential for MSC differentiation. Oil Red O and Von Kossa micrographs aquired with a Zeiss Axiovert 40 C microscope (Carl Zeiss, Singapore) fitted with a 20×/0.3 NA objective. Alcian Blue micrographs were acquired with a 10×/0.25 NA objective on the same microscope. All images were captured on a 7.1 megapixel Canon Powershot A620 camera (Canon, Singapore) and the contrast was adjusted using the auto-contrast feature in Picasa (Google, Mountain View, CA).
β-FGF, TGF-β, and PDGF signaling is also important for MSC growth
Since growth and differentiation are very tightly linked biologic processes, we investigated the importance of these 3 signaling pathways on MSC survival and growth. Figure 4 shows the effect of these inhibitors on cell survival. At 20 μM concentration, tyrphostin showed significant cell toxicity. Adding higher concentrations of tyrphostin resulted in extensive cell death (Figure 4A). The fold increase in viable cell number was only 5.5-fold for MSCs cultured in the presence of 20 μM tyrphostin, compared with 10-fold for control MSCs. Treatment with SB431542 did not result in extensive toxicity or altered morphology of MSCs. However, MSC growth slowed down significantly, since the increase in viable cell number after 14 days of treatment with SB431542 was only 3.5-fold. Treatment with SU5402 resulted in altered cell morphology (thinner cells), and the viable cell number increased by only 2.8-fold after 15 days in culture. Thus, all these inhibitors slowed the growth of MSCs in culture.
Activin-mediated TGF-β, PDGF, and FGF signaling are also important for MSC growth.Marrow-derived MSCs were cultured for 7 days in the presence of inhibitors of these pathways. Three doses (0.2× IC50 dose, IC50 dose, and 2× IC50 dose) were used for each inhibitor. Panel A shows the morphology of undifferentiated MSCs in the presence of these inhibitors. Panel B shows the fold increase in number of viable cells in these cultures. Fold increase in viable cells was calculated as ratio of viable cells in the culture on day 7 or day 14 compared with the starting number of viable cells that were seeded in these cultures. Micrographs were acquired by imaging with a Zeiss Axiovert 40 C microscope fitted with a 20×/0.3 NA objective and a Canon Powershot A620 camera. Image contrast was adjusted using the Auto Contrast function in Picasa.
Activin-mediated TGF-β, PDGF, and FGF signaling are also important for MSC growth.Marrow-derived MSCs were cultured for 7 days in the presence of inhibitors of these pathways. Three doses (0.2× IC50 dose, IC50 dose, and 2× IC50 dose) were used for each inhibitor. Panel A shows the morphology of undifferentiated MSCs in the presence of these inhibitors. Panel B shows the fold increase in number of viable cells in these cultures. Fold increase in viable cells was calculated as ratio of viable cells in the culture on day 7 or day 14 compared with the starting number of viable cells that were seeded in these cultures. Micrographs were acquired by imaging with a Zeiss Axiovert 40 C microscope fitted with a 20×/0.3 NA objective and a Canon Powershot A620 camera. Image contrast was adjusted using the Auto Contrast function in Picasa.
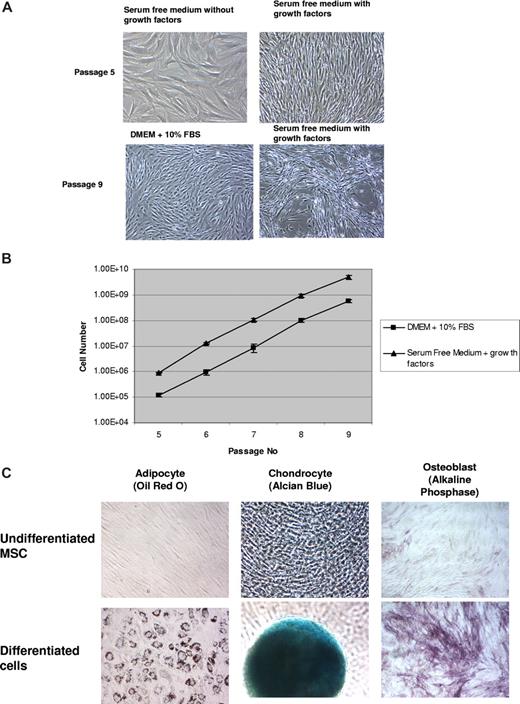
To determine whether these 3 signaling pathways are sufficient for MSC growth, we cultured MSCs in the presence of β-FGF, TGF-β, and PDGF in serum-free medium. Figure 5A shows the spindle-shaped morphology of MSCs cultured in the presence of these growth factors could be maintained for 5 passages. MSCs cultured in serum-free medium in the absence of these growth factors, however, showed a more flattened morphology. MSC cultured in serum-free medium in the presence of 3 growth factors displayed similar increases in cell number to those cultured in DMEM containing 10% FBS (Figure 5B). MSCs cultured in serum-free medium in the presence of these 3 growth factors also retained their ability to differentiate normally into adipogenic, chondrogenic, and osteogenic lineages (Figure 5C).
β-FGF, TGF-β, and PDGF are sufficient to support MSC growth in serum-free medium. (A) MSCs grown under serum-free conditions without growth factors displayed a flattened, fibroblastic morhpology, while cells grown in the presence of growth factors displayed a distinct spindle-shaped morphology, which was closer to the morphology displayed by MSCs. Micrographs were acquired with a Nikon Diaphot TMD microscope (Nikon, Melville, NY) fitted with a 20×/0.4 NA objective and a Moticam 2000 camera (Motic Instruments, Richmond, BC). (B) Human MSCs (passage 5) grown under serum-free conditions displayed robust expansion in the presence of the 3 growth factors, which was comparable with MSCs grown in DMEM containing 10% FBS. The initial seed number for MSCs in serum-containing cultures was 28 800, whereas in serum-free cultures the seed number was 96 600. This difference in initial seeding density was necessary, since the optimal cell density for serum-containing cultures was 3000 cells/cm2, whereas cells in serum-free medium grew best at a density of 10 000 cells/cm2. The total number of cells in each passage was calculated as a ratio of total number of cells harvested to total number of cells seeded multiplied by the total number of cells from the previous passage. Although 28 800 cells and 96 600 cells were seeded, respectively, in serum-containing and serum-free cultures at every passage, the total cell number was calculated as described above. This enabled us to compute the total numbers of cells we would have harvested if we had cultured all cells from each passage between passages 5 and 9. It is important to note that since the cultures started with different seed numbers, the final number of cells at each passage were proportionately different. (C) MSCs grown in serum-free medium retain their ability to differentiate into all 3 lineages. To induce differentiation, hMSC grown for 5 passages in serum-free medium were seeded under respective differentiation conditions, cultured for 14 days, and stained with Oil Red O (Adipocytes), Alcian Blue (Chondrocytes), and Alkaline Phosphatase (Osteoblasts). Adipocytes and chondrocytes at 10× magnification; osteoblasts at 4× magnification. Micrographs were acquired with a Nikon Diaphot TMD microscope fitted with a 10×/0.25 NA (Adipocyte & Chondrocyte) and a 4×/0.13 NA objective (Osteoblast) and a Moticam 2000 camera. Brightness and contrast of all images were adjusted using Adobe Photoshop Elements 5.0 (Adobe Systems, San Jose, CA).
β-FGF, TGF-β, and PDGF are sufficient to support MSC growth in serum-free medium. (A) MSCs grown under serum-free conditions without growth factors displayed a flattened, fibroblastic morhpology, while cells grown in the presence of growth factors displayed a distinct spindle-shaped morphology, which was closer to the morphology displayed by MSCs. Micrographs were acquired with a Nikon Diaphot TMD microscope (Nikon, Melville, NY) fitted with a 20×/0.4 NA objective and a Moticam 2000 camera (Motic Instruments, Richmond, BC). (B) Human MSCs (passage 5) grown under serum-free conditions displayed robust expansion in the presence of the 3 growth factors, which was comparable with MSCs grown in DMEM containing 10% FBS. The initial seed number for MSCs in serum-containing cultures was 28 800, whereas in serum-free cultures the seed number was 96 600. This difference in initial seeding density was necessary, since the optimal cell density for serum-containing cultures was 3000 cells/cm2, whereas cells in serum-free medium grew best at a density of 10 000 cells/cm2. The total number of cells in each passage was calculated as a ratio of total number of cells harvested to total number of cells seeded multiplied by the total number of cells from the previous passage. Although 28 800 cells and 96 600 cells were seeded, respectively, in serum-containing and serum-free cultures at every passage, the total cell number was calculated as described above. This enabled us to compute the total numbers of cells we would have harvested if we had cultured all cells from each passage between passages 5 and 9. It is important to note that since the cultures started with different seed numbers, the final number of cells at each passage were proportionately different. (C) MSCs grown in serum-free medium retain their ability to differentiate into all 3 lineages. To induce differentiation, hMSC grown for 5 passages in serum-free medium were seeded under respective differentiation conditions, cultured for 14 days, and stained with Oil Red O (Adipocytes), Alcian Blue (Chondrocytes), and Alkaline Phosphatase (Osteoblasts). Adipocytes and chondrocytes at 10× magnification; osteoblasts at 4× magnification. Micrographs were acquired with a Nikon Diaphot TMD microscope fitted with a 10×/0.25 NA (Adipocyte & Chondrocyte) and a 4×/0.13 NA objective (Osteoblast) and a Moticam 2000 camera. Brightness and contrast of all images were adjusted using Adobe Photoshop Elements 5.0 (Adobe Systems, San Jose, CA).
Discussion
Transcriptional profiling of MSCs can yield useful information on markers to reliably identify MSCs and their differentiated progeny. Illumina bead array has been shown to be a cost-effective method for profiling whole genome expression in different embryonic stem cell lines and distinguishing these cells from differentiated embryoid bodies as well as fibroblast feeder cells.41 Using a similar approach, we sought to identify genes that are specific for a particular lineage from a set of genes that exhibit significant differences in expression between lineages. Our high relative stringency (genes with MSI > 1000, confidence > 0.99) enabled us to identify candidate putative markers for MSCs as well as their differentiated progeny.
Of course the validity of these markers for enumerating MSCs in fresh tissue and from a larger pool of donors remains to be proven. The specificity of these markers will also need to be examined in the context of other stem cells present in the same tissue like hematopoietic stem cells. Nevertheless, this list of putative markers is a good starting point for such analyses.
In this study we have also identified markers specific for differentiated progeny of MSCs applying the same criteria as those for MSC markers. Our list differs from the one published by Liu et al.43 This could be attributed to a number of parameters like donor variation, differences in culture conditions, etcetera, which could affect the transcriptome. An important difference is also the use of different statistics to analyze differential gene expression. We have used ANOVA, whereas Liu et al have used the t-statistic to generate lists of differentially expressed genes. The strength of ANOVA lies in the simultaneous testing of multiple hypotheses. Compared with the commonly used pairwise t test, performing ANOVA provides more accurate results than conducting multiple pairwise t tests between all possible pairs of lineages. ANOVA breaks down the sources of variation in the data and reports genes where the variation is due to differences in cell type. These genes are statistically significant with P values less than or equal to .05 and with the false discovery rate kept at a maximum of .05. Thus, our approach of using ANOVA allows us to identify differentially expressed genes that are specific classifiers for the selected lineage only. This is demonstrated well in Figure 1, where differentially expressed genes in one lineage are poor discriminators of cells from other lineages. This specificity is not a feature of signature genes identified by Liu et al. For example, in their study FKBP is part of a molecular signature of MSC-derived osteocytes and chondrocytes. While Liu et al have focused on identifying genes functionally critical in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages, these may not be ideal markers since a gene may be critical for differentiation without necessarily be highly differentially expressed in differentiated MSCs. We rely only on differential expression and do not make any claims on the functional role these genes play in MSC differentiation.
This study proves that it is possible to predict signaling pathways active in cellular differentiation using transcriptional profiling and to experimentally verify these predictions. From the microarray data and comparison with reported literature, we identified 3 signaling pathways that seemed to be critical to MSC differentiation and growth. It is important to note that while such an approach enabled us to identify these pathways, it does not give us any information about the mechanisms these pathways use for influencing MSC differentiation and growth. Genes of the PDGF pathway are expressed strongly in undifferentiated MSCs. Fresh frozen pooled plasma (FFPP), which is rich in PDGF, has been used to replace serum for MSC culture.44 Our results give further evidence for the importance of PDGF signaling in MSC growth. The PDGF signaling inhibitor tyrphostin was toxic to MSCs at 2× IC50 concentration. Inhibiting the PDGF receptor-α did not result in complete inhibition of adipogenic or chondrogenic differentiation in our perturbation experiments. Although PDGF signaling is important in MSC migration,45 there is little evidence of the importance of PDGF in MSC differentiation, apart from inhibition of osteogenesis.46,47 We also observed inhibition of osteogenesis in tyrphostin-treated cultures.
The TGF-β pathway has been shown to be important in MSC differentiation into the osteogenic and chondrogenic lineages.48,49 Also in our study activin receptor was very strongly expressed on undifferentiated MSCs. The activin/nodal pathway, which signals through the TGF-β pathway, cooperates with FGF signaling in maintaining the pluripotency of embryonic stem cells.50 Therefore, in this study, we investigated if activin signaling plays a similar role in MSC differentiation. Treatment with ALK-5 inhibitor SB431542 did not result in extensive cell death but did slow down MSC growth. This observation further supports the fact that TGF-β signaling is an important event in MSC proliferation in a Smad-3–dependent manner.51 TGF-β is also an inhibitor of adipogenesis via Smad 3 signaling.52 Our results provide further evidence to this observation, since blocking Smad 3-mediated TGF-β signaling resulted in enhanced adipogenic differentiation.
FGF is an important growth factor for MSC expansion. Addition of β-FGF has been shown to increase the growth rate and life span of MSCs from different species.40,53 Surprisingly, the FGFR1 transcript was very weakly expressed in undifferentiated MSCs and was not up-regulated during differentiation. Therefore, we decided to test the importance of FGF receptor in a blocking assay. Our results reconfirm the fact that FGF signaling is essential for MSC proliferation. FGFR1 is a key regulator of osteoblast maturation in osteogenesis.54,55 Therefore, it is not surprising that inhibition of FGF signaling leads to an abrogation of osteogenic differentiation. FGF-1 has also been shown to play a role in chondrogenesis.39 Human MSCs treated with β-FGF gave rise to larger chondrogenic pellets with higher proteoglycan production.56 Our results show that FGF signaling, though not critical for chondrogenesis (since we did not observe a complete inhibition of chondrogenesis), is important and blocking this pathway results in reduced chondrogenic differentiation.
Our study provides the first direct evidence of inhibition of these signaling pathways on MSC differentiation. We have used a different approach to this problem compared with Liu et al.43 While they have assessed differential gene expression and then tried to look for pathways in which these genes are involved in, we have tried to mine information about known canonical signaling pathways from the microarray data. Thus, we were able to immediately compare this information with available literature to select the pathways for inhibition studies. Also, this approach resulted in better prediction of the inhibition experiments by the bioinformatics analysis. For example, the informatics analysis of the array data predicted that TGF-β is down-regulated in adipogenesis and up-regulated in chondrogenesis (Figure S4), which was experimentally validated since we saw increased adipogenesis and inhibition of chondrogenesis when TGF-β signaling was blocked. Also, this approach allows us to block pathways at the receptor level using small molecules to control differentiation. This information will be useful when developing media for targeted differentiation as well as drugs to direct the fate of transplanted cells toward a particular lineage compared with blocking selected genes using siRNA.
Our approach to identify signaling mechanisms also differs from that used by Song et al.57 While both approaches use gene expression data, Song et al use a differentiation de-differentiation system to identify genes important in MSC differentiation. While this is a good approach to identify “stemness genes,” our approach is better suited to identify pathways involved in specific lineage differentiation of MSCs, since it is difficult to gain information about cellular function based only on differential gene expression. We have based our approach on known signaling pathways as units of gene expression, and since these pathways are well studied, it is easier to assess their effect on cellular function. However, it is interesting to note that TGF-β signaling was also identified by Song et al as an important pathway, and they also observed strong expression of activin receptor 2 gene. Since our approach does not require a differentiation de-differentiation system, this may be useful in studying differentiation of other stem cells, where such a differentiation de-differentiation system may not be available.
Further, we are also able to show that these 3 pathways are sufficient for expanding MSCs in serum-free media. It will be interesting to determine whether we can achieve robust MSC expansion and targeted differentiation through activating one pathway while blocking another one. Such an approach could then be used to identify combinations of growth factors and small molecules for clinical use of MSCs, where robust expansion and targeted differentiation is desirable.
In summary, this study profiles the MSC transcriptome through cells cultured under carefully controlled conditions, identifies putative markers for MSCs and their differentiated progeny, and elucidates the signaling pathways active in MSC differentiation and proliferation. This study also shows the importance of bioinformatics tools to study cell function. For example, in the absence of specific markers, global array analysis can be used to classify MSC state. The results from this study should form a basis for further studies examining specific MSC markers or signaling pathways active in MSC proliferation and differentiation in more detail.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Eng Hin Lee and Associate Prof James Hui for providing bone marrow samples for this study. Technical assistance of Ms Rafidah Suleman and Ms Sharon Goh from Laboratory of Stem Cell Biology, Singapore Stem Consortium, is also acknowledged. Finally, the authors are grateful to Ingenuity Systems Pvt for providing an extended complimentary trial license access to the IPA platform. The pathway analysis described in this study was carried out using the trial license.
This work was supported by the Biomedical Research Council of A*STAR (Agency for Science Technology and Research), Singapore and by Invitrogen.
Authorship
Contribution: F.N. and S.K. analyzed the data and helped draft the final manuscript; S.B., S.K., K.S.S., U.L., L.C., and M.C.V. conducted the experiments; C.C. provided laboratory infrastructure for the research, analyzed data, and participated in the discussion of experimental results; Z.Y. provided marrow samples, standardized protocols for MSC differentiation and staining; V.T. designed research, analyzed data, and wrote the paper; and M.S.R. analyzed the data, suggested experiments, and provided editorial advice. All authors have read and approved the final version of the manuscript.
Conflict-of-interest disclosure: S.B., U.L., M.C.V., L.C., and M.S.R. are employees of Invitrogen, whose products were used in this study. The other authors declare no competing financial interests.
Correspondence: Vivek Tanavde, Bioinformatics Institute, Agency for Science Technology and Research (A*STAR), 30 Biopolis Street, Matrix #07-01, Singapore 138671; e-mail: vivek@bii.a-star.edu.sg.
References
Author notes
*F.N. and S.B. contributed equally to this manuscript.