The fireworks from the spectacular emergence of Notch as the protagonist in the etiology of T-ALL are not likely to end any time soon. Indeed, in this issue of Blood, Sulis and colleagues describe yet another sophisticated mechanism by which ligand-independent activation of NOTCH1 is achieved in T-ALL. NOTCH1-activating mutations mark more than half of all T-ALL cases, underscoring the fundamental role of aberrant NOTCH1 signaling in this disease.
Activation of NOTCH signaling by Delta-like ligands is required for commitment of early thymic progenitors to the T-cell lineage as well as for the double-negative 3a (DN3a) to DN3b thymocyte transition, and entry to β-selection.1 It is therefore not surprising that the aberrant, ligand-independent activation of Notch is a hallmark of human T-cell lymphoblastic leukemia (T-ALL).
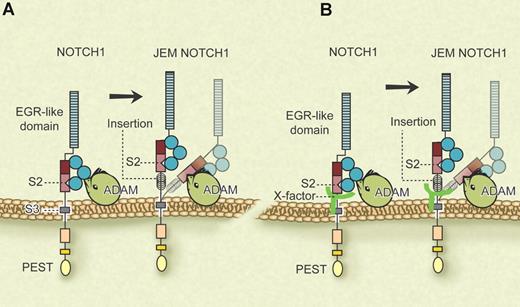
NOTCH receptors are type-I transmembrane glycoproteins that regulate cell growth, differentiation, and tissue homeostasis in multiple systems. NOTCH signaling is tightly regulated by the members of the Delta, LAG-2 Serrate family of NOTCH ligands binding to the extracellular domain of the mature receptor. Initial proteolytic cleavage of the NOTCH precursor protein within the extracellular domain yields an extracellular and a transmembrane subunit that are held together by a heterodimerization (HD) domain (see figure). NOTCH receptor heterodimers further stabilize themselves in a resting state through 3 LIN-12/NOTCH repeats (LNRs), which seem to engulf the HD domain and shield the S2 site from proteolytic cleavage by ADAM-type metalloproteases.2 S2 cleavage is triggered when the receptor binds to its ligand, and is a prerequisite for subsequent cleavage by γ-secretase at the S3 site. This final step releases intracellular Notch (ICN) from the membrane and allows it to translocate to the nucleus, where it converts CSL repressor into activator complexes by displacing corepressors and attracting coactivators.
Possible models for the manner in which JEM mutations may alter receptor processing. (A) By placing the HD-LNR complex away from the membrane, conformational changes allow for increased cleavage of S2. (B) One or several membrane-linked X-factor(s) lend additional stability to the HD-LNR complex that is lost in JEM mutants due to the increased distance of the HD-LNR complex from the membrane. Professional illustration by Debra T. Dartez.
Possible models for the manner in which JEM mutations may alter receptor processing. (A) By placing the HD-LNR complex away from the membrane, conformational changes allow for increased cleavage of S2. (B) One or several membrane-linked X-factor(s) lend additional stability to the HD-LNR complex that is lost in JEM mutants due to the increased distance of the HD-LNR complex from the membrane. Professional illustration by Debra T. Dartez.
In human T-ALL, activating NOTCH1 mutations are concentrated in exons 26 and 27, encoding the HD domain, and in exon 34, encoding the PEST domain, which is important for proteasomal degradation of ICN1. Thus far, the described mutations can be grouped into 3 categories. HD mutations comprise single amino acid substitutions and small in-frame deletions and insertions that allow for ligand-independent ADAM-mediated S2 and γ-secretase–dependent S3 cleavage, ultimately releasing ICN1. This can be achieved either by destabilizing the HD-LNR interaction, resulting in increased dissociation of the heterodimer (class 1 mutants), or by displacing the S2 site out of the protective reach of the HD-LNR complex and exposing it to proteolytic cleavage (class 2 mutants). PEST mutations, in contrast, encode premature stop codons. Loss of the PEST domain results in increased ICN1 levels due to reduced proteasomal degradation.3,4
Sulis and colleagues have now identified a fourth family of NOTCH1-activating mutations in T-ALL accounting for a subset of leukemias that show elevated ICN1 but none of the previously characterized mutations in exons 26, 27, or 34. In an elegant biochemical approach, the authors characterize a novel group of mutations in the extracellular juxtamembrane domain of NOTCH1. These juxtamembrane extension mutants (JEMs) distance the entire HD-LNR complex from the membrane (see figure), and presumably allow ligand-independent proteolytic processing of S2. They are generated by internal tandem duplications in the 3′ end of intron 27 and/or the proximal region of exon 28. Thus, they do not alter the spacing within the HD-LNR complex, but rather shift the entire complex away from the membrane. In fact, the authors show that JEMs do not alter HD-LNR complex stability. Moreover, the activity of JEMs is critically dependent on the length of the inserted sequence, that is, the increased distance from the membrane rather than the sequence of the insertion per se.
The authors offer 2 possibilities to explain why the maintenance of the HD-LNR complex in close proximity to the membrane is strictly required for avoiding spontaneous metalloprotease cleavage of NOTCH1. They hypothesize that “the HD-LNR complex interacts with membrane proteins that contribute to hold this structure together and that this interaction is disrupted upon displacement of the HD-LNR complex away from the membrane” (page 739; see figure panel B). They also suggest the alternative possibility that “the proximity to the membrane restrains the ability of proteases to access the S2 cleavage site located within the HD-LNR complex, and that displacement from the cell surface makes this sequence more accessible for protease cleavage” (page 739; see figure panel A). In conclusion, the authors have identified a novel family of NOTCH1-activating mutations that result in aberrant levels of ICN1, further expanding our understanding of the important molecular parameters that control Notch activity.
Conflict-of-interest disclosure: The authors declare no competing financial interests. ■
REFERENCES
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal