Abstract
In vitro studies indicate that Cul4A ubiquitin ligases target for ubiquitin-mediated proteolysis regulators of cell-cycle progression, apoptosis, development, and DNA repair. In hematopoietic cell lines, studies by our group and others showed that Cul4A ligases regulate proliferation and differentiation in maturing myeloid and erythroid cells. In vivo, Cul4A-deficient embryos die in utero. Cul4A haploinsufficient mice are viable but have fewer erythroid and primitive myeloid progenitors. Yet, little more is known about Cul4A function in vivo. To examine Cul4A function in adults, we generated mice with interferon-inducible deletion of Cul4A. Cul4A deficiency resulted in DNA damage and apoptosis of rapidly dividing cells, and mutant mice died within 3 to 10 days after induction with dramatic atrophy of the intestinal villi, bone marrow, and spleen, and with hematopoietic failure. Cul4A deletion in vivo specifically increased cellular levels of the Cul4A ligase targets Cdt1 and p27Kip1 but not other known targets. Bone marrow transplantation studies with Cul4A deletion in engrafted cells specifically isolated analysis of Cul4A function to hematopoietic cells and resulted in hematopoietic failure. These recipients died within 9 to 11 days, demonstrating that in hematopoietic cells, Cul4A is essential for survival.
Introduction
Ubiquitin-mediated proteolysis is a key regulatory mechanism by which eukaryotic cells control critical processes, including cell-cycle progression, differentiation, apoptosis, development, signal transduction, and gene expression. In particular, the ubiquitin pathway controls the degradation of critical regulators of hematopoiesis, including p27Kip1 (p27), HoxA9, IκB, c-kit, Bcl-2, cyclin D, AML1, GATA-2, Gfi1, Hif1α, p53, and c-myc.1 Dysregulation of the ubiquitin pathway has been linked to oncogenesis and tumor development,1-4 and the proteasome inhibitor, bortezomib, is being used to treat relapsed multiple myeloma.5,6
Ubiquitination of targets destined for degradation at the 26S proteasome involves 3 classes of enzymes. The ubiquitin-activating enzyme (E1) requires ATP to activate ubiquitin. Activated ubiquitin is transferred to the ubiquitin-conjugating enzyme (E2), which transfers ubiquitin to the target protein with the ubiquitin ligase (E3).7,8 The modular cullin-RING ligases (CRLs) represent a large and diverse superfamily of E3 ubiquitin ligases that generate a multitude of targeting complexes through assembly of different subunits. The cullin subunit acts as a scaffold to bring together the target substrate and activated ubiquitin.9,10 Modularity derives from an N-terminal domain in the cullin that interacts with a specificity factor to recruit target substrates instead of directly binding the target. The specificity factor can be composed of one or more subunits.
Among vertebrates there are 7 related cullins (Cul1, Cul2, Cul3, Cul4A, Cul4B, Cul5, and Cul7). Cul4A and Cul4B are paralogs of the CUL-4 cullin, which is highly conserved through evolution and has orthologs in the fission yeast, Arabidopsis thaliana, Caenorhabditis elegans, and Drosophila melanogaster. Silencing experiments demonstrate Cul4 is important for regulating DNA replication in C elegans.11 Loss of Cul4 in Drosophila cells leads to G1 arrest with an increase in the CDK inhibitor Dacapo.12 In human cells, CUL4A is a core subunit of a CRL composed of CUL4A, the RING finger protein ROC1 and damaged DNA binding protein 1 (DDB1). DDB1 interacts with WD-40 repeat motif containing proteins to form specificity factors for the CUL4A ubiquitin ligase complex.13 CUL4A-DDB1 ligases ubiquitinate the following important targets: (1) the CDK inhibitor p27,12,14,15 (2) the homeodomain protein HOXA9, regulator of hematopoiesis and embryonic development,16 (3) CDT1, licensing factor required for DNA replication initiation,17 (4) the proto-oncogenic transcription factor c-Jun,18 (5) p53,19,20 and (6) DDB2, required for nucleotide excision repair.21-23
In vivo, we demonstrated that Cul4A is required for murine embryonic development, as Cul4A-null mutant embryos did not survive past day 7.5.24 In hematopoietic cell lines, we showed that Cul4A overexpression promotes proliferation and interferes with myeloid and erythroid terminal differentiation. With Cul4A haploinsufficient mice, we found Cul4A regulates the homeostasis of myeloid and erythroid progenitors.15,25 More recently, we demonstrated that Cul4A haploinsufficiency leads to reduced stem cell engraftment and self-renewal.26 Although these Cul4A+/− hematopoietic defects are clear, these animals are healthy, and these phenotypes are relatively subtle compared with the dramatic Cul4A-deficient phenotypes described in this report.
Although the requirement of CUL-4 in C elegans has been studied in vivo,11,27 besides our earlier studies15,25,26 the in vivo function of the Cul4A paralog in more complex metazoans, including vertebrates, has not been addressed. This report is the first to establish the requirement of Cul4A in an adult vertebrate. We generated mice with a Cul4A conditional null allele, where deletion was catalyzed by Cre recombinase expressed from an interferon-inducible transgene (Mx-Cre).28 Upon induction of Cre to delete Cul4A, mutant mice died 3 to 10 days after induction with severe atrophy in the bone marrow, spleen, and small intestine, all tissues with rapidly dividing cells, and apoptosis and DNA damage were detected. Although the Cul4A ubiquitin ligase has been shown to target for proteolysis multiple regulators, its relative contribution to the protein level of each has not been examined in vivo. We determined in bone marrow that Cul4A deletion in vivo led to marked increases in Cdt1 and p27 but little or no change in other known targets, suggesting that increased Cdt1 and p27 triggered apoptosis. Lastly, analysis of Cul4A function was isolated to hematopoietic cells by transplantation of Cul4Aflox/floxMx-Cre bone marrow into wild-type recipients to demonstrate that Cul4A in hematopoietic cells is essential for survival.
Methods
Animal studies were approved by the Indiana University Institutional Animal Care and Use Committee (IACUC).
Mice and treatments
Embryonic stem (ES) cells hemizygous for the Cul4A-T allele were generated (Figure 1A) and used to generate chimeric males with germ-line transmission (identified by Southern analysis).24 EIIa-Cre(B6.FVB-Tg(EIIa-cre)C5379Lmgd/J), Mx-Cre(B6.Cg-Tg(Mx-cre)1Cgn/J), and C57Bl/6J mice were from The Jackson Laboratory (Bar Harbor, ME).
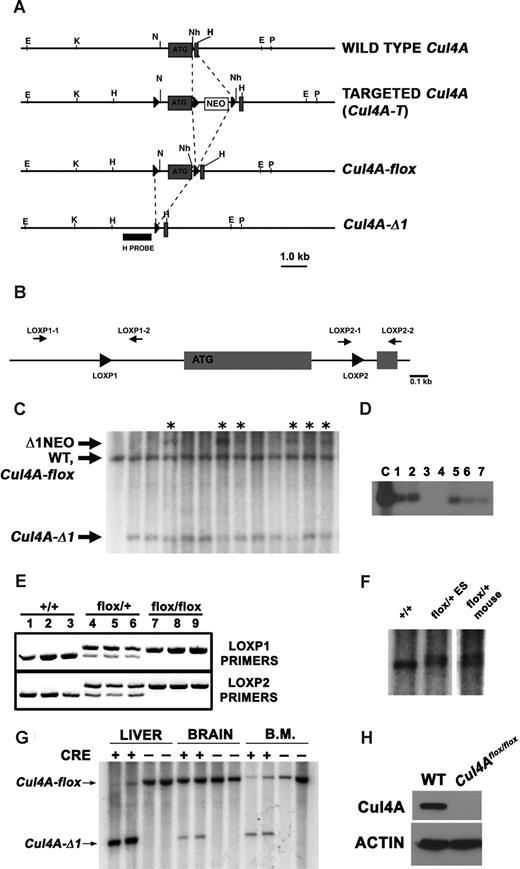
Generation of mice with an inducible deletion of Cul4A. (A) Restriction maps of the wild-type Cul4A, targeted allele (Cul4A-T), conditional deletion (Cul4A-flox), and Cul4A deletion (Cul4A-Δ1) are shown. Gray boxes denote exons; “ATG” denotes the first coding exon. E indicates EcoRI; K, KpnI; N, NotI; Nh, NheI; P, PstI; and H, HindIII. “H probe” was used for Southern analysis. Dashed lines indicate insertions and deletions. (B) PCR primers used for genotyping. PCR primers (black arrows) flank loxP sites (black triangles) that flank the first coding exon in the conditional Cul4A. (C) Genomic blot to identify Cul4A alleles in mosaic mice. Mutant and wild-type Cul4A alleles are indicated: Δ1NEO indicates deletion of the first coding exon, retention of the floxed Neo (3.7 kb); WT, wild-type Cul4A (3.3 kb); Cul4A-flox, Cul4A conditional deletion (3.3 kb); and Cul4A-Δ1, Cul4A deletion (2.0 kb). Asterisks indicate samples analyzed further by PCR for Cul4A-flox. (D) PCR screen to detect mosaics with Cul4A-flox. Genomic DNA from mosaics identified in the previous screen (C) were screened with PCR primers LOXP2–1 and LOXP2–2. A PCR fragment containing loxP was detected by DNA blot with an oligonucleotide probe. The positive control template (C) was genomic DNA from a Cul4Aflox/+ ES cell line. For panels C and D, representative sample blots are shown. (E) PCR screen to genotype Cul4A-flox mice. PCR primers flanking each loxP site were used to identify wild-type (+/+), Cul4Aflox/+ (flox/+), and Cul4Aflox/flox (flox/flox) mice. (F) Southern analysis to identify Cul4Aflox/+ mice. Genomic DNA from a wild-type mouse (+/+), a Cul4Aflox/+ ES cell line (flox/+ES), and a Cul4Aflox/+ mouse (flox/+mouse) was analyzed by Southern analysis. The doublet in the last 2 lanes consists of the Cul4A wild-type (smaller band) and the Cul4A-flox allele, 116 bp larger with 2 loxP sites. (G) Cul4A deletion is induced in liver and bone marrow. Mutant and control mice were treated with pIpC, and 5 days after induction, DNA from liver, brain, and bone marrow was analyzed by Southern analysis for Cul4A-flox and Cul4A-Δ1. (H) Cul4A protein is not detected in mutant liver after induction of Cre. Ten days after induction, Cul4A was detected by immunoblot in lysates from mutant and control mice. Actin was a loading control.
Generation of mice with an inducible deletion of Cul4A. (A) Restriction maps of the wild-type Cul4A, targeted allele (Cul4A-T), conditional deletion (Cul4A-flox), and Cul4A deletion (Cul4A-Δ1) are shown. Gray boxes denote exons; “ATG” denotes the first coding exon. E indicates EcoRI; K, KpnI; N, NotI; Nh, NheI; P, PstI; and H, HindIII. “H probe” was used for Southern analysis. Dashed lines indicate insertions and deletions. (B) PCR primers used for genotyping. PCR primers (black arrows) flank loxP sites (black triangles) that flank the first coding exon in the conditional Cul4A. (C) Genomic blot to identify Cul4A alleles in mosaic mice. Mutant and wild-type Cul4A alleles are indicated: Δ1NEO indicates deletion of the first coding exon, retention of the floxed Neo (3.7 kb); WT, wild-type Cul4A (3.3 kb); Cul4A-flox, Cul4A conditional deletion (3.3 kb); and Cul4A-Δ1, Cul4A deletion (2.0 kb). Asterisks indicate samples analyzed further by PCR for Cul4A-flox. (D) PCR screen to detect mosaics with Cul4A-flox. Genomic DNA from mosaics identified in the previous screen (C) were screened with PCR primers LOXP2–1 and LOXP2–2. A PCR fragment containing loxP was detected by DNA blot with an oligonucleotide probe. The positive control template (C) was genomic DNA from a Cul4Aflox/+ ES cell line. For panels C and D, representative sample blots are shown. (E) PCR screen to genotype Cul4A-flox mice. PCR primers flanking each loxP site were used to identify wild-type (+/+), Cul4Aflox/+ (flox/+), and Cul4Aflox/flox (flox/flox) mice. (F) Southern analysis to identify Cul4Aflox/+ mice. Genomic DNA from a wild-type mouse (+/+), a Cul4Aflox/+ ES cell line (flox/+ES), and a Cul4Aflox/+ mouse (flox/+mouse) was analyzed by Southern analysis. The doublet in the last 2 lanes consists of the Cul4A wild-type (smaller band) and the Cul4A-flox allele, 116 bp larger with 2 loxP sites. (G) Cul4A deletion is induced in liver and bone marrow. Mutant and control mice were treated with pIpC, and 5 days after induction, DNA from liver, brain, and bone marrow was analyzed by Southern analysis for Cul4A-flox and Cul4A-Δ1. (H) Cul4A protein is not detected in mutant liver after induction of Cre. Ten days after induction, Cul4A was detected by immunoblot in lysates from mutant and control mice. Actin was a loading control.
Southern and PCR analyses
To identify Cul4A alleles, Southern analyses were performed with H probe (Figure 1A) to a HindIII fragment that encompasses all of the mutations generated for this study.24 Polymerase chain reaction (PCR) was used to identify regions of the conditional Cul4A allele and for genotyping. To identify the loxP site upstream of the first coding exon, primers LOXP1-1 (caacagtgtttgcttgtgccactcccaggc) and LOXP1-2 (tgacagctgcccccaaagaagtgctctcac) were used to generate a 581-bp fragment from the mutant allele and a 523-bp fragment from the wild-type. To identify the loxP site downstream of the first coding exon, primers LOXP2-1 (cgacttgtccctgcacctc) and LOXP2-2 (gtacagctcctccaggttgtacc) were used to generate a 313-bp and 255-bp fragment from mutant and wild-type alleles, respectively. When the 313-bp fragment with this downstream loxP site was not detectable by ethidium bromide, it was detected by Southern analysis with end-labeled loxP oligonucleotide probe (ggccgcataacttcgtataatgtatgctatacgaagttatc).
Hematopoietic cell analyses
Spleen cell suspensions were prepared by flushing spleens with RPMI 1640 plus 10% fetal bovine serum (FBS) and 100 U/mL penicillin/streptomycin. Low-density mononuclear cells (LDMNCs) were prepared and progenitors were assayed.25 Peripheral blood counts were determined using a Hemavet hematology analyzer (Drew Scientific, Oxford, CT).
For transplants, bone marrow was isolated from donors, Cul4Aflox/flox Mx-Cre mice (mutant) or Cul4Aflox/flox mice without Mx-Cre (controls), and 3 × 106 LDMNCs were transplanted by tail vein injection into 6- to 12-week-old wild-type littermate lethally irradiated recipients (1100 cGy split dose).26
Histology
Tissues were collected in 10% neutral-buffered formalin. Femur, tibia, and sternum were decalcified 3 days prior to embedding (Formic Decalcifying Solution; US Biotex, Webbville, KY). Tissues were dehydrated, embedded, sectioned at 6 μm, and stained with hematoxylin and eosin.
Cell culture
For LDMNC in vitro culture, 1 × 106 cells/mL were plated on 3.5 cm non–tissue culture dishes in IMDM plus 20% FBS 100 ng/mL TPO, 100 ng/mL SCF, 50 ng/mL Flt-3 ligand, and 233 U/mL IL-6 (Peprotech, Rocky Hill, NJ). For immunoblots, cells were treated with 2.7 mM diisopropylfluorophosphate (Sigma-Aldrich, St Louis, MO) for 10 minutes on ice and lysed in 8 M urea buffer (8 M urea, 10 mM Tris-HCl, 100 mM NaH2PO4, pH 8.0).
Cell cycle, apoptosis, and DNA damage
Annexin V and cell-cycle distribution analyses were performed as described.15,25 For TUNEL analysis, 1 × 105 cells were washed and slide preparations (Cytospin3; Shandon, Pittsburgh, PA) were fixed with 4% paraformaldehyde for 1 hour and permeabilized with 0.1% TritonX-100 for 2 minutes on ice, and apoptotic cells were detected according to the manufacturer's protocol (Roche, Indianapolis, IN). Nuclei were stained with Hoechst 33342 (Invitrogen, Carlsbad, CA) for 2 minutes, followed by fluorescent microscopy. Cells with DNA damage were detected by comet assay.29
Immunoblots
Immunoblots were prepared as described.25 Loading was normalized to GAPDH (CB1001; Calbiochem, San Diego, CA) or β-actin. Cul4A protein was detected with anti-Cul4A antiserum.25 Other antibodies used were anti-Cdt1 (sc-28262), -p27 (sc-776), -p53 (sc-1312), –c-Jun (sc-45), -DDB2 (sc-25367), –β-actin (sc-47778), –goat-HRP (sc-2020), –rabbit-HRP (sc-2357), and –mouse-HRP (sc-2005) from Santa Cruz Biotechnology (Santa Cruz, CA) and anti-HoxA9 (07-178; Upstate, Temecula, CA).
Results
Generation of Cul4A conditional deletion mice
Deletion of Cul4A in the mouse is embryonic lethal,24 so to study further its in vivo function, we generated a mouse strain with a Cul4A conditional deletion allele. A targeting construct with loxP sites flanking the first coding exon and floxed NeoR in the downstream intron was constructed and mouse ES cells hemizygous for this mutant allele (Cul4A-T) were generated as described (Figure 1A).24 Transient transfection of these cells with a Cre recombinase plasmid24 yielded a subclone that lacked NeoR but retained the floxed exon (Cul4A-flox), but multiple attempts to obtain germ-line transmission with this cell line failed, perhaps because it lost totipotency.
Holzenberger et al encountered this technical difficulty when deriving conditional deletions of the insulin-like growth factor type I receptor locus and devised a solution whereby partial Cre recombination to remove the selectable marker was caused to occur in vivo.30 Following this approach, the Cul4A-T ES cell line was used to obtain males exhibiting germ-line transmission, and Cul4A-T, EIIa-Cre double transgenic offspring were derived. EIIa-Cre is expressed only in oocytes and during preimplantation stages of the embryo. Therefore, in male double transgenic offspring, Cre was expressed only during the morula and blastocyst stages. Mosaic offspring with a variety of Cre recombination-derived deletion alleles resulted. DNA samples from tail biopsies from 112 mosaic males were analyzed by Southern analysis (Figure 1C). Because the desired Cul4A-flox allele was not easily distinguished from the wild-type Cul4A, mice with 3 visible Cul4A bands were chosen for further screening. It was reasoned that if the other recombined alleles were detected, the conditional allele might also be present. Genomic DNA was analyzed by PCR followed by DNA blot to detect alleles where the second loxP site (LOXP2 in Figure 1B) was only 78 bp from the second coding exon (Figure 1D) indicating deletion of the floxed Neo and uniquely characteristic of the conditional allele. Eight mosaic mice with this gene were backcrossed. Offspring were screened by PCR for the transmission of the Cul4A-flox allele (Figure 1E), and genomic DNA was analyzed by Southern analysis to confirm that the overall structure of this conditional deletion allele was the same as the wild type (Figure 1F). In addition, PCR and DNA sequence analysis confirmed the position of each loxP site (D.L.W., unpublished results, October 2007).
The interferon-inducible Mx-Cre transgene was used to induce deletion of Cul4A-flox.28 Homozygous mutant mice with and without Mx-Cre were generated, and Cre expression was induced (“Methods”). Five days after the beginning of induction, liver, brain, and bone marrow cells were analyzed by Southern analysis (Figure 1G). Mx-Cre was reported to cause deletion of floxed genes in liver, spleen, and duodenum (70%-100%), deletion to a lesser extent in heart, lung, uterus, thymus, and kidney (40%-50%), even less in muscle and tail (20%), and very little in brain (less than 10%).28 Comparing the relative amounts of Cul4A-flox and Cul4A-Δ1 alleles within a sample, most of the Cul4A-flox alleles were deleted in mutant liver cells after 5 days, and no deletion was detected in controls. Under these conditions, little deletion was observed in brain, whereas robust deletion was observed in bone marrow. In liver, Cul4A protein expression was reduced to an undetectable level (Figure 1H). Overall, these findings indicate that Cul4A deletion was effectively induced in tissues previously reported for Mx-Cre.
Deletion of Cul4A in adult mice causes a dramatic loss of bone marrow cellularity and spleen and villus atrophy, and mutant mice die within 3 to 10 days
To determine the physiologic effect of Cul4A deficiency, Cul4Aflox/flox Mx-Cre (mutant) mice and Cul4Aflox/flox without Mx-Cre (controls) were induced with pIpC and monitored. At 3 days after induction, mutant mice began to die, and none survived longer than 10 days (n = 7, Figure 2A). Control mice remained healthy and sur-vived (n = 4). Wild-type mice with the Mx-Cre transgene and Cul4Aflox/wtMx-Cre mice also remained healthy following induction with pIpC, confirming that Cre alone was not toxic under these conditions and the presence of only a single Cul4A allele does not lead to death (N.J., unpublished results, April-May 2006).
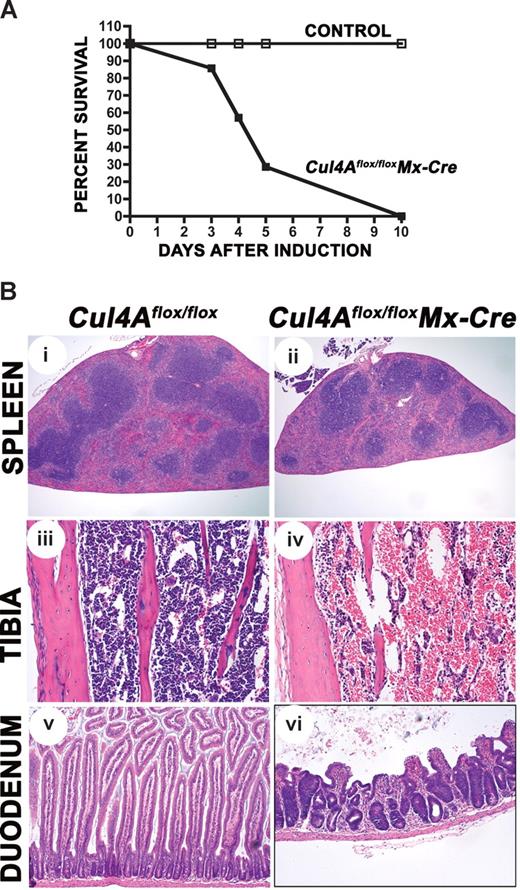
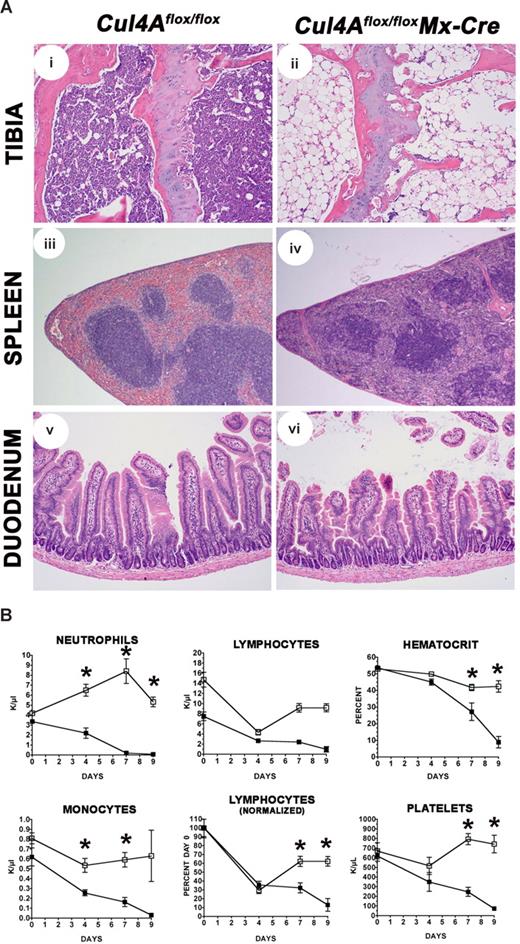
Cul4A deletion is lethal. (A) Mutant (■) and controls (□) were induced with pIpC (n = 7 and n = 4, respectively) and monitored daily. Results from 3 independent experiments were combined. Percentage of the total surviving is graphed versus days after the initiation of induction. (B) Mutant and control mice were induced with pIpC. Four days after induction, sections from spleen, tibia for bone marrow, and duodenum were prepared. For each tissue, mutant and control images are shown at the same magnification. An Olympus BX41 microscope (Center Valley, PA) with Plan N 4×/0.10, Plan 10×/0.25, and Plan N 20×/0.40 objectives were used. Images were captured with a SPOT Insight 2MP Firewire Color Mosaic camera (Diagnostic Instruments, Sterling Heights, MI) and Spot Imaging software. Student t test was used for all statistical analyses. Mean and standard error of the mean are reported. Cul4Aflox/flox without Mx-Cre mice were controls in all experiments. In addition, wild-type mice with Mx-Cre were also controls for experiments for Figures 2–3 and Table 1 (N.J., unpublished results, April-May 2006). For these experiments, following pIpC induction, viability, changes in peripheral blood counts, and bone marrow total cellularity were the same in both control strains, and bone marrow progenitor frequency was slightly higher in wild-type Mx-Cre mice, indicating that Cre expression alone does not contribute to the lethality and hematopoietic failure resulting from Cul4Adeletion.
Cul4A deletion is lethal. (A) Mutant (■) and controls (□) were induced with pIpC (n = 7 and n = 4, respectively) and monitored daily. Results from 3 independent experiments were combined. Percentage of the total surviving is graphed versus days after the initiation of induction. (B) Mutant and control mice were induced with pIpC. Four days after induction, sections from spleen, tibia for bone marrow, and duodenum were prepared. For each tissue, mutant and control images are shown at the same magnification. An Olympus BX41 microscope (Center Valley, PA) with Plan N 4×/0.10, Plan 10×/0.25, and Plan N 20×/0.40 objectives were used. Images were captured with a SPOT Insight 2MP Firewire Color Mosaic camera (Diagnostic Instruments, Sterling Heights, MI) and Spot Imaging software. Student t test was used for all statistical analyses. Mean and standard error of the mean are reported. Cul4Aflox/flox without Mx-Cre mice were controls in all experiments. In addition, wild-type mice with Mx-Cre were also controls for experiments for Figures 2–3 and Table 1 (N.J., unpublished results, April-May 2006). For these experiments, following pIpC induction, viability, changes in peripheral blood counts, and bone marrow total cellularity were the same in both control strains, and bone marrow progenitor frequency was slightly higher in wild-type Mx-Cre mice, indicating that Cre expression alone does not contribute to the lethality and hematopoietic failure resulting from Cul4Adeletion.
To identify which organ(s) failed, mutant and control animals were induced with pIpC and necropsies were performed 4 days after induction. Histologic survey of tissues where Mx-Cre was reported to be expressed (liver, spleen, small intestine, heart, lung, uterus, kidney, and bone marrow) and the brain, where little Mx-Cre was expressed,28,31-33 showed that all tissues except the spleen, bone marrow, and small intestine appeared normal. Mutant spleens were clearly smaller (Figure 2Bi,ii; Table 1), with a 2-fold reduction in weight (45.6 ± 4.4 mg for mutants and 87.2 ± 4.4 mg for controls). The spleen capsule was irregular and the red pulp was reduced in size with marked loss of hematopoietic cells. There was a moderate reduction of the white pulp with increased apoptosis of lymphocytes compared with controls. The bone marrow of the femur, tibia, and sternum was markedly hypocellular, and large, cavernous, blood-filled sinuses occupied most of the bone marrow cavity (Figure 2Biii,iv). The thymus was small in both the induced and control mice, presumably a result of spontaneous interferon production as previously described28 (and H.H., unpublished results, October 2006). Intestinal pathology was mostly limited to the small intestine and characterized by marked villus atrophy (Figure 2Bv,vi). The number of apoptotic enterocytes was increased at the base and along the sides of the crypts. Scattered crypts were dilated with necrotic cellular debris and lined by flattened epithelial cells. The majority of crypts contained hyperchromatic enterocytes with enlarged nuclei and loss of polarity. Villus enterocytes were swollen with vacuolated cytoplasm and small pycnotic nuclei. In other places, remnants of villi were covered by flattened epithelial cells. In the large intestine, a few apoptotic epithelial cells were present in crypts (H.H., unpublished results, October 2006).
Cul4A deletion causes a dramatic loss of hematopoietic cells
| Genotype . | Weight, mg . | Total cellularity, ×106 . | Progenitors . |
|---|---|---|---|
| Bone marrow (femur) | |||
| Control | 11.62 ± 1.24 | 45 200 ± 5200 | |
| Cul4Aflox/flox Mx-Cre | 1.11 ± 0.27 | 12 ± 12 | |
| Spleen | |||
| Control | 87.2 ± 4.4 | 57.79 ± 7.6 | 9660 ± 2920 |
| Cul4Aflox/flox Mx-Cre | 45.6 ± 4.4 | 31.09 ± 8.1 | 121 ± 121 |
| Genotype . | Weight, mg . | Total cellularity, ×106 . | Progenitors . |
|---|---|---|---|
| Bone marrow (femur) | |||
| Control | 11.62 ± 1.24 | 45 200 ± 5200 | |
| Cul4Aflox/flox Mx-Cre | 1.11 ± 0.27 | 12 ± 12 | |
| Spleen | |||
| Control | 87.2 ± 4.4 | 57.79 ± 7.6 | 9660 ± 2920 |
| Cul4Aflox/flox Mx-Cre | 45.6 ± 4.4 | 31.09 ± 8.1 | 121 ± 121 |
Four days after induction of Cre with pIpC, Cul4Aflox/flox Mx-Cre mice and controls were killed (n=8 and n=7, respectively). Spleens were weighed, and the number of splenocytes per spleen was determined, as well as the total number of LDMNCs per femur. Splenocytes and LDMNCs were then cultured in progenitor cell methylcellulose colony assay, and after 7 days total colonies were counted, and the number of progenitors per spleen or femur was calculated. The results from 2 independent experiments were combined. Means and standard error of the mean are reported. P ≤ .03.
These results show Cul4A is important in the small intestine, so further studies are of interest. However, because we observed marked changes in hematopoietic tissues, and because our previous findings indicated Cul4A is required for normal hematopoiesis,15,25,26 we investigated the effects of Cul4A deletion on cells of the bone marrow, spleen, and peripheral blood.
Cul4A deficiency in hematopoietic tissues
Cul4A deletion caused a 2-fold reduction in spleen cellularity and a 10-fold decline in bone marrow cellularity 4 days after induction (Table 1). In vitro functional assays for hematopoietic progenitors determined that these cells were drastically reduced to essentially zero, 3800-fold in the bone marrow and 80-fold in mutant spleens compared with controls. Peripheral blood counts of mature hematopoietic cells were also reduced in mutant animals (Figure 3). Neutrophil counts in mutant mice declined rapidly and were near zero by 4 days. For monocytes and lymphocytes, both mutant and control counts declined initially. However, monocyte counts in mutants declined to a significantly lower level by day 4 and did not recover in contrast to controls. The lymphoid counts were normalized to day 0 due to differences in the steady-state levels of lymphoid counts in uninduced mutant and control animals. This difference is likely due to a low level of leakiness from the Mx-Cre transgene as was previously reported.34 As with monocyte counts, control animal lymphocyte counts were consistently observed to drop following induction due to interferon response, as previously observed,35,36 but these always recovered compared with the mutant animal counts, which continued to decline. Hematocrits in mutants were similar to controls, except for the single mutant mouse that survived 10 days, with a hematocrit of only 0.14 (14%) compared with 0.43 (43%) for the controls. Platelet counts initially declined in both mutant and control animals. For the single mutant mouse that survived 10 days, its platelet count at 276 × 109/L was much less than the control counts (an average of 2180 × 109/L), which had recovered. A common feature of the mature blood cells whose counts declined rapidly is that these cells have the shortest lifespan and is consistent with Cul4A deletion affecting hematopoietic progenitors (Table 1).
Cul4A loss causes a decline in peripheral blood hematopoietic cells. Mutant (■) and controls (□) were induced with pIpC, and peripheral blood counts were determined and plotted versus days after induction. For panel D, lymphocyte counts were normalized to the counts on day 0. Asterisks denote time points where mutant and control count differences were statistically significant (P ≤ .03). Results from 6 independent experiments were combined.
Cul4A loss causes a decline in peripheral blood hematopoietic cells. Mutant (■) and controls (□) were induced with pIpC, and peripheral blood counts were determined and plotted versus days after induction. For panel D, lymphocyte counts were normalized to the counts on day 0. Asterisks denote time points where mutant and control count differences were statistically significant (P ≤ .03). Results from 6 independent experiments were combined.
Cul4A deficiency causes apoptosis in the bone marrow
The dramatic loss of cells in the bone marrow and of mature hematopoietic cells in the peripheral blood and the observation of apoptotic cells in the small intestine suggested that Cul4A deficiency caused apoptosis in the bone marrow. Since apoptotic cells in the bone marrow are rapidly phagocytosed by macrophages,37 Cul4A deletion was induced in vivo, followed by isolation and analysis of mutant bone marrow in vitro. Mutant and control mice were induced with pIpC and 2 days later bone marrow was isolated and cultured for 2 to 5 days prior to staining for apoptotic cells (annexin V–positive, 7AAD-negative staining cells). As shown in Figure 4A, at 4 and 5 days after induction, the frequency of apoptotic cells was significantly greater for the mutant bone marrow cells than for the controls (P = .01 and P = .03, respectively). In addition, TUNEL assay was performed to detect cells at a later stage of apoptosis. At 5 days after induction, the frequency of apoptotic cells was 4.5-fold greater in the mutant cells (Figure 4B). In addition, cell-cycle distribution was quantified during cell culture. Under homeostatic conditions, most bone marrow cells are in G0/G1. However, in vitro culturing of these cells required the addition of cytokines to support survival, and these conditions are known to drive wild-type cells to proliferate. Consequently, the proportion of cells in S-phase increased as the proportion in G0/G1 declined (Figure 4C). However, bone marrow cells derived from in vivo–induced mutant mice did not proliferate in vitro and fewer entered S-phase (Figure 4C). Although the failure of mutant cells to respond to cytokines could have caused this failure to proliferate, this result is also consistent with these cells instead undergoing apoptosis, as demonstrated in Figure 4A,B.
Cul4A protein loss in bone marrow following in vivo Cul4A deletion leads to apoptosis and altered cell cycle distribution. Mutant mice (closed bars or symbols) and controls (open bars or symbols) were induced with pIpC, and 48 hours later LDMNCs were harvested from femurs and plated in vitro. At each time point, cells were collected and analyzed by (A) annexin V–positive, 7AAD-negative staining, (B) TUNEL assay, and (C) DNA content for cell-cycle distribution. Asterisks denote time points where mutant and control differences were statistically significant (P ≤ .04). Cells for these assays were cultured under identical conditions or were assayed simultaneously for apoptosis (A,B) or cell-cycle distribution (C), so results for corresponding time points are comparable. For panel C, ■ and □ represent average percentage cells in G0/G1-phase, and ▴ and ▵ represent average percentage cells in S-phase. Results from independent experiments were combined: 4 for panel A, 2 for panel B, and 3 for panel C. M indicates mutant; C, control.
Cul4A protein loss in bone marrow following in vivo Cul4A deletion leads to apoptosis and altered cell cycle distribution. Mutant mice (closed bars or symbols) and controls (open bars or symbols) were induced with pIpC, and 48 hours later LDMNCs were harvested from femurs and plated in vitro. At each time point, cells were collected and analyzed by (A) annexin V–positive, 7AAD-negative staining, (B) TUNEL assay, and (C) DNA content for cell-cycle distribution. Asterisks denote time points where mutant and control differences were statistically significant (P ≤ .04). Cells for these assays were cultured under identical conditions or were assayed simultaneously for apoptosis (A,B) or cell-cycle distribution (C), so results for corresponding time points are comparable. For panel C, ■ and □ represent average percentage cells in G0/G1-phase, and ▴ and ▵ represent average percentage cells in S-phase. Results from independent experiments were combined: 4 for panel A, 2 for panel B, and 3 for panel C. M indicates mutant; C, control.
Cul4A deficiency causes the accumulation of Cdt1 and p27
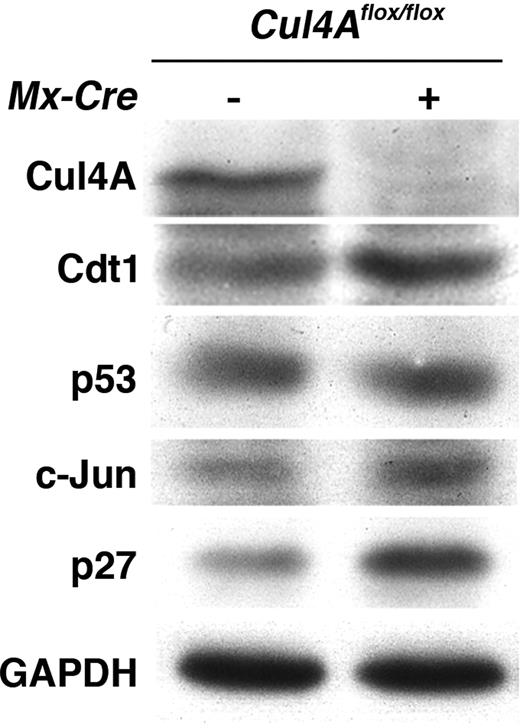
It was shown that knockdown of Cul4A protein in tissue culture and of CUL-4 in C elegans leads to the accumulation of target substrates. Here we show for the first time in primary cells the efficient knockdown of Cul4A protein via Cul4A deletion and analysis of target substrates. Two well-defined targets of the Cul4A ubiquitin ligase are the DNA replication licensing factor, Cdt1, and the cyclin-dependent kinase inhibitor, p27. Stabilization of Cdt1 protein leads to DNA damage, cell-cycle arrest, and apoptosis, while stabilization of p27 protein leads to G0/G1 arrest and apoptosis.38,39 To determine whether these and any of the other known Cul4A ligase complex targets are overexpressed in Cul4A-deficient cells, mutant mice and controls were induced with pIpC, 2 days later bone marrow LDMNCs were purified, and the expression levels of Cul4A targets was determined by immunoblot (Cdt1, p27, p53, c-Jun, HoxA9, and DDB2). As shown in Figure 5, Cul4A protein expression was efficiently knocked down following induction. Concomitantly, Cdt1 and p27 protein levels increased markedly, while c-Jun protein levels were only slightly increased, and p53 protein level remained unchanged. HoxA9 and DDB2 proteins were not detected under these conditions. These results suggest a mechanistic basis for the apoptosis observed in primary culture and the changes in hematopoietic tissues and intestine following deletion of Cul4A.
Cul4A deletion and protein loss in vivo leads to Cdt1 and p27 protein accumulation in bone marrow. Mutant mice and controls were induced with pIpC, and 48 hours later LDMNCs were harvested from femurs. Total cellular protein was analyzed by immunoblot. GAPDH was a loading control. Representative results are shown for at least 3 independent experiments.
Cul4A deletion and protein loss in vivo leads to Cdt1 and p27 protein accumulation in bone marrow. Mutant mice and controls were induced with pIpC, and 48 hours later LDMNCs were harvested from femurs. Total cellular protein was analyzed by immunoblot. GAPDH was a loading control. Representative results are shown for at least 3 independent experiments.
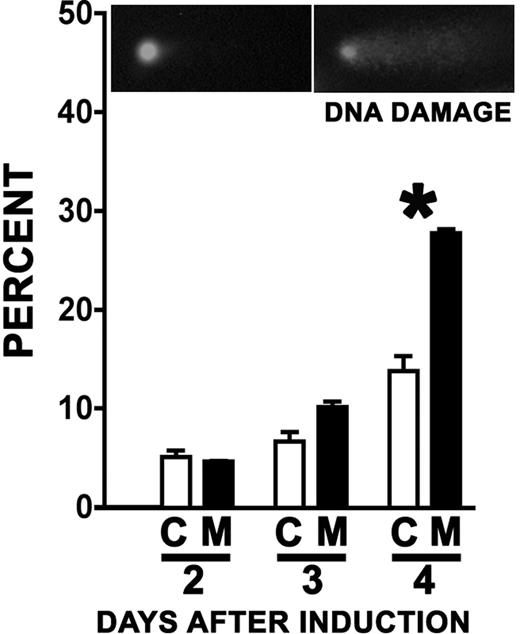
Because a failure to regulate Cdt1 would be expected to cause DNA damage, mutant and control mice were induced in vivo, bone marrow was isolated and cultured in vitro (as described for Figure 4), and cells with damaged DNA were detected by comet assay. A clear increase in the frequency of mutant cells with damaged DNA compared with controls was observed 4 days after induction (Figure 6, P = .01), supporting the notion that the hematopoietic failure resulting from Cul4A deletion is mediated in part by Cdt1.
Cul4A deletion leads to an increase in cells with DNA damage. Mutant mice (■) and controls (□) were induced and bone marrow LDMNCs were harvested and cultured in vitro as described for Figure 4. At 2 to 4 days after induction, cells were collected and analyzed for DNA damage by comet assay. The asterisk denotes a statistically significant difference (P = .01). A representative result of 3 independent experiments is shown. Inset images are of representative cells with ethidium bromide–stained DNA; a cell exhibiting no DNA damage (left) and one with damaged DNA (right).
Cul4A deletion leads to an increase in cells with DNA damage. Mutant mice (■) and controls (□) were induced and bone marrow LDMNCs were harvested and cultured in vitro as described for Figure 4. At 2 to 4 days after induction, cells were collected and analyzed for DNA damage by comet assay. The asterisk denotes a statistically significant difference (P = .01). A representative result of 3 independent experiments is shown. Inset images are of representative cells with ethidium bromide–stained DNA; a cell exhibiting no DNA damage (left) and one with damaged DNA (right).
Cul4A in hematopoietic cells is essential for viability
To specifically isolate the effect of Cul4A deletion to hematopoietic cells, we used transplantation of mutant bone marrow into wild-type recipient mice. Bone marrow was isolated from mutant or control mice and transplanted separately into lethally irradiated wild-type recipients. The donor cells were allowed to engraft for 2 months, and then Cre-mediated deletion was induced with pIpC. Induced mice were monitored closely and when mutants appeared sick and near death, they were killed for necropsy. Of 5 recipients of mutant bone marrow, all died 9 to 11 days after induction. The 6 controls, which did not become sick, were killed and necropsied at the same time as mutants.
Necropsies of the mutant engrafted mice near death indicated a failure of hematopoiesis. The bone marrow, composed of mostly hematopoietic cells but also containing cells of the microenvironment, was nearly devoid of cells (Figure 7Ai,ii). However, because the bone marrow microenvironment was expected to have been host derived and therefore wild type, the expectation was that these cells would remain after Cul4A deletion in mutant bone marrow–derived hematopoietic cells. The presence of adipocytes, part of the microenvironment, is consistent with this. However, more specific analyses for cells of the microenvironment would be required to establish its status. The weight of the spleen was only 29% of the weight of controls (43 ± 6 mg compared with 150 ± 16 mg, P < .001), with much of this decrease corresponding with a dramatic loss of red pulp, as well as a lesser, but marked loss of white pulp (Figure 7Aiii,iv). However, unlike the Cul4Aflox/floxCre mice, the small intestine of the recipients of mutant bone marrow had no lesions (Figure 7Av,vi). This is consistent with the intestine in these animals being wild type and the mutant phenotype in these transplant recipients being caused by Cul4A-deficient hematopoietic cells. The decline of neutrophil, monocyte, and lymphocyte counts in these mutant transplant recipients was similar to that observed in Cul4Aflox/floxCre mice after Cre induction (Figures 3,7B). Most of these induced mutant animals died before changes in hematocrit and platelet counts became apparent. But the transplant recipients of mutant bone marrow survived longer, and the decline in these counts was more severe, with hematocrits falling to 21% of the controls and platelet counts to 10% of the controls.
Loss of Cul4A in the bone marrow is sufficient to cause hematopoietic failure and is lethal. (A) Cul4A deletion in transplant recipients of mutant bone marrow causes a dramatic loss of bone marrow and spleen cellularity. Recipients of either mutant or control bone marrow LDMNCs were induced with pIpC and monitored daily. Sick, near-death recipients of mutant cells were killed along with a control recipient and sections of tibia, spleen, and duodenum were prepared. For each tissue, mutant and control images are shown at the same magnification. The same microscope and camera were used as described for Figure 2. (B) For transplant recipients described in panel A, blood counts from recipients of mutant (■) and control (□) bone marrow were graphed versus days after induction. Asterisks denote time points where mutant and control count differences were statistically significant (P ≤ .03). Results from 3 independent experiments were combined.
Loss of Cul4A in the bone marrow is sufficient to cause hematopoietic failure and is lethal. (A) Cul4A deletion in transplant recipients of mutant bone marrow causes a dramatic loss of bone marrow and spleen cellularity. Recipients of either mutant or control bone marrow LDMNCs were induced with pIpC and monitored daily. Sick, near-death recipients of mutant cells were killed along with a control recipient and sections of tibia, spleen, and duodenum were prepared. For each tissue, mutant and control images are shown at the same magnification. The same microscope and camera were used as described for Figure 2. (B) For transplant recipients described in panel A, blood counts from recipients of mutant (■) and control (□) bone marrow were graphed versus days after induction. Asterisks denote time points where mutant and control count differences were statistically significant (P ≤ .03). Results from 3 independent experiments were combined.
Six transplant recipients of mutant bone marrow survived after induction and appeared healthy, but 3 to 4 weeks after induction, Southern analysis to genotype their bone marrow revealed these cells were wild type. PCR analysis confirmed these cells lacked loxP sites and therefore, must have arisen from wild-type host bone marrow cells that had not been ablated before transplantation and were able to engraft and rescue the loss of Cul4A-deficient cells. This result supports the likeli-hood that lethality results from hematopoietic cell–intrinsic Cul4A deficiency.
Discussion
Cullins were initially identified as core components of multisubunit ubiquitin ligases.40 Since then, it has become clear that these proteins can assemble with different specificity factors, where each determines a distinct target of ubiquitination. Consequently, each cullin is required for targeting multiple substrates. Numerous in vitro studies examined how a Cul4A ligase ubiquitinates a particular target and the affected pathway, but little is known about its function in vivo. In this report, we examine the relative importance of these functions for Cul4A ubiquitin ligases in hematopoietic cells in vivo by generating and characterizing a Cul4A conditional deletion mouse. Our findings demonstrate that in the bone marrow, spleen, and small intestine, all tissues with proliferating cells, Cul4A deletion causes apoptosis. Mutant mice died within 3 to 10 days, indicating Cul4A is essential for viability. Among known Cul4A targets, only Cdt1 and p27 protein levels increased markedly upon Cul4A deletion, suggesting that in bone marrow cells, Cul4A ubiquitin ligases contribute more to Cdt1 and p27 degradation than to that of other known targets and that the absence of this function leads to DNA damage and apoptosis. We focused on the requirement of Cul4A for hematopoiesis and showed that its deletion in bone marrow–derived cells results in hematopoietic failure, and these mice died within 9 to 11 days, demonstrating that Cul4A in hematopoietic cells specifically is essential for viability.
Cul4A deficiency caused severe atrophy of the small intestine and hematopoietic tissues. The lesions in the intestine resemble those observed following irradiation and are likely caused by a block in cell proliferation in the crypts.41,42 In the small intestine, stem cells in the base of the crypts give rise to progenitor cells that differentiate into enterocytes that migrate toward the tip of the villus and are shed. Diminished renewal of enterocytes leads to a reduction in the length of the villi. Interference with enterocyte production also compromises the intestinal epithelial barrier and may lead to sepsis. It is likely that susceptibility of the mice to sepsis was augmented by the loss of white blood cells, and together these defects brought about the death of Cul4A-deficient mice.
Focusing on hematopoietic cells, we observed in Cul4A-deficient bone marrow cells that apoptosis followed Cdt1 and p27 accumulation (Figures 4,5), consistent with what is known about Cdt1 and p27 functions. Cdt1 is required to initiate DNA replication and its degradation by a Cul4A-ubiquitin ligase ensures the genome is replicated only once during a cell cycle.17 In C elegans, without the CUL-4 cullin, CDT-1 protein levels increased, multiple rounds of DNA replication were initiated in a single cell cycle, and development arrested at the L2 larval stage.11 Because there are 2 mammalian paralogs for CUL-4 (Cul4A and Cul4B), it was not known which or whether both perform this function in more complex metazoans. Our in vivo findings implicate the Cul4A cullin alone. Studies using cell lines and in vitro approaches indicate that Cdt1 overexpression causes rereplication of DNA, DNA damage, activation of G2/M checkpoints, and apoptosis.43 However, whether checkpoints function in transformed cells in the same manner as nontransformed cells is not clear. Our findings in vivo and with primary cells establish that in nontransformed cells DNA damage and apoptosis result when Cul4A is depleted (Figures 4,6) and indicate that a Cul4A ubiquitin ligase is required to down-regulate Cdt1.
We also observed marked accumulation of p27 in Cul4A-deficient primary cells, suggesting that its overexpression might also contribute to apoptosis. Recently, it was shown in C elegans that depletion with RNAi of CKI-1, a p27 homolog, partially suppressed the cul-4 mutant phenotype.44 However, although DNA rereplication was suppressed, development still arrested and CDT-1 protein still accumulated, indicating that CKI-1 accumulation contributes to the rereplication phenotype but not CDT-1 accumulation or the arrest in development. Therefore, our findings indicate that in vertebrates both Cdt1 and p27 are important targets of Cul4A-mediated proteolysis, that the inappropriate accumulation of both leads to apoptosis, and that these pathways are evolutionarily conserved. In hematopoietic cells, the fundamental, essential nature of these pathways is underscored by the failure of hematopoiesis following Cul4A deletion.
Although both myeloid and erythroid progenitors essentially disappeared by 4 days after induction of Cul4A deletion (Table 1), mature cells in the peripheral blood were lost at different rates, and all were lost more slowly than progenitors. Neutrophil counts in mutant animals were approximately half the control before 4 days after induction, and monocyte counts declined to a similar level shortly thereafter (Figures 3,7B). Platelet, lymphocyte, and hematocrit levels in mutants dropped to about half control levels by 5 to 8 days. Because platelets and erythrocytes lack nuclei, undoubtedly these cells were indifferent to Cul4A deletion. Nevertheless, the relative rates with which mature blood cells were lost approximately correspond with the relative period of time each cell type normally circulates in the peripheral blood, suggesting that mature cells were indirectly affected by Cul4A deletion.45-49 It is likely that the loss of immature hematopoietic cells in the bone marrow halted generation of new mature hematopoietic cells, so peripheral blood counts declined as aging mature cells were not replaced. In fact, the rate of erythrocyte loss mirrors that observed after wild-type mice were treated with 5-fluorouracil, which kills rapidly proliferating cells, including hematopoietic progenitors.15
We also observed a variable requirement for Cul4A in different tissues. Although Cul4A deletion caused efficient depletion of Cul4A protein in liver, these cells appeared normal at the same time the bone marrow was nearly empty, the spleens were half the size of controls, and the small intestine enterocytes were undergoing apoptosis (Figures 1G,H,2B, and H.H., unpublished results, October 2006). For the tissues where there was marked atrophy following Cul4A deletion, normally these cells are proliferating. Taken together, these findings suggest that the failure of a Cul4A ubiquitin ligase to regulate Cdt1 and p27 degradation causes apoptosis in proliferating cells. In fact, Cang et al recently showed that loss of DDB1 (which forms part of the Cul4A ubiquitin ligase core) from mouse brain, lens, or epidermis caused apoptosis in proliferating cells.50,51 Similarities between these Cul4A- and DDB1-deficient phenotypes likely correspond with their encoded proteins being components of the same Cul4A ubiquitin ligases.
In general, the relative impact of either Cul4A or DDB1 deficiency on the expression of some known Cul4A-DDB1 ubiquitin ligase targets is different for different tissues. From our findings, Cdt1 and p27 protein levels increase markedly in Cul4A-deficient bone marrow cells, while c-Jun increase is modest (Figure 5). In murine embryonic fibroblasts, DDB1 deficiency causes marked up-regulation of all 3 proteins, but in mutant keratinocytes, c-Jun level increases, but p27 and Cdt1 protein levels remain unchanged.50,51 These findings illustrate how examination of mutant phenotypes in vivo can reveal varied functions in different cell types.
Although the common requirement for Cul4A and DDB1 as components of the same ubiquitin ligases and similarities between their null phenotypes are apparent, because DDB1-null embryos die much later than Cul4A-null embryos (embryonic day 12.5 compared with earlier than day 7.5), these 2 genes must also have distinct functions.24,50 The fact that Cul4A-null embryos die earlier indicates there are requirements for Cul4A that are independent of DDB1. Additional investigation is required to further elucidate common and distinct functions.
Because the Mx-Cre transgene is not expressed ubiquitously, Cul4A function in tissues where this Cre was not expressed or poorly expressed was not evaluated. In addition, for tissues where a mutant phenotype would require longer than 10 days to become apparent, our mutant mice died before their phenotypes could be examined. Therefore, Cre recombinase transgenes with different tissue specificities will be needed to assess Cul4A function in other tissues.
In general, ubiquitin-mediated proteolysis has been implicated in oncogenesis and tumorigenesis, and our findings along with those of others suggest that Cul4A could also play a role. Successful treatment of relapsed multiple myelomas with bortezomib (a proteasome inhibitor) validated targeting the ubiquitin-proteasome pathway for chemotherapy.5,6 It is thought that bortezomib inhibits degradation of ubiquitinated inhibitor of kappa B (IκB) and thereby promotes apoptosis of myeloma cells via NF-κB signaling.6 However, targeting the proteasome in other cancers could block vital proteasome-dependent pathways. Targeting specific ubiquitin ligases could yield more specific therapies. Cul4A is amplified in hepatocellular carcinomas and breast cancer, suggesting Cul4A might be a good chemotherapy target. However, our findings indicate complete ablation of Cul4A activity is lethal. Nevertheless, it remains possible that malignant cells might be more sensitive to Cul4A inhibition. Alternatively, targeting specific Cul4A ubiquitin ligases might be more effective. The findings presented in this report help provide a basis for studies to address these possibilities.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dave Skalnik and Merv Yoder for insightful suggestions and Bill Carter and the Indiana University (IU) Simon Cancer Center Transgenic and Knock-Out Mouse Core (directed by Loren Field) for technical expertise. Desiree Lane contributed important technical support.
This work was supported by the Indiana University School of Medicine (IUSM) Core Centers of Excellence in Molecular Hematology (NIH P30DK49218), the National Institutes of Health (R01 DK66603), the Lilly Endowment, and the Riley Children's Fund (K.T.C.). D.L.W. was supported, in part, by the National Institutes of Health, NRSA T32 DK07519-Regulation of Hematopoietic Cell Production. Y.N. was supported by a National Institutes of Health predoctoral fellowship (F31 HL079889).
National Institutes of Health
Authorship
Contribution: D.L.W., B.L., N.J., Y.N., H.H., and K.T.C. performed research and analyzed data; W.S.G. performed research; D.L.W., B.L., N.J., H.H., and K.T.C. designed research; D.L.W. wrote the paper with contributions from H.H. and K.T.C.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kristin T. Chun, Cancer Research Institute R4, Room 474, 1044 W Walnut St, Indianapolis, IN 46202; e-mail: kchun@iupui.edu.