Abstract
Cyclin D1 (CCND1) is a well-known regulator of cell-cycle progression. It is overexpressed in several types of cancer including breast, lung, squamous, neuroblastoma, and lymphomas. The most well-known mechanism of overexpression is the t(11;14)(q13;q32) translocation found in mantle cell lymphoma (MCL). It has previously been shown that truncated CCND1 mRNA in MCL correlates with poor prognosis. We hypothesized that truncations of the CCND1 mRNA alter its ability to be down-regulated by microRNAs in MCL. MicroRNAs are a new class of abundant small RNAs that play important regulatory roles at the posttranscriptional level by binding to the 3′ untranslated region (UTR) of mRNAs blocking either their translation or initiating their degradation. In this study, we have identified the truncation in CCND1 mRNA in MCL cell lines. We also found that truncated CCND1 mRNA leads to increased CCND1 protein expression and increased S-phase cell fraction. Furthermore, we demonstrated that this truncation alters miR-16-1 binding sites, and through the use of reporter constructs, we were able to show that miR-16-1 regulates CCND1 mRNA expression. This study introduces the role of miR-16-1 in the regulation of CCND1 in MCL.
Introduction
Mantle cell lymphoma (MCL) represents 5% to 10% of all non-Hodgkin lymphomas.1,2 It is a relatively uncommon but particularly difficult form of lymphoma to treat. It has the worst prognosis among the B-cell lymphomas, with median survival of 3 years with no standard effective therapy.3 The genetic hallmark of MCL is the t(11;14)(q13;q32) translocation that displaces the CCND1 gene on chromosome 11 downstream to the enhancer region of the IgH gene on chromosome 14 and causes its overexpression.4 CCND1 is a well-known cell-cycle regulator and promotes G1 to S-phase cell-cycle progression.5,6 Although CCND1 overexpression has unified and simplified the diagnostic approach to MCL, no therapeutic advances have been made to target this particular pathway. In fact, controversy remains regarding the oncogenic potential of CCND1. Some have speculated that wild-type CCND1 requires another cooperating oncogenic event such as Ras activation to cause tumorigenesis.7 Others such as Diehl et al8 and Lu et al9 have reported a CCND1 variant (cyclin D1b), the nuclear localization of which represents a critical step in oncogenesis. The most recent finding reported by Wiestner et al10 showed that point mutations and genomic deletions in CCND1 created a truncated mRNA and result in increased proliferation and shorter survival. Although this report associates important clinical outcomes with the truncated mRNA, it does not delineate a definitive molecular mechanism or rationale. In their discussion, Wiestner et al alluded to the possibility of microRNAs as regulators of CCND1.10 We hypothesize that CCND1 mRNA is normally under the regulation of microRNAs, and that truncated CCND1 mRNA escapes this regulation by deletion of the microRNA target binding sequences.
MicroRNAs (miRNAs) are small RNAs that play important roles in posttranscriptional regulation of genes such as oncogenes and tumor suppressor genes. They are located throughout the genome but are often found at fragile sites and other genomic regions involved in the development of cancer.11 They are initially transcribed as noncoding primary miRNA (pri-miRNAs) transcripts in the nucleus. The pri-miRNAs are processed into precursor miRNA (pre-miRNA) and finally into the mature and functional miRNA of 21 to 25 nucleotides in the cytoplasm.12 The mature miRNAs function by binding to the 3′ untranslated region (3′ UTR) of multiple target mRNAs and either inhibit the translation of target protein or degrade the target mRNA.13-17 Numerous recent studies have identified altered expression of miRNAs in a variety of cancer sites such as B-cell chronic lymphocytic leukemia (CLL)18-20 and have confirmed that this is yet another cellular component capable of functioning as tumor suppressors or oncogenes.15-17
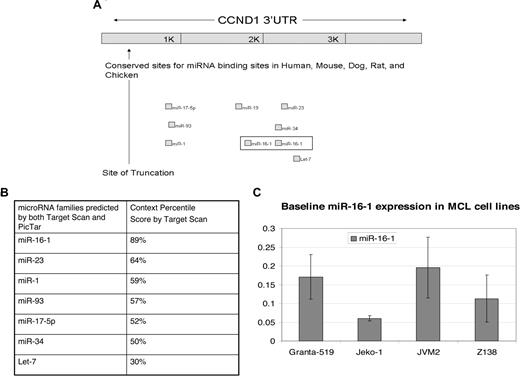
The numerous reports of alterations in miRNAs in different tumors led us to question whether altered miRNA regulation plays a role in MCL. We believe that altered regulation by miRNAs could explain the correlation between truncated CCND1 mRNA and poor prognosis in patients. We initially conducted a survey of postulated microRNAs that can bind to CCND1 mRNA using bioinformatics databases such as TargetScan21 and PicTar.22 Both databases predicted essentially the same families of microRNAs (Figure 1B). MiR-16-1 was identified as a strong candidate due to its multiple binding sites and its high context score percentile of 89% (Figure 1B). It has also been reported to function as a cell-cycle regulator.23 We undertook studies to confirm the predicted role of miR-16-1 in the regulation of CCND1. In addition, we identified a truncation in CCND1 mRNA in MCL cell lines that deletes miR-16-1 binding sites in the 3′ UTR of CCND1.
Methods
miRNA computational predictions
The public-access database Target Scan21 was consulted to identify putative miRNAs that could bind to CCND1. Another database, Pictar22 was used for confirmation of the microRNAs predicted by Target Scan.21 MiRNA binding predictions were confirmed by manually analyzing the CCND1 3′ UTR for binding sites of miR-16-1.
Cell lines and growth conditions
The human MCL cell lines Granta-519, Jeko-1, JVM2, and Z138 were used. Granta-519 was obtained from Dr Witzig at the Mayo Clinic. Jeko-1 was purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). JVM2 and Z138 were obtained from Dr Catherine Tucker at the British Columbia Cancer Agency (BCCA, Vancouver, BC). All 4 cell lines have been previously confirmed to harbor the t(11;14) translocation.24 Granta-519, JVM-2, and Z138 were cultured in RPMI media supplemented with 10% FBS, 1% penicillin/streptomycin, and 50 mM l-glutamine at 37°C and 5% CO2. Jeko-1 was cultured in RPMI media supplemented with 20% FBS, 1% penicillin/streptomycin, and 50 mM l-glutamine at 37°C and 5% CO2.
Western blot analysis
Cells were harvested by rinsing with phosphate-buffered saline and lysed using Laemmli buffer (Bio-Rad, Hercules, CA) as directed. The samples were sonicated briefly, and centrifuged at 14 000 rpm for 15 minutes at 4°C. The antibodies used in this study included CCND1 (556470; BD Biosciences, San Jose, CA) and alpha tubulin (2144; Cell Signaling, Beverly, MA). Protein samples were resolved on a 10% SDS polyacrylamide gel, transferred to a nitrocellulose membrane, and blocked in 5% dried milk in phosphate-buffered saline/0.0025% Tween-20. Membranes were incubated in the appropriate primary antibody overnight, followed by incubation with the appropriate secondary antibody. Proteins were detected by enhanced luminol/peroxide buffers SuperSignal West Femto (Pierce, Rockford, IL) on a Chemidoc (Bio-Rad) imaging system and quantitated using the Quantity One software (4.5.0 basic; Bio-Rad).
Cell-cycle analysis
Cells were harvested, washed once with PBS, and stained at 4°C overnight with Krishan stain containing 3.8 mM trisodium citrate (Sigma-Aldrich, St Louis, MO), 70 μM propidium iodide (Sigma-Aldrich), 0.01% Nonidet P-40 (Sigma-Aldrich), and 0.01 mg RNase A (Boehringer Mannheim, Indianapolis, IN) per milliliter. Cell-cycle analysis was performed using a Beckman Coulter FC500 flow cytometer (Beckman-Coulter, Hialeah, FL). Alignment of the instrument was verified daily using DNA FlowCheck beads (Beckman Coulter). Data were collected for 10 000 events. The Modfit LT program (Verity Software House, Topsham, ME) was used for cell-cycle modeling and doublet exclusion.
RNA isolation, cDNA synthesis, and qRT-PCR
To assess truncations within the CCND1 mRNA, RNA was isolated from cell lines using the Qiagen AllPrep DNA/RNA Mini kit (Qiagen, Valencia, CA). cDNA was synthesized using the SuperScript II Reverse Transcriptase Kit (Invitrogen, Carlsbad, CA). Three primer sets were used: proximal, distal, and cross exon. The proximal primer set for CCND1 was forward, 5′-TCCCTCCTCTCCGGAGCATTT-3′ and reverse, 5′-ATCATCTGTAGCACAACCCTCCTC-3′. The distal primer set for CCND1 was forward, 5′-TCTCAATGAAGCCAGCTCACA-3′ and reverse, 5′-CAAGCTGCCGAACCAAAAG-3′. The cross exon set for CCND1 was forward, 5′-AGTTCATTTCCAATCCGCCCTCCA-3′ and reverse, 5′-AATGCTCCGGAGAGGAGGGACT-3′. All values were normalized to beta-actin. The primer set for beta-actin was forward, 5′-ATCCACGAAACTACCTTCAACTC-3′ and reverse, 5′-GAGGAGCAATGATCTTGATCTTC-3′. Quantitative reverse-transcription–polymerase chain reaction (qRT-PCR) was performed under the following thermocycler conditions: 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds, and 57°C for 60 seconds. qRT-PCR was used to determine the expression levels of CCND1 and beta-actin as an internal control.15
RNA isolation, cDNA synthesis, and qRT-PCR for microRNA
Total RNA for miRNA analysis was isolated using the miRNeasy Mini Kit (Qiagen). cDNA was synthesized using the NCode miRNA First-Strand cDNA Synthesis Kit (Invitrogen). Briefly, this step adds a poly(A) tail to the miRNA in the total RNA samples. The amount of miRNA was monitored using Platinum SYBR Green qPCR SuperMix-UDG reagent (Invitrogen). Quantitative PCR was performed under the following thermocycler conditions: 50°C for 2 minutes, 95°C for 10 minutes, and 40 cycles of 95°C for 15 seconds, and 57°C for 60 seconds. Expression of the mature miR-16-1 and U6 as an internal control was conducted using the NCode kit (Invitrogen). Results are expressed as relative quantitation as described previously. Primers were as follows: mature miR-16, 5′-TAGCAGCACGTAAATATTGGCG-3′; U6 forward, 5′-CGC AAG GAT GAC ACG CAA ATT CGT-3′.
GFP-CCND1-3′ UTR reporter constructs
Three GFP reporter plasmids were constructed. A cDNA encoding the distal 3236 bp of the 3′ UTR of CCND1 ((IMAGE Clone ID: 5521686) was purchased from ATCC (Manassas, VA), and naturally occurring EcoRI and Xho1 sites from the purchased plasmid were used to purify a 2574–base pair fragment that was inserted into the pEGFP-C1 plasmid (Clontech Laboratories, Mountain View, CA) at the EcoRI and SalI sites. The resulting reporter construct is called GFP-L and contains the distal 3′ UTR of CCND1 including both miR-16-1 binding sites. The second reporter construct called GFP-T contains the last 10 amino acids of CCND1 in frame with GFP and 331 base pairs of the 3′ UTR of CCDN1, similar to that which was cloned from the Jeko-1 cell line using the NCode method. The primers used to make this construct were as follows: CCND1 truncated forward, 5′-GTG GAC CTG AAT TCC ACA CCC ACC-3′ and CCND1 truncated reverse, 5′-GCC CGG GAT CCT TAA TAT AAA AAC GAG TTG A-3′. The third reporter construct, GFP-M, is a double mutant with mutations in both the first and second miR-16-1 binding sites. Mutation of the miR-16-1 binding sites in the 3′ UTR of CCND1 was created using the Quick Change Stratagene (La Jolla, CA) method. Primers for quick change mutations were as follows: CCND1 mutant first site forward, 5′-TTC TTA TTG CGC TTC GAC CGT TGA CTT C-3′ and reverse, 5′-GAA GTC AAC GGT CGA AGC GCA ATA AGA A-3′; CCND1 mutant second site forward, 5′-ACA TTG TTT GCT TCG ATT GGA GGA TCA G and reverse, 5′-CTG ATC CTC CAA TCG AAG CAA ACA ATG T-3′. H157 cells were then transfected using the lipophilic reagent Effectene (Qiagen). The different reporter constructs were cotransfected with RFP vector and 5 nM of a mimic of miR-16-1 (Ambion, Austin, TX) or 5 nM of a negative control random miRNA (Ambion). GFP expression was monitored by flow cytometry and normalized to RFP expression.
Results
In initial studies, we used computational methods to identify miRNAs with putative binding sites in CCND1 3′ UTR. miR-16-1 was predicted to have 2 binding sites in this location (Figure 1A). In addition, it was predicted to have the highest context score, which was 89% (Figure 1B). As noted, miR-16-1 is considered an important regulator of cell cycle23 and has been found to be misregulated in CLL.18,19 miR-16-1 is located on chromosome 13q14, an area commonly deleted in CLL, and has been reported to be deleted in approximately in 25% to 43% of MCL clinical samples.25,26 However, based on a paper by Camps et al, 13q14 deletion is not found in 5 MCL cell lines studied (JVM-2, Granta-519, REC-1, Jeko-1, and NCEB-1).27 We used qRT-PCR to assess the endogenous levels of miR-16-1 in 4 MCL cell lines and found adequate expression levels (Figure 1C), which is consistent with the notion that miR-16-1 is not deleted in these 4 MCL cell lines.
CCND1 is a target of miR-16-1 as predicted by Target Scan. (A) Shown are the seed binding regions located in the distal region of the CCND1 3′ UTR. The 2 binding sites for miR-16-1 are boxed. The truncation (shown by ↑) deletes CCND1 all miRNA binding sites. (Adapted from Target Scan.21 ) (B) Table showing the context percentile score of each microRNA family predicted to target CCND1 by Target Scan.21 The higher the context score, the higher the probability that the microRNA will be able to inhibit CCND1 translation. (C) qRT-PCR to show the endogenous miR-16-1 expression in MCL cell lines. Error bars represent SD.
CCND1 is a target of miR-16-1 as predicted by Target Scan. (A) Shown are the seed binding regions located in the distal region of the CCND1 3′ UTR. The 2 binding sites for miR-16-1 are boxed. The truncation (shown by ↑) deletes CCND1 all miRNA binding sites. (Adapted from Target Scan.21 ) (B) Table showing the context percentile score of each microRNA family predicted to target CCND1 by Target Scan.21 The higher the context score, the higher the probability that the microRNA will be able to inhibit CCND1 translation. (C) qRT-PCR to show the endogenous miR-16-1 expression in MCL cell lines. Error bars represent SD.
Correlations between CCND1 truncation, total CCND1 mRNA expression, and CCND1 protein expression
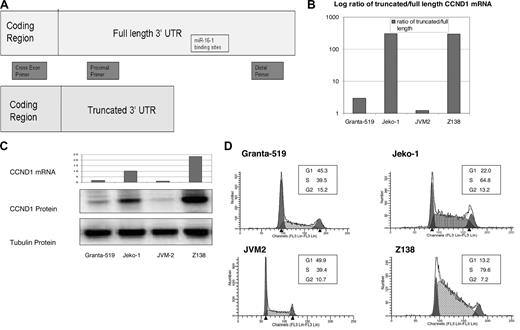
Previous reports showed that MCL tumor samples harbor truncations within their CCND1 mRNA 3′ UTR.10 However, no correlation had been made between CCND1 protein expression and truncated CCND1 mRNA in MCL cell lines, nor was the exact site of truncation cloned and sequenced. We used qRT-PCR to compare the ratio of intact CCND1 mRNA 3′ UTR with truncated CCND1 mRNA 3′ UTR among 4 MCL cell lines (Figure 2A). Specifically, we designed a distal primer set to amplify the distal region of the 3′ UTR (2685-2749 bp) downstream of both miR-16-1 binding sites. We also designed a proximal primer set to amplify region 72 to 285 bp on the 3′ UTR. We then compared the total CCND1 mRNA between 4 MCL cell lines using cross exon primers to eliminate contamination by genomic DNA. Cells from the same plates were also collected to examine CCND1 protein expression and cell-cycle progression. We found that although all 4 cell lines contained both the truncated and full-length CCND1 mRNA, Jeko-1 and Z138 had much higher levels of truncated message than full length compared with Granta-519 and JVM2 (data not shown). We also found that cell lines with increased ratio of truncated CCND1 also had increased total CCND1 mRNA levels (Z138: 19.9-fold; Jeko-1: 8.74-fold; Granta-519: 1.51-fold; JVM2: 1-fold), increased CCND1 protein expression, and increased S-phase cell fraction (Figure 2B-D).
Truncated CCND1 leads to increased CCND1 mRNA, increased CCND1 protein expression, and cell-cycle progression. (A) Primer design for qRT-PCR to assess relative values of truncated versus full-length CCND1 mRNA. The cross exon primer set will amplify only mRNA in the coding region, thus avoiding genomic contamination. The proximal primer set will amplify both the full-length and truncated mRNA. The distal primer set will amplify only the full-length mRNA. (B) qRT-PCR showing the log ratio of truncated to full-length CCND1 mRNA in MCL cell lines. mRNA is expressed as relative quantitation equalized to beta-actin. Jeko-1 and Z138 had much higher quantitation values for truncated message and lower values for full-length message compared with Granta-519 and JVM2. Similarly, the ratio of truncated to full-length CCND1 mRNA is much higher in Jeko-1 and Z138. (C) Relative expression of total amount of CCND1 mRNA and corresponding CCND1 protein expression in 4 MCL cell lines. Cross exon primers were designed to amplify total CCND1 mRNA to avoid genomic contamination. Jeko-1 and Z138 also have increased total amount of CCND1 mRNA compared with Granta-519 and JVM-2. Similarly, Jeko-1 and Z-138 have increased CCND1 protein expression compared with Granta-519 and JVM-2. (D) Cell cycle in MCL cell lines. Jeko-1 and Z138 have increased percentage of cells in S-phase compared with Granta-519 and JVM-2.
Truncated CCND1 leads to increased CCND1 mRNA, increased CCND1 protein expression, and cell-cycle progression. (A) Primer design for qRT-PCR to assess relative values of truncated versus full-length CCND1 mRNA. The cross exon primer set will amplify only mRNA in the coding region, thus avoiding genomic contamination. The proximal primer set will amplify both the full-length and truncated mRNA. The distal primer set will amplify only the full-length mRNA. (B) qRT-PCR showing the log ratio of truncated to full-length CCND1 mRNA in MCL cell lines. mRNA is expressed as relative quantitation equalized to beta-actin. Jeko-1 and Z138 had much higher quantitation values for truncated message and lower values for full-length message compared with Granta-519 and JVM2. Similarly, the ratio of truncated to full-length CCND1 mRNA is much higher in Jeko-1 and Z138. (C) Relative expression of total amount of CCND1 mRNA and corresponding CCND1 protein expression in 4 MCL cell lines. Cross exon primers were designed to amplify total CCND1 mRNA to avoid genomic contamination. Jeko-1 and Z138 also have increased total amount of CCND1 mRNA compared with Granta-519 and JVM-2. Similarly, Jeko-1 and Z-138 have increased CCND1 protein expression compared with Granta-519 and JVM-2. (D) Cell cycle in MCL cell lines. Jeko-1 and Z138 have increased percentage of cells in S-phase compared with Granta-519 and JVM-2.
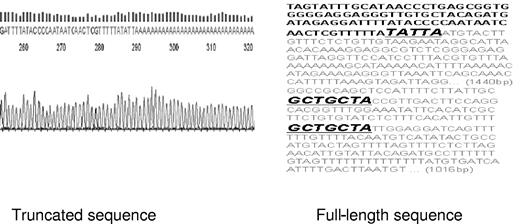
To prove that this truncation alters both miR-16-1 binding sites within the CCND1 mRNA, we cloned the truncated version from all 4 MCL cell lines. We adopted the NCode method initially designed to examine miRNA expression to clone the truncated mRNA. Specifically, total RNA was isolated from all 4 MCL cell lines. Poly A tails were added to all RNA. The CCND1 proximal forward primer and the universal reverse RT primer were used to amplify CCND1 truncated message and generate cDNA. It is expected that the full-length CCND1 mRNA could not be amplified due to its approximate length of 3000 bp. qRT-PCR product was then sequenced and all 4 MCL cell lines were found to express the same form of truncated CCND1 mRNA. This truncation occurs 328 bp immediately downstream of the stop codon and eliminates 2863 bp of the distal CCND1 3′ UTR including both miR-16-1 binding sites (Figure 3).
Site of CCND1 mRNA truncation. NCode was used to clone the truncated CCND1 mRNA from MCL cell lines as previously described (“GFP-CCND1-3′ UTR reporter constructs”). Truncation occurs 345 bp downstream of the stop codon and eliminates both miR-16-1 binding sites. Note that the poly A tails immediately follow the sequence TATTA in sequencing data. The letters in gray reflect the truncated sequences, and GCTGCTA denotes miR-16-1 binding sites.
Site of CCND1 mRNA truncation. NCode was used to clone the truncated CCND1 mRNA from MCL cell lines as previously described (“GFP-CCND1-3′ UTR reporter constructs”). Truncation occurs 345 bp downstream of the stop codon and eliminates both miR-16-1 binding sites. Note that the poly A tails immediately follow the sequence TATTA in sequencing data. The letters in gray reflect the truncated sequences, and GCTGCTA denotes miR-16-1 binding sites.
Functional analysis of miR-16-1 through GFP constructs
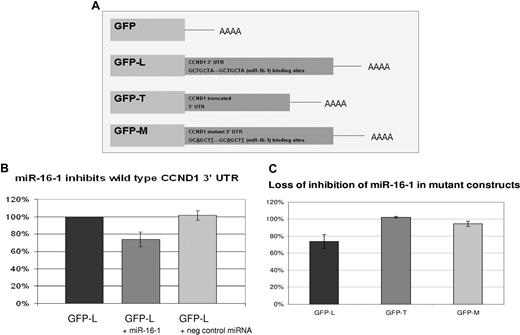
To confirm the predicted regulation of CCND1 by miR-16-1 through its 3′ UTR, a reporter plasmid was constructed. The reporter plasmid consisted of a GFP expression vector with 3′ UTR of CCND1 in place of the UTR in the pEGFP-C1 plasmid (Figure 4A). In this construct, binding of miR-16-1 to the 3′ UTR of CCND1 in GFP-L would lead to suppression of GFP expression. Both of the miR-16-1 binding sites were included in the GFP-L construct. The GFP-L construct was transfected into lung cancer cell line H157 along with a transfection control plasmid expressing RFP. In addition, the H157 cells were cotransfected with a mimic of miR-16-1 or negative control random miRNA sequence. H157 cell line was used due to its high transfection efficiency. In these studies the GFP protein expression was reduced by 24% when transfected with mimics of miR-16-1 compared with empty transfection and negative control miRNA transfection (Figure 4B).
miR-16-1 regulates CCND1 through its 3′ UTR. (A) Three GFP reporter constructs were made as previously described (“Methods”). GFP-L contains the distal CCND1 3′ UTR including both miR-16-1 binding sites. GFP-T contains the truncated CCND1 3′ UTR. GFP-M is a double mutant where both miR-16-1 binding sites were mutated. Notice the mutation changes the T from the third base pair to an A and the A in the seventh base pair to a T. (B) H157 cells were transfected with GFP-L and cotransfected with a mimic of miR-16-1 or negative control random miRNA. Shown is the GFP expression measured by flow cytometry. RFP-expressing plasmids were cotransfected to control for transfection efficiency. Error bars represent SD.(C) H157 cells were transfected with either GFP-L, GFP-T, or GFP-M. They were also cotransfected with either a mimic of miR-16-1 or negative control random miRNA. GFP expression of cells transfected by a mimic of miR-16-1 is first normalized to the RFP expression and then normalized to cells transfected with negative control random miRNA. This shows that miR-16-1 mimic cannot regulate a truncated or mutated CCND1 3′ UTR.
miR-16-1 regulates CCND1 through its 3′ UTR. (A) Three GFP reporter constructs were made as previously described (“Methods”). GFP-L contains the distal CCND1 3′ UTR including both miR-16-1 binding sites. GFP-T contains the truncated CCND1 3′ UTR. GFP-M is a double mutant where both miR-16-1 binding sites were mutated. Notice the mutation changes the T from the third base pair to an A and the A in the seventh base pair to a T. (B) H157 cells were transfected with GFP-L and cotransfected with a mimic of miR-16-1 or negative control random miRNA. Shown is the GFP expression measured by flow cytometry. RFP-expressing plasmids were cotransfected to control for transfection efficiency. Error bars represent SD.(C) H157 cells were transfected with either GFP-L, GFP-T, or GFP-M. They were also cotransfected with either a mimic of miR-16-1 or negative control random miRNA. GFP expression of cells transfected by a mimic of miR-16-1 is first normalized to the RFP expression and then normalized to cells transfected with negative control random miRNA. This shows that miR-16-1 mimic cannot regulate a truncated or mutated CCND1 3′ UTR.
A GFP-T construct was also made that included the truncated CCND1 mRNA 3′ UTR. This construct was transfected into H157 cells and cotransfected with a mimic of miR-16-1 and RFP. In contrast to the GFP-L experiment, the mimic of miR-16-1 could not shut down GFP-T expression when the reporter construct lacked the binding sites for miR-16-1 (Figure 4C). Further confirmation that miR-16-1 regulates CCND1 by binding to the 3′ UTR was accomplished by mutating both miR-16-1 binding sites in the 3′ UTR. The mimic to miR-16-1 could not shut down GFP-M (double mutant) reporter construct (Figure 4C).
Endogenous function of miR-16-1
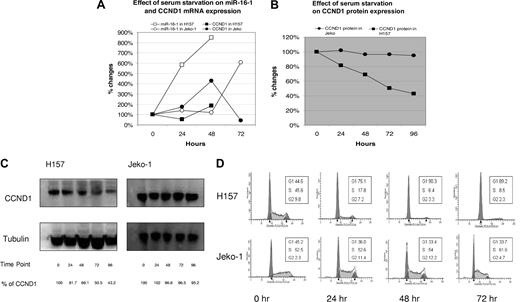
In the next experiment, we found that endogenous miR-16-1 is able to down-regulate CCND1 protein expression, similar to the findings of the previous reporter construct. As described above, transfection with a mimic to miR-16-1 down-regulates CCND1 through its 3′ UTR. Because the level of knockdown was only 24% with the mimic of miR-16-1 and the level of endogenous CCND1 mRNA was high in MCL cell lines, it was unlikely that CCND1 protein translation could be reduced with a mimic of miR-16-1 in MCL cell lines. In addition, lymphoma cells lines are notoriously difficult to transfect and we were unable to find a consistent method of transfection. Thus we used serum starvation to suppress CCND1 mRNA levels to magnify the effects of miR-16-1 mimic. Surprisingly, miR-16-1 levels were increased by serum starvation. We were able to demonstrate an increase in the endogenous miR-16-1 levels in both the H157 and Jeko-1 cell lines with serum starvation. In the H157 cell line, serum starvation increased endogenous mature miR-16-1 by 800% and CCND1 mRNA was increased by 200% at 48 hours (Figure 5A). However, CCND1 protein expression was decreased by 50%, suggesting that the increase in endogenous miR-16-1 decreased translation of CCND1 mRNA (Figure 5B,C). In the Jeko-1 cell line, serum starvation increased endogenous mature miR-16-1 by 600% and decreased the CCND1 mRNA by 50% at 72 hours. However, the CCND1 protein expression remained unchanged, showing that miR-16-1 was not able to down-regulate the translation of a truncated CCND1 mRNA (Figure 5B,C). As expected, serum starvation decreased CCND1 protein expression in H157 and was able to induce cell-cycle arrest (Figure 5D). In contrast, serum starvation was unable to decrease CCND1 protein expression in Jeko-1 and therefore could not induce cell-cycle arrest (Figure 5D). These data are consistent with the hypothesis that truncated CCND1 3′ UTR is unresponsive to regulation by miR-16-1.
Endogenous miR-16-1 inhibits CCND1 translation in H157. (A) Serum starvation increases miR-16-1 by approximately 800% in H157 and approximately 600% in Jeko-1. Serum starvation also increases CCND1 mRNA approximately 200% in H157 but decreases CCND1 mRNA in Jeko-1. (B) Serum starvation decreases CCND1 protein expression in H157 by approximately 30% at 48 hours and has no effect on CCND1 protein expression in Jeko-1 even at 96 hours. (C) Western blot analysis showing down-regulation of CCND1 protein expression in H157, while no changes occurred in Jeko-1 with serum starvation. (D) Cell-cycle analysis showing cell-cycle progression in H157 and Jeko-1 with serum starvation. The accumulation of cells in G1-phase in H157 at 48 hours indicates that serum starvation causes cell-cycle arrest. Notice that cell-cycle progression does not change dramatically in Jeko-1 even with serum starvation.
Endogenous miR-16-1 inhibits CCND1 translation in H157. (A) Serum starvation increases miR-16-1 by approximately 800% in H157 and approximately 600% in Jeko-1. Serum starvation also increases CCND1 mRNA approximately 200% in H157 but decreases CCND1 mRNA in Jeko-1. (B) Serum starvation decreases CCND1 protein expression in H157 by approximately 30% at 48 hours and has no effect on CCND1 protein expression in Jeko-1 even at 96 hours. (C) Western blot analysis showing down-regulation of CCND1 protein expression in H157, while no changes occurred in Jeko-1 with serum starvation. (D) Cell-cycle analysis showing cell-cycle progression in H157 and Jeko-1 with serum starvation. The accumulation of cells in G1-phase in H157 at 48 hours indicates that serum starvation causes cell-cycle arrest. Notice that cell-cycle progression does not change dramatically in Jeko-1 even with serum starvation.
Discussion
MiRNAs are of ever increasing importance as regulators of gene expression after transcription. To date, hundreds of human miRNAs have been described and more are being added to databases each day.28 They have been shown to have key regulatory roles during embryogenesis, cell development, cell-cycle progression, and differentiation.16,20,23,29 With this in mind, miRNAs have recently been investigated in a variety of cell types and tissues including human neoplasms. In this study, we began by searching for miRNAs that were predicted to bind to CCND1, a gene commonly altered in MCL.
Alterations in CCND1 causing overexpression represent the pathognomonic feature of MCL. However, MCL is a heterogeneous disease, and the translocation of CCND1 is not the only molecular mechanism that determines the pathophysiology and clinical outcome. Studies of patient samples have shown that truncations exist within the CCND1 mRNA 3′ UTR and the truncation leads to a worse prognosis. We hypothesized that truncations in CCND1 exist in MCL cell lines and alter miRNA binding sites. miR-16-1 is a classic 22-bp miRNA proposed to bind at 2 sites in the 3′ UTR of CCND1. We were able to show by multiple methods that this miRNA regulates CCND1 protein expression.
In the present studies, we have confirmed, as predicted, that miR-16-1 regulates CCND1 protein expression and that truncation of the 3′ UTR occurs in MCL cell lines preventing proper miR-16-1 regulation of CCND1. It is likely that miR-16-1 is not the only miRNA regulating CCND1. Bioinformatics calculations indicate that there are at least 7 other miRNAs that may be important in this regard. This can explain the relatively low level of knockdown in our reporter construct experiments. How all of these miRNAs interact in the regulation of this and other genes remains to be determined, but it is likely that there are multiple miRNA signals regulating each gene, particularly those involved in cell cycle such as CCND1. We also recognize that miRNA regulation is not the only mechanism responsible for posttranscriptional regulation of CCND1. Wiestner et al have demonstrated that the half-life of the truncated CCND1 mRNA is longer than the full-length mRNA.10 One possible explanation is that the truncation eliminates an ARE, which is an AU-rich element responsible for rapid degradation.
Neither we nor others have determined the exact mechanisms leading to the truncated message. In the Z138 cell line, we found a point mutation in the genomic sequence described previously as a possible mechanism for truncation.10 Yet we were able to find the truncated message in 3 cell lines that did not contain this point mutation, suggesting that the point mutation is not required for the truncation. Whether the truncation is due to splicing variants, genetic polymorphisms, or another mechanism remains to be answered and will be important both for understanding the disease and for future therapies.
How the t(11;14) translocation and CCND1 overexpression leads to lymphomagenesis is also not well understood. In normal cells, CCND1 activation is necessary for cell-cycle progression and is stimulated by growth factors and mitogens. It has not been proven that MCL cell lines can undergo cell-cycle progression without growth factor support. In our studies, both Granta-519 and Jeko-1 were able to maintain cell-cycle progression after serum starvation. Granta-519 was able to maintain cell-cycle progression for 72 hours, Jeko-1 for 120 hours. However, H157, a lung cancer cell line, shows complete cell-cycle arrest after 48 hours of serum starvation. It also seems that the stronger the CCND1 protein expression, the longer the cell line can maintain cell-cycle progression without serum support.
These studies have added significantly to furthering our understanding of the molecular pathways involved in the development and progression of mantle cell lymphoma. This understanding is beginning to point toward new therapeutic strategies in this disease, which is so difficult to treat with current therapies. If overexpression of CCND1, particularly overexpression in the truncated form, is pivotal in this disease process, then it follows that therapeutic targets should be aimed toward this pathway. Exactly how the findings noted here will play into new strategies remains to be determined. However, therapeutics are being developed such as interfering RNAs directed against a number of genes involved in the development of cancer.30-32 For example, knowing that the truncated form of CCND1 is unresponsive to the usual miRNA regulators, one could design interfering RNAs specifically inhibiting the truncated form. Novel biologic agents such as bortezomib, suberoylanilide hydroxamic acid (SAHA), and mTOR inhibitors have shown single-agent activity in MCL cell lines and patients.33-37 SAHA, in particular, is able to decrease CCND1 protein translation while not altering the CCND1 mRNA levels. Kawamata et al34 recently reported that SAHA can down-regulate CCND1 protein expression in Jeko-1. Because we have found a high level of truncated CCND1 mRNA in Jeko-1, it is possible that SAHA would be a good choice of agent for patients expressing high levels of truncated CCND1 mRNA transcript.
In summary, truncated CCND1 mRNA expression leads to increased total CCND1 mRNA, increased CCND1 protein expression, and increased S-phase fraction in MCL cell lines. This truncation deletes miR-16-1 binding sites within the CCND1 mRNA 3′ UTR and alters miRNA regulation of MCL. The data presented here highlight the importance of miRNA in determining prognosis and predicting therapeutic response, and its role as a possible therapeutic target in the future.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank the DNA sequencing Core, Flow Cytometry Core, and Tissue Culture Core at the University of Colorado Cancer Center Core for technical support.
This work was sponsored by National Institutes of Health (Bethesda, MD) T-32 grant.
National Institutes of Health
Authorship
Contribution: R.W.C. designed and performed research, collected and analyzed data, and drafted the paper; L.T.B. designed and performed research, and drafted the paper; C.M.A. performed research and collected and analyzed data; H.M. designed research and drafted the paper; H.T. and D.K.B. performed research; S.G.E. analyzed data and drafted the paper; and W.A.R. designed research, analyzed data, and drafted the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert W. Chen, PO Box 6500, Mail Stop 8117, Aurora, CO 80045; e-mail: robert.chen@uchsc.edu.