Abstract
To identify novel genes involved in the molecular pathogenesis of chronic lymphocytic leukemia (CLL) we performed a serial analysis of gene expression (SAGE) in CLL cells, and compared this with healthy B cells (nCD19+). We found a high level of similarity among CLL subtypes, but a comparison of CLL versus nCD19+ libraries revealed 55 genes that were over-represented and 49 genes that were down-regulated in CLL. A gene ontology analysis revealed that TOSO, which plays a functional role upstream of Fas extrinsic apoptosis pathway, was over-expressed in CLL cells. This finding was confirmed by real-time reverse transcription–polymerase chain reaction in 78 CLL and 12 nCD19+ cases (P < .001). We validated expression using flow cytometry and tissue microarray and demonstrated a 5.6-fold increase of TOSO protein in circulating CLL cells (P = .013) and lymph nodes (P = .006). Our SAGE results have demonstrated that TOSO is a novel over-expressed antiapoptotic gene in CLL.
Introduction
Chronic lymphocytic leukemia (CLL) is characterized by the accumulation of small mature B cells, mostly in early G1-phase,1 that display typical B-cell surface markers in addition to CD5. Two subtypes of CLL are defined by either mutated or unmutated IgVH genes and this has prognostic significance.2 Studies have revealed surrogate markers for the IgVH status in CLL cases,3-5 but microarray studies demonstrated relatively few differences in gene expression between the 2 subtypes,6,7 suggesting that the cells arise from a common cell of origin
CLL pathogenesis has been linked to a failure of the malignant cells to undergo apoptosis. Apoptosis resistance stems from a combination of microenvironmental survival signals (extrinsic pathway) as well as intrinsic alterations of the apoptotic machinery within the CLL cell.8,9 BCL-2 is present at increased levels in more than 80% of CLL cases. The mechanism for this was recently elucidated by the demonstration that miR15 and miR16 inhibit BCL-2 at a posttranscriptional level in CLL.10 The extrinsic apoptosis pathway is regulated by Fas membrane activation, and CLL cells display resistance to apoptosis driven by anti-Fas treatment. Although CLL cells express low levels of Fas, an upstream event might be responsible for this phenotype.8,9
The search for differentially expressed genes has mainly been performed by DNA-array studies6,7,11 and, although powerful, this methodology investigates only known genes. To evaluate for new mechanisms involved in apoptosis resistance of CLL cells, we performed a serial analysis of gene expression (SAGE) in CLL samples and identified that TOSO (regulator of FAS-induced apoptosis) is aberrantly expressed in CLL cells.
Methods
Patients and controls
Peripheral blood (PB) was obtained from 78 CLL patients, who met the diagnostic criteria for CLL.12 Eight untreated and early stage (Binet A/Rai I) CLL patients (4 mutated and 4 unmutated) where selected for construction of SAGE libraries from their cells. PB of 12 healthy blood donors was used as a source of normal CD19+ cells (nCD19+). Informed consent was obtained prior to all sample collections in accordance with the Declaration of Helsinki. The study was approved by the North East London Health Authority National Health Service Research Ethics Committee.
RNA extraction, SAGE procedure, and library comparison
Total RNA was extracted from PB mononuclear cells (PBMNC) for all CLL samples and from CD19+purified cells from 4 mutated and 4 unmutated CLL patients and from 12 normal controls (more than 95% purity). For SAGE analysis, mRNA pools from mutated and unmutated CLL cases were generated and SAGE was performed and analyzed as previously described,13 and compared against a CD19+ public Blood SAGE library14 and other nonlymphoid libraries.15 The top 1000 expressed tags in CLL and nCD19+ libraries were analyzed using Ingenuity Pathways Analysis (IPA), software from Ingenuity Systems (Redwood City, CA).
RT-PCR and real-time quantitative PCR
Reverse transcription real-time (RT) polymerase chain reaction (PCR) was performed for 7 representative genes (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) for 6 CLL patient samples: nCD19+, PBMCs, normal bone marrow, skeletal muscle, cardiac muscle, and liver. Expression of TOSO, BCL-2, and FLIP was assessed by TaqMan real-time quantitative (RQ)–PCR for all 78 CLL samples and 12 nCD19+ cells.
Tissue microarray construction and TOSO immunohistochemistry
Tissue microarrays (TMAs) were constructed in triplicate of 1-mm cores of patient tissue taken from representative areas of CLL and from reactive tonsil and appendix controls, and stained with monoclonal antibodies (MoAb) as previously described.16 Anti-TOSO MoAb (Abnova Corporation, Taiwan, China) was used at 1:50 dilution. TMAs were scored independently by 2 examiners (AL and RPS) as 0, 1+, 2+, and 3+,and the percentage of positive cells was evaluated.
Quantitative flow cytometric analysis of TOSO
PBMCs from 12 CLL samples and 5 healthy individuals were analyzed by quantitative flow cytometry for TOSO and CD19 compared with unlabeled IgG1 isotype control (Dako, Glostrup, Denmark). Cells (106) were analyzed using 5μL of anti-TOSO MoAb and GAM-PE (BD, Franklin Lakes, NJ). TOSO expression was quantified using normalized median fluorescence intensity (nMFI). These experiments were also performed gating on CD5+/CD19+ B cells from healthy donors.
Results and discussion
We performed SAGE in CLL cases to identify new genes and novel mechanisms of apoptosis resistance in CLL pathogenesis. A total of 104 057 and 100 260 tags were obtained by sequencing both CLL libraries, corresponding to approximately 26 000 unique tags, of which 60% matched known genes in the Cancer Genomic Anatomy Project (http://cgap.nci.nih.gov/). A list of all tags is freely available on our website.17
To identify new genes involved in CLL pathogenesis, we compared our CLL SAGE libraries to a nCD19 library and identified 55 overrepresented and 49 down-regulated genes in CLL (fold change higher than 20×, P < .001; Table S2). SAGE results were corroborated by quantitative PCR for 7 randomly selected genes that correlated with tag numbers in ours and other publicly available SAGE libraries (data not shown). Comparison of mutated and unmutated cases of CLL by SAGE revealed only 27 genes with significant (P < .001) differences, in keeping with gene expression profiling studies demonstrating a common gene expression “signature,” irrespective of the IgVH mutational status.6,7
To identify other genes that might be related to CLL pathogenesis, we used a functional analysis pathway (IPA) performed among the top 1000 uniquely expressed tags of CLL and nCD19+ SAGE libraries. IPA analysis revealed 450 and 479 genes assigned to eligible networks (P < .05, Fisher exact test) in CLL and nCD19+ cells, respectively. Genes were mostly classified as protein synthesis, cellular growth and proliferation, cell death, cell signaling and cellular development (Figure S1), the majority of which are similar, but not fully overlapping with previous reports.6,7,18,19 Of note, analysis revealed 19 genes associated with apoptosis inhibition, 6 of which (CD27, ANXA1, FAIM3, DAD1, HSPB1, PRDX2 and IFI6) were represented exclusively in CLL (Figure S2). Among the antiapoptotic genes, the IPA analysis identified a 4-fold over-expression of FAIM3 (TOSO), a transmembrane protein that colocalizes with Fas and binds specifically to FADD, inhibiting procaspase 8 and inducing resistance to Fas-mediated apoptosis.20-23 Previous studies have demonstrate that CLL cells are resistant to Fas mediated apoptosis.8,23
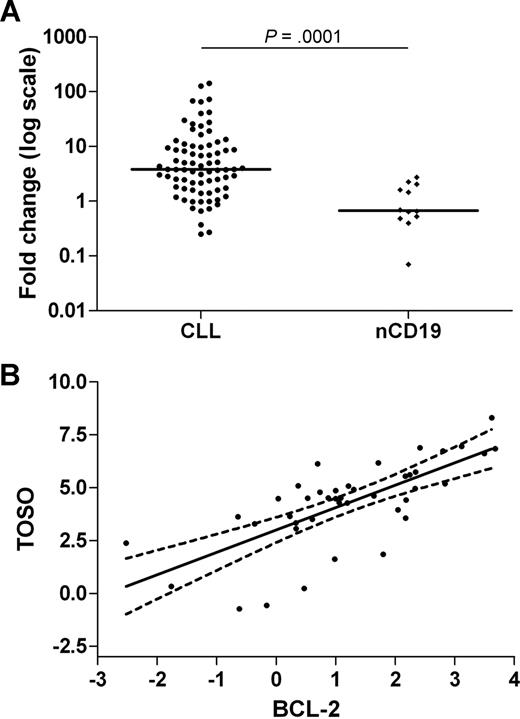
BCL-2 overexpression is a known mechanism for intrinsic apoptosis resistance in CLL.10 We postulated that TOSO might be associated with BCL-2 deregulation and evaluated FLIP as a possible up-stream regulator of TOSO expression.18 We quantified expression of TOSO, BCL-2, and FLIP mRNA by RQ-PCR in a larger set of clinical samples (78 CLL cases and 12 nCD19+ cells). Expression of TOSO was higher in CLL than in nCD19+ (median fold 4.4, range: 0.8-141, Mann-Whitney test P < .001; Figure 1A). FLIP (fold 5.3, P = .009) and BCL-2 (fold 4.8, P = .002) were also overexpressed. A positive correlation was observed between TOSO and BCL-2 (r = 0.7427, P < .001; Figure 1B), but not between TOSO and FLIP (data not shown). TOSO expression correlated with the number of peripheral lymphocytes in CLL cases (r = 0.3750, P = .038) but there was no correlation between TOSO expression and prognostic factors, including IgVH mutational status, cytogenetics or previous treatment. We postulate that overexpression of TOSO and BCL-2 might be responsible for inhibition of BAX21 leading to suppression of apoptosis. Recently it has been demonstrated that BAX degradation activity is associated with poor prognosis in CLL,24 and that bortezomib blocks BAX degradation in malignant B cells. TOSO might therefore be considered as a possible target for small molecule therapy in combination with newer proapoptotic drugs such as bortezomib25 and lumiliximab.26
Corroboration of SAGE results by RQ-PCR quantification of TOSO and BCL-2. (A) Relative real-time quantification of TOSO. The results are shown as fold changes, where we used a pool of 12 cDNA from nCD19+ cells to calculate 2−ΔΔCt. The results are plotted in log scale, with bars representing the median values; the P value was calculated using Mann-Whitney test. (B) Graphic correlation between TOSO and BCL-2 expression. The results of real-time PCR for 42 CLL samples are demonstrated as ΔCt values, showing a positive correlation between TOSO and BCL-2 (P < .001, and Spearman r = 0.7427).
Corroboration of SAGE results by RQ-PCR quantification of TOSO and BCL-2. (A) Relative real-time quantification of TOSO. The results are shown as fold changes, where we used a pool of 12 cDNA from nCD19+ cells to calculate 2−ΔΔCt. The results are plotted in log scale, with bars representing the median values; the P value was calculated using Mann-Whitney test. (B) Graphic correlation between TOSO and BCL-2 expression. The results of real-time PCR for 42 CLL samples are demonstrated as ΔCt values, showing a positive correlation between TOSO and BCL-2 (P < .001, and Spearman r = 0.7427).
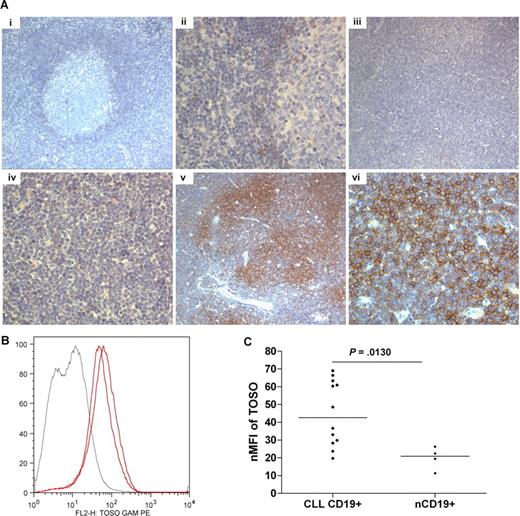
To validate these findings, we examined protein expression in tissue microarray (TMA) and by flow cytometry. TMA demonstrated that more than 50% of CLL cases strongly express TOSO (2+, 3+) with a range of 10% to 95% of positive tumor cells (P = .006, chi-square test). In contrast, control samples were negative for TOSO except the mantle cells from one tonsil and one appendix which scored weakly positive (1+; Figure 2A)
Protein analysis of TOSO using CLL tissue microarray and quantitative flow cytometry. (A) Immunohistochemistry for TOSO in tissue microarray cores. The entire 1-mm core was analyzed using a Leica DM2500 microscope, and images were acquired by a Leica DFC320 camera and processed using Leica IM50 software (Meyer Instruments, Houston, TX) at low (10×/0.30, HC PL FLUOTAR) and high (40×/0.75, HCX PL FLUOTAR) power objective magnification (OM) in each case, and the consensus of 2 cores was accepted. Results shown demonstrate (i) positive cells in mantle zone of tonsil OM ×10; (ii) positive cells in mantle zone of tonsil OM ×40; (iii) negative CLL sample OM ×10; (iv) negative CLL sample OM ×40; (v) positive CLL sample OM ×10; (vi) positive CLL sample OM ×40. A total of 44 CLL samples were assessed for TOSO. (B) Quantitative flow cytometry demonstrating a shift in TOSO expression in 2 CLL cases (red lines) against a single healthy individual (gray line). (C) Graphic comparison for normalized median fluorescence intensity (nMFI) for TOSO in CLL and nCD19+ cells (nMFI = 44.98 ± 18.31 in CLL vs 7.96 ± 11.45 in nCD19+ cells, P = .31, Mann-Whitney test). Bars represent the median values.
Protein analysis of TOSO using CLL tissue microarray and quantitative flow cytometry. (A) Immunohistochemistry for TOSO in tissue microarray cores. The entire 1-mm core was analyzed using a Leica DM2500 microscope, and images were acquired by a Leica DFC320 camera and processed using Leica IM50 software (Meyer Instruments, Houston, TX) at low (10×/0.30, HC PL FLUOTAR) and high (40×/0.75, HCX PL FLUOTAR) power objective magnification (OM) in each case, and the consensus of 2 cores was accepted. Results shown demonstrate (i) positive cells in mantle zone of tonsil OM ×10; (ii) positive cells in mantle zone of tonsil OM ×40; (iii) negative CLL sample OM ×10; (iv) negative CLL sample OM ×40; (v) positive CLL sample OM ×10; (vi) positive CLL sample OM ×40. A total of 44 CLL samples were assessed for TOSO. (B) Quantitative flow cytometry demonstrating a shift in TOSO expression in 2 CLL cases (red lines) against a single healthy individual (gray line). (C) Graphic comparison for normalized median fluorescence intensity (nMFI) for TOSO in CLL and nCD19+ cells (nMFI = 44.98 ± 18.31 in CLL vs 7.96 ± 11.45 in nCD19+ cells, P = .31, Mann-Whitney test). Bars represent the median values.
We evaluated TOSO protein expression by flow cytometry in 12 CLL cases and in 5 nCD19+ PB cells and demonstrated a5.6 fold increase of TOSO in PB CLL cells (P = .013 Mann-Whitney test; Figure 2B,C). No expression of TOSO was seen in CD5+/CD19+ healthy peripheral blood B cells (data not shown).
In summary, SAGE identified a new gene involved in CLL pathogenesis, and has corroborated results from previous expression arrays demonstrating similar profiles between the 2 CLL subtypes. Our results are the first to describe the over expression of TOSO in CLL. TOSO protein plays a functional role upstream of Fas in the extrinsic apoptosis pathway and studies are ongoing to assess the mechanisms whereby TOSO might mediate resistance of CLL cells to anti-Fas treatment and new studies should be directed to elucidate TOSO as a potential therapeutic target.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank, for expert technical support, Amélia G. Araújo, Marli H. Tavela, Anemari Santos, Cristiane Ayres, Aglair B. Garcia, Dalila L. Zannete, Samir Agrawal, Jerome Giustatini, Catherin Wiseman, Elio Stolz, and Ruth Segara; and all patients and healthy blood donors who agreed to participate in this study.
This work was supported by a grant from the State of São Paulo Research Foundation (FAPESP) and by grant PO1 CA 81538 from the National Cancer Institute (J.G.G.) to the CLL Research Consortium.
National Institutes of Health
Authorship
Contribution: R.P-S., designed and performed research, analyzed data, and wrote the paper; R.A.P., F.P.C., A.L., A.C., K.M., E.G.R., and W.A.S. performed research and analyzed data; C.O. edited the manuscript, R.P.F. provided vital materials; and M.A.Z. and J.G.G.designed research, analyzed data, wrote the paper and supervised the study.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: John G. Gribben, Barts and The London School of Medicine, Charterhouse Square, London, EC1M6BQ; e-mail: john.gribben@cancer.org.uk.