Abstract
We tested the possibility that immune complexes formed following platelet factor 4 (PF4/CXCL4) binding to anti-PF4 antibody can stimulate neutrophil activation, similar to previous reports with platelets. Monoclonal Abs against PF4 and IgG from a heparin-induced thrombocytopenia (HIT) patient were applied. We observed that although PF4 or anti-PF4 antibody alone did not alter neutrophil function, costimulation with both reagents resulted in approximately 3-fold increase in cell surface Mac-1 expression, enhanced cell adhesion via L-selectin and CD18 integrins, and degranulation of secondary and tertiary granules. The level of Mac-1 up-regulation peaked at an intermediate PF4 dose, suggesting that functional response varies with antigen-antibody stoichiometry. PF4 binding to neutrophils was blocked by chondroitinase ABC. Cell activation was inhibited by both chondroitinase ABC and anti-CD32/FcγRII blocking mAb, IV.3. Confocal microscopy demonstrated that immune complexes colocalize with CD32a. Studies with HIT IgG demonstrated that neutrophils could be activated in the absence of exogenous heparin. These data, together, show that leukocyte surface chondroitin sulfates promote neutrophil activation by enhancing immune-complex binding to CD32a. Studies with recombinant PF4 suggest a role for arginine 49 in stabilizing PF4-chondroitin binding. Neutrophils activated via this mechanism may contribute to thrombosis and inflammation in patients mounting an immune response to PF4-heparin.
Introduction
Platelet factor 4 (PF4/CXCL4) is an ELR− tetrameric, cationic chemokine that constitutes 25% of the protein in platelet α-granules.1,2 It is also found bound to the luminal vascular endothelial surface.3,4 Although platelets represent the primary source of PF4, a recent report5 also suggests that the protein is expressed at lower levels in other cells of the immune system including cultured T cells, monocytes, and endothelial and smooth muscle cells. Treatment of patients with heparin results in PF4-heparin complex formation, and a dramatic increase in blood concentration of PF4. Heparin-PF4 binding is facilitated by the multivalent nature of both heparin and PF4. Whereas PF4 binds heparin with relatively high apparent affinity (Kd, ∼ 4-20 nM),6 binding interactions with glycosaminoglycans including those on the neutrophil surface are weaker (Kd, ∼ 650 nM).7 PF4 is typically at less than 1 nM in normal serum, and this level rises to 0.4 to 2.5 μM upon platelet activation.1,2
One to five percent of patients receiving unfractionated heparin suffer from heparin-induced thrombocytopenia (HIT).8,9 In these cases, PF4-heparin complexes trigger an immune response and the generation of anti-PF4 antibodies. Macromolecular antibody–heparin–PF4 immune complexes then bind FcγRIIa (CD32a) on platelets and induce platelet activation.4,10,11 Using chondroitinase ABC to cleave cell surface glycosaminoglycans, Rauova et al12 suggest a role for platelet chondroitin sulfates in cell activation. Platelet activation in turn results in more PF4 release into circulation, enhanced immune-complex formation, exaggerated platelet activation, and cell clearance.
Ten to fifty percent of HIT patients experience thrombosis in the arterial and venular circulation. This can lead to limb- and life-threatening complications. Although the precise mechanism(s) of thrombosis is yet unestablished, evidence in literature supports a role for activated platelets, resulting procoagulants, and microparticles.13 Immune complexes also bind to endothelial cells3,4 and they may up-regulate adhesion molecules that are typically associated with inflammatory ailments including E-/P-selectin, VCAM-1, and ICAM-1.14 The release of tissue factor and interleukin-8 by monocytes is an additional feature that is thought to contribute to thrombosis.15,16 Finally, plasma from HIT patients has been shown to activate neutrophils and enhance platelet-neutrophil adhesion via yet unidentified mechanisms.17
In the current paper, we examined the effect of PF4, heparin, and anti-PF4 antibodies (monoclonals and polyclonal HIT patient IgG) on neutrophil activation. Mac-1 was used as a marker in many experiments because its expression is prominently up-regulated following cell activation. Its role in the progress of inflammation and thrombosis is also well established. Our study demonstrates the following: (1) Neutrophils are activated by immune complexes via the FcγRIIa receptor. (2) Leukocyte chondroitin sulfates play an important role in enhancing immune-complex binding to the cell surface and subsequent cell activation. (3) Neutrophil activation leads to enhanced cell adhesion via selectins and CD18 integrins. Since activated neutrophils adherent on the vessel wall may damage tissue and initiate the process of thrombosis and vasculitis, our observation may be relevant to the pathogenesis of thrombosis in HIT patients.
Methods
Materials
All protocols used in this paper are approved by the University at Buffalo (Buffalo, NY) IRB/IACUC. Human neutrophil activating peptide-2 (NAP-2) and chondroitinase ABC were from Sigma-Aldrich (St Louis, MO). Unfractionated heparin was from Elkins-Sinn (Cherry Hill, NJ), and protamine sulfate (PS) was from Spectrum Chemicals (Gardena, CA). Human platelet PF4 (hPF4) from ChromaTec (Greifswald, Germany) was high-performance liquid chromatography (HPLC) purified and sequenced by the manufacturer prior to use in this study.
All antihuman monoclonal antibodies (mAbs) were raised in mice unless otherwise mentioned. These include PE-conjugated anti-CD11b clone D12 (BD Biosciences, San Jose, CA), anti–CD162/PSGL-1 KPL-1 (BD Biosciences), anti-CD16/FcγRIII 3G8 F(ab′)2 (Ancell, Bayport, MN), and anti-CD32/FcγRII mAbs IV.3 (StemCell Technologies, Vancouver, BC) and CIKm5 (Chemicon, Temecula, CA). Blocking mAbs against β2-integrin/CD18 IB4 and L-selectin/CD62L DREG-56 were produced as described previously.18 Anti-BSA mAb A00837.01 (IgG1) and polyclonal rabbit anti-BSA IgG were from Genscript (Piscataway, NJ) and Invitrogen (Carlsbad, CA), respectively. Isotype control mAbs P3 (IgG1) and eBMG2b (IgG2b) were from eBiosciences (San Diego, CA). Flow cytometric control antibodies were from BD Biosciences.
Anti-PF4 antibodies
Twelve hybridoma-producing mAbs against human PF4 were obtained from the fusion of splenocytes from mice immunized with either purified human PF4 (197 series including mAbs 197.2 and 197.3) or heparin-PF4 complex (196 series including 196.2 and 196.8) at the Blood Research Institute of the Blood Center of Wisconsin (Milwaukee, WI). Results are presented primarily for 2 of these anti-PF4 mAbs, 197.2 (IgG1) and 197.3 (IgG2b), and these are representative of at least 4 clones in the panel of reagents. The reactivity of these mAbs was determined by enzyme-linked immunosorbent assay (ELISA) and immunoblotting versus human PF4 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Antibodies 197.2 and 197.3 bind PF4 in the absence of heparin. Purified polyclonal rabbit anti–human PF4 antibody used in some assays was from Cell Sciences (Canton, MA). Polyclonal IgG was purified from one HIT patient plasma (patient 1 in Suh et al19 ).
Recombinant human PF4 (rPF4) and rat PF4
Wild-type rPF4 and mutant PF4 (PTA37–39AVP, R49S, L55R, L11V, and E4S) were expressed in Escherichia coli as described elsewhere.4,20,21 The original cDNA for this was kindly provided by Dr Mortimer Poncz (University of Pennsylvania, Philadelphia). Rat PF4 was purified from blood of male Sprague Dawley (CD) rats drawn into acid-citrate-dextrose (ACD) following cardiac puncture.
All PF4 preparations were endotoxin free as determined using the limulus amebocyte lysate (LAL) assay (Cambrex, Walkersville, MD). Protein concentrations were determined using micro-BCA assay (Pierce, Rockford, IL). PF4 concentrations are expressed in micromolar units (7.8 μg/mL = 1 μM monomeric PF4). Document S1 provide details on protein expression, purification, and characterization steps.
Neutrophil assays
Human polymorphonuclear cells (PMNs) were isolated as described previously22,23 using gradient centrifugation of blood obtained by venipuncture from healthy, nonsmoking, adult volunteers in 10 U/mL heparin. The human subject protocol used in this paper was approved by State University of New York (SUNY) Institutional Review Board, and all human participants provided written informed consent in accordance with the Declaration of Helsinki. EDTA (5 mM) was used as anticoagulant in some runs performed with HIT patient IgG. Isolated cells were maintained in calcium-free HEPES buffer22,23 containing 0.1% human serum albumin (HSA) at 4°C until use. Neutrophils constituted more than 90% of the PMNs. Thus, we refer to these isolated PMNs as neutrophils. All experiments were completed within 2 hours of neutrophil isolation. Cells were diluted into HEPES buffer containing 0.1% HSA and 1.5 mM CaCl2 just prior to functional studies.
All functional assays with isolated neutrophils were performed under uniform conditions. Neutrophils (1.5-2 × 106/mL) were equilibrated at 37°C for 6 minutes in HEPES buffer containing 1.5 mM CaCl2. Blocking reagents (1 U/mL chondroitinase ABC, heparin, 5 mM EDTA, or 10 μg/mL mAbs) were added in some runs during this incubation step. Following this, live samples were withdrawn for flow cytometric or functional analysis (described below). Samples were also fixed in ice-cold 1% paraformaldehyde for postexperiment analysis. Various types of PF4 (0-20 μM) were then added either in the presence or absence of anti-PF4 mAbs (0-30 μg/mL). Both PF4 and anti-PF4 were added simultaneously in all runs. The time of PF4 addition is designated t = 0, unless otherwise mentioned. Samples were withdrawn at various time points following stimulation. All end-point assays were performed at 9 minutes.
Flow cytometry
Mac-1 expression, PF4 binding to neutrophils, and anti-PF4 mAb binding to these cells were quantified using cell samples that were fixed overnight at 4°C. These samples were washed thrice in HEPES buffer prior to 20-minute incubation with PE-conjugated Mac-1 mAb at room temperature (RT) in the case of Mac-1 expression assay, polyclonal rabbit anti–human PF4 antibody followed by Alexa 488 goat anti–rabbit IgG in the case of bound PF4 detection, and Alexa 488–conjugated F(ab′)2 fragment of goat anti–mouse IgG in the case of anti-PF4 detection. In studies with HIT patient IgG, Alexa 488–conjugated F(ab′)2 fragment of goat anti–human IgG was used to detect immune-complex binding to leukocytes. Following incubation, samples were washed and read in a FACSCalibur flow cytometer (BD-Biosciences). Geometric mean fluorescence intensity (MFI) was recorded. Data were normalized, in some cases, with respect to control run performed without stimulus. Independent controls were performed to verify that sample fixation did not result in artifacts that affect our interpretation.
Neutrophil degranulation assays
Cells were rapidly centrifuged and supernatant was harvested at specified time. Myeloperoxidase release was quantified using o-dianisidine dihydrochloride (ODD) as substrate in phosphate-citrate buffer containing sodium perborate (Sigma-Aldrich).24 Here, following 10-minute incubation of sample with 0.16 mg/mL ODD at RT, the reaction was stopped using 50 μL of 1 N HCl, and light absorbance was measured at 405 nm using a plate reader. Lactoferrin release was measured using a sandwich ELISA. Here, plates coated overnight at 4°C with 10 μg/mL mouse anti–human lactoferrin mAb (QED Biosciences, San Diego, CA) were blocked with PBS containing 1% BSA prior to addition of either lactoferrin standards or lysis/experiment supernatant. After 30-minute incubation at RT with mixing, plates were washed and bound lactoferrin was detected using horseradish peroxidase (HRP)–conjugated rabbit anti–human lactoferrin polyclonal antibody (US Biological, Swampscott, MA) for 30 minutes at RT. ODD buffer was used as substrate for HRP detection. Neutrophil gelatinase B or MMP-9 (gelatinase granule) release was measured using human MMP-9 immunoassay kit (R&D Systems, Minneapolis, MN). The total granule content of unstimulated neutrophils was also determined and this was used to normalize data from individual runs. To quantify total neutrophil content, the same number of unstimulated cells as in the functional assays were lysed in buffer containing 20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 2% NP-40 with Complete protease inhibitor cocktail (Roche, Indianapolis, IN) at 4°C for 15 minutes. Biochemical assays were then performed with supernatant obtained following lysate centrifugation at 10 000g for 10 minutes.
Confocal microscopy
Anti-PF4 antibody (197.2) was labeled with Alexa Fluor 488, and 2 CD32 mAbs IV.3 and CIKm5 were labeled with Alexa Fluor 647 using kits from Invitrogen.
For microscopy experiments, fixed neutrophil samples were washed thrice and resuspended in HEPES buffer. Samples were then incubated with fluorescently labeled antibodies on ice for 20 minutes, washed, and then mounted using ProLong Gold antifade reagent (Invitrogen). After overnight curing at RT, confocal microscopic examination was performed using a Carl Zeiss Laser Scanning Systems LSM 510 (Heidelberg, Germany). Objective used was Zeiss 63× PlanApochomat/1.4 NA oil.
Neutrophil homotypic aggregation
Cell aggregation studies were performed using a cone-plate viscometer.22,23 Briefly, neutrophils were labeled with 10 ng/mL acridine orange during the 6-minute incubation step described in “Neutrophil assays.” Stimulus was introduced either right before initiation of shear at 1077/s (at t = 0) or 5 minutes prior to shear. Samples were withdrawn from the viscometer at different time intervals, and fixed in 1% paraformaldehyde at 4°C. Following overnight fixation, samples were read in the flow cytometer. Neutrophil population was gated based on forward versus side scatter. Percentage of neutrophil homotypic aggregation was quantified based on the depletion of singlets: % Aggregation = [1 −S/(S + 2D + 3T + 4Q+) × 100], where S, D, T, and Q+ represent single neutrophil, doublets, triplets, and aggregates with 4 or more cells, respectively.
Statistics
All data are presented as means plus or minus SEM. Statistical significance was assessed using Student t test for dual comparisons, and using ANOVA along with Student-Newman-Keuls test for multiple comparisons. P less than .05 was considered significant.
Results
Costimulation of human neutrophils by PF4 and anti-PF4 antibody induces dose-dependent Mac-1 up-regulation
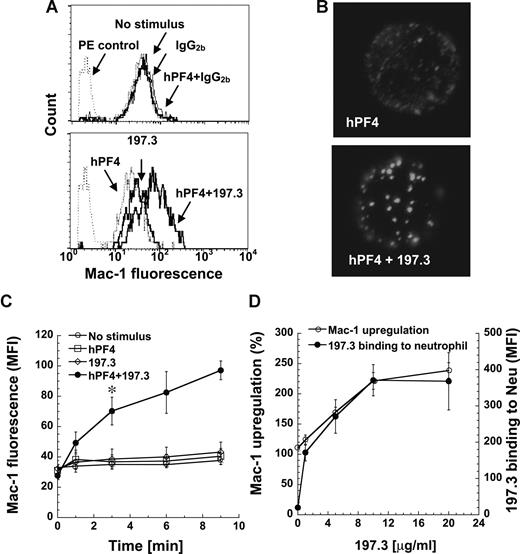
We measured neutrophil activation by monitoring changes in leukocyte shape and cell surface Mac-1 (CD11b/CD18) expression following addition of human platelet PF4 (hPF4), either in the presence or absence of anti-PF4 mAb. Data are presented for anti-PF4 clone 197.3 in Figure 1, and similar cell activation was observed with 197.2, 196.2, and 196.8 (data not shown). A distinct change in neutrophil shape was detected in the cytometer forward- versus side-scatter plot upon addition of both hPF4 and mAb simultaneously, but not upon addition of either one reagent alone (Figure S2). Addition of hPF4 along with 197.3 resulted in a 240% increase in cell surface Mac-1 expression at 9 minutes (Figure 1A,C). Addition of hPF4 and 197.3 alone resulted in only 23% to 30% increase in Mac-1 expression, which is similar to the no-stimulus control (14%). Such small increase in Mac-1 expression under no stimulus conditions is not unusual. Addition of isotype-matched control antibody, instead of 197.3, in these runs did not result in either shape change or Mac-1 up-regulation. Negative control immune complex formed by BSA interaction with anti-BSA also did not result in neutrophil activation (Figure S3).
Costimulation with human PF4 (hPF4) and anti-PF4 mAb induces dose-dependent Mac-1 up-regulation. Human neutrophils were incubated with hPF4 in the presence or absence of 197.3 or IgG2b isotype control. Cell surface Mac-1 expression and 197.3 binding to cells was quantified in fixed samples using PE-conjugated anti–Mac-1 mAb and Alexa 488–conjugated goat anti–mouse F(ab′)2 IgG, respectively. (A) Mac-1 expression 9 minutes after addition of 2 μM hPF4, 10 μg/mL 197.3, 10 μg/mL IgG2b, or their combinations. PE control represents nonbinding IgG control. Costimulation with hPF4 and anti-PF4 resulted in enhanced Mac-1 expression. (B) In the top panel, neutrophils were incubated with 2 μM hPF4 for 9 minutes and fixed. PF4 distribution on fixed cells was determined by incubation with 197.3 followed by Alexa 488 F(ab′)2 goat anti–mouse IgG. In the bottom panel, neutrophils were incubated with 2 μM hPF4 and 10 μg/mL 197.3 prior to fixation. Fixed sample was then incubated with Alexa 488 F(ab′)2 goat anti–mouse IgG. Clustering of PF4 was observed in right panel only. (C) Time kinetics of Mac-1 surface expression following various treatments: no stimulus (○), 2 μM hPF4 alone (□), 10 μg/mL 197.3 alone (◇), and 2 μM hPF4 plus 10 μg/mL 197.3 costimulation (●). Absolute geometric mean fluorescence intensity (MFI) data are presented. Costimulation resulted in significant Mac-1 expression beyond 3 minutes (*P < .01). (D) In studies where 197.3 dose was varied and PF4 dose was fixed at 2 μM, Mac-1 expression (left axis) was observed to vary in proportion to 197.3 binding to neutrophils (right axis). Mac-1 up-regulation (%) represents percentage change in Mac-1 levels on neutrophils in treatment runs versus no-stimulus controls. Error bars represent SEM (n ≥ 3).
Costimulation with human PF4 (hPF4) and anti-PF4 mAb induces dose-dependent Mac-1 up-regulation. Human neutrophils were incubated with hPF4 in the presence or absence of 197.3 or IgG2b isotype control. Cell surface Mac-1 expression and 197.3 binding to cells was quantified in fixed samples using PE-conjugated anti–Mac-1 mAb and Alexa 488–conjugated goat anti–mouse F(ab′)2 IgG, respectively. (A) Mac-1 expression 9 minutes after addition of 2 μM hPF4, 10 μg/mL 197.3, 10 μg/mL IgG2b, or their combinations. PE control represents nonbinding IgG control. Costimulation with hPF4 and anti-PF4 resulted in enhanced Mac-1 expression. (B) In the top panel, neutrophils were incubated with 2 μM hPF4 for 9 minutes and fixed. PF4 distribution on fixed cells was determined by incubation with 197.3 followed by Alexa 488 F(ab′)2 goat anti–mouse IgG. In the bottom panel, neutrophils were incubated with 2 μM hPF4 and 10 μg/mL 197.3 prior to fixation. Fixed sample was then incubated with Alexa 488 F(ab′)2 goat anti–mouse IgG. Clustering of PF4 was observed in right panel only. (C) Time kinetics of Mac-1 surface expression following various treatments: no stimulus (○), 2 μM hPF4 alone (□), 10 μg/mL 197.3 alone (◇), and 2 μM hPF4 plus 10 μg/mL 197.3 costimulation (●). Absolute geometric mean fluorescence intensity (MFI) data are presented. Costimulation resulted in significant Mac-1 expression beyond 3 minutes (*P < .01). (D) In studies where 197.3 dose was varied and PF4 dose was fixed at 2 μM, Mac-1 expression (left axis) was observed to vary in proportion to 197.3 binding to neutrophils (right axis). Mac-1 up-regulation (%) represents percentage change in Mac-1 levels on neutrophils in treatment runs versus no-stimulus controls. Error bars represent SEM (n ≥ 3).
hPF4 was distributed uniformly on the neutrophil surface when it was added alone, and protein clusters were noted upon addition of hPF4 and 197.3 (Figure 1B). Mac-1 levels increased with time and were significantly higher than other treatments by 3 minutes (Figure 1C). hPF4 and anti-PF4 Abs were added simultaneously to neutrophils in all studies presented in this paper. This resulted in greater neutrophil activation compared with runs where either PF4–anti-PF4 complex was formed prior to neutrophil addition or neutrophils were incubated with PF4 prior to addition of anti-PF4 mAb (Figure S4).
Neutrophil activation was maximal when 2 μM hPF4 was added along with 10 μg/mL 197.3. When hPF4 concentration was fixed at 2 μM and 197.3 concentration was varied from 0 to 20 μg/mL, neutrophil activation increased in proportion to mAb 197.3 binding and it saturated at 10 μg/mL (Figure 1D). In other studies where hPF4 was varied from 0 to 20 μM in the presence of fixed 197.3 concentration (10 μg/mL), maximum cell activation was noted at 2 to 4 μM. Overall, these results show that PF4–anti-PF4 immune-complex binding to neutrophil may trigger cell activation.
Clustered PF4 activates neutrophils via CD32a
We investigated the possibility that tetrameric PF4 may form a multimeric immune complex with anti-PF4 mAb, and that these complexes may stimulate neutrophil exocytosis via Fc receptors. Since neutrophils have 2 types of Fc receptors, CD32a and CD16b, blocking antibodies against both receptors were tested. Here, whereas IV.3 (anti-CD32) effectively blocked Mac-1 up-regulation (Figure 2A) and inhibited cell shape change, 3G8 F(ab′)2 (anti-CD16) and isotype control reagents did not inhibit either feature. 3G8 F(ab′)2 augmented CD11b expression in costimulation runs, possibly by preventing immune-complex interaction with CD16b and enhancing the efficiency of CD32a clustering.
FcγRIIa (CD32a) mediated neutrophil Mac-1 up-regulation. The ability of 10 μg/mL anti-CD32/FcγRII (IV.3), anti-CD16 (3G8 F(ab′)2), and isotype control for IV.3 (IgG2b) to block Mac-1 up-regulation induced by 2 μM hPF4 and 10 μg/mL 197.3 was measured. Blocking reagents were added 6 minutes prior to stimulation. (A) Mac-1 expression was measured 9 minutes after stimulation in fixed samples. (B) Total mouse antibody bound to cells was measured using Alexa 488–conjugated goat anti–mouse F(ab′)2 IgG. IV.3 significantly reduced Mac-1 up-regulation without dramatically altering the binding of PF4 to cells. *P < .05, with respect to positive control with hPF4 plus 197.3. (C-D) Confocal microscopy images showed clustering and colocalization of FcγRIIa and anti-PF4 antibody. Two Alexa 647–conjugated anti-CD32 monoclonal antibodies were used for these runs, IV.3 (Ci-Civ) and CIKm5 (Di-Div). In panels Ci and Di, CD32 distribution on unstimulated neutrophils determined using IV.3–Alexa 647 and CIKm5–Alexa 647, respectively, was seen to be uniform. Cells were stimulated with hPF4 and Alexa 488–conjugated 197.2. After fixation, these cells were labeled with either IV.3–Alexa 647 or CIKm5–Alexa 647. Panels Cii and Dii show signal from 197.2–Alexa 488 channel, panels Ciii and Diii show data from IV.3–Alexa 647 and CIKm5–Alexa 647, respectively, and panels Civ and Div show merged fluorescence images along with differential interference contrast (DIC) image. Error bars represent SEM (n ≥ 3).
FcγRIIa (CD32a) mediated neutrophil Mac-1 up-regulation. The ability of 10 μg/mL anti-CD32/FcγRII (IV.3), anti-CD16 (3G8 F(ab′)2), and isotype control for IV.3 (IgG2b) to block Mac-1 up-regulation induced by 2 μM hPF4 and 10 μg/mL 197.3 was measured. Blocking reagents were added 6 minutes prior to stimulation. (A) Mac-1 expression was measured 9 minutes after stimulation in fixed samples. (B) Total mouse antibody bound to cells was measured using Alexa 488–conjugated goat anti–mouse F(ab′)2 IgG. IV.3 significantly reduced Mac-1 up-regulation without dramatically altering the binding of PF4 to cells. *P < .05, with respect to positive control with hPF4 plus 197.3. (C-D) Confocal microscopy images showed clustering and colocalization of FcγRIIa and anti-PF4 antibody. Two Alexa 647–conjugated anti-CD32 monoclonal antibodies were used for these runs, IV.3 (Ci-Civ) and CIKm5 (Di-Div). In panels Ci and Di, CD32 distribution on unstimulated neutrophils determined using IV.3–Alexa 647 and CIKm5–Alexa 647, respectively, was seen to be uniform. Cells were stimulated with hPF4 and Alexa 488–conjugated 197.2. After fixation, these cells were labeled with either IV.3–Alexa 647 or CIKm5–Alexa 647. Panels Cii and Dii show signal from 197.2–Alexa 488 channel, panels Ciii and Diii show data from IV.3–Alexa 647 and CIKm5–Alexa 647, respectively, and panels Civ and Div show merged fluorescence images along with differential interference contrast (DIC) image. Error bars represent SEM (n ≥ 3).
The effect of Fc receptor inhibiting antibodies on PF4 binding to neutrophils was evaluated (Figure 2B). In these studies, the binding of an Alexa 488–conjugated anti–mouse Ab to fixed cells was quantified since this reagent recognizes both the anti-PF4 mAb 197.3 and the anti-Fc mAbs. As seen, the binding of anti–mouse Ab to cells treated with both 197.3 and IV.3 was roughly equal to the sum of Ab binding in the presence of either Ab alone. The observation suggests that IV.3 does not markedly reduce the binding of PF4 to neutrophils. 3G8 F(ab′)2 also did not alter PF4 or 197.3 binding to neutrophils. Similar observations as in Figure 2B were made when 197.3 was directly conjugated with FITC and costimulation experiments were performed. Together, these experiments suggest that Fc-receptor independent binding sites for PF4 complex exist on leukocytes.
Confocal microscopy investigations were undertaken to assess the colocalization of PF4 complexes on neutrophils with CD32. Two antibodies, IV.3 in Figure 2C and CIKm5 in Figure 2D, that bind different sites on CD32 were applied. As seen in Figure 2Ci and Di, in the absence of stimulation, neutrophil CD32 is evenly distributed on cells. Clustering and colocalization (overlap coefficient = 0.9) of CD32a with the PF4-antibody complex were observed in runs where live cells were costimulated with PF4 and Alexa 488–conjugated 197.2 and fixed, prior to staining of cells, with either IV.3–Alexa 647 (Figure 2Cii-iv) or CIKm5–Alexa 647 (Figure 2Dii-iv). In control studies, we confirmed the specificity of anti-CD32 mAbs since excess unconjugated mAbs blocked the binding of labeled reagents. Isotype-matched, nonbinding control mAb Alexa 488–conjugated HAE-1f (IgG1) also did not bind neutrophils in microscopy runs. Further in functional studies, we noted that 10 μg/mL 197.2 even after Alexa 488 conjugation up-regulated Mac-1 to a similar extent as unconjugated mAb. Confocal image stacks in movie format are provided as Video S1.
Overall, these observations are consistent with the proposition that PF4–anti-PF4 immune complexes activate neutrophils via cell surface CD32a.
Degranulation of secondary and tertiary granules by immune complex
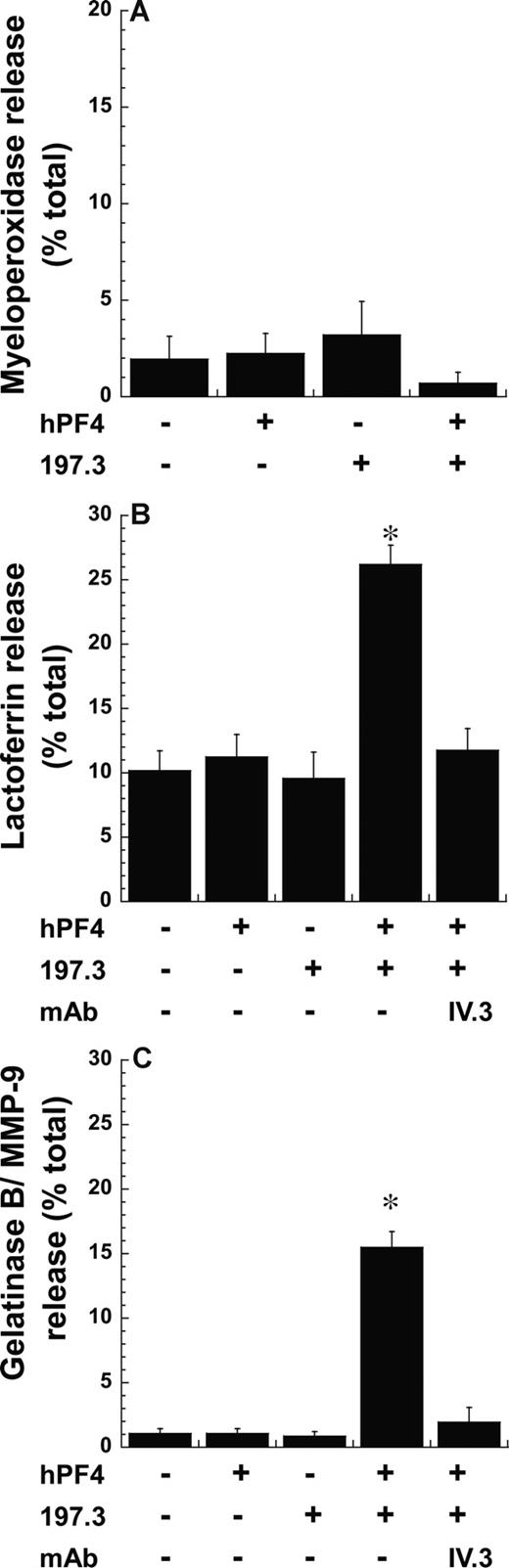
Mac-1 is stored in neutrophil secondary and tertiary granules, and in secretory vesicles. To investigate the cell compartments that were released following stimulation with PF4 complex, we measured cellular myeloperoxidase (Figure 3A), lactoferrin (Figure 3B), and gelatinase B (Figure 3C) release. These molecules are markers of primary/azurophil, secondary/specific, and tertiary/gelatinase granules, respectively. Here, we observed the release of statistically higher levels of lactoferrin (26%) and gelatinase (15%) but not myeloperoxidase upon costimulation with PF4 and anti-PF4, compared with runs with either reagent alone. IV.3 inhibited lactoferrin and gelatinase release induced upon costimulation. These observations are consistent with the finding that azurophil/primary granules are most resistant to exocytosis,24 and that Mac-1 in this study may emerge from secondary and tertiary granules.
Exocytosis of secondary and tertiary but not primary granules following costimulation. Neutrophil degranulation 9 minutes after costimulation with 2 μM PF4 and 10 μg/mL anti-PF4 Ab (197.3) was measured by quantifying the release of (A) myeloperoxidase (primary granule), (B) lactoferrin (secondary granule), and (C) gelatinase B/MMP-9 (tertiary granule). Degranulation (mean ± SEM; n = 3) is expressed as percentage release of total neutrophil granule content. IV.3 blocks lactoferrin and gelatinase B release. *P < .01, with respect to all other treatments.
Exocytosis of secondary and tertiary but not primary granules following costimulation. Neutrophil degranulation 9 minutes after costimulation with 2 μM PF4 and 10 μg/mL anti-PF4 Ab (197.3) was measured by quantifying the release of (A) myeloperoxidase (primary granule), (B) lactoferrin (secondary granule), and (C) gelatinase B/MMP-9 (tertiary granule). Degranulation (mean ± SEM; n = 3) is expressed as percentage release of total neutrophil granule content. IV.3 blocks lactoferrin and gelatinase B release. *P < .01, with respect to all other treatments.
PF4 binding to chondroitin sulfates on neutrophils is necessary for cell activation
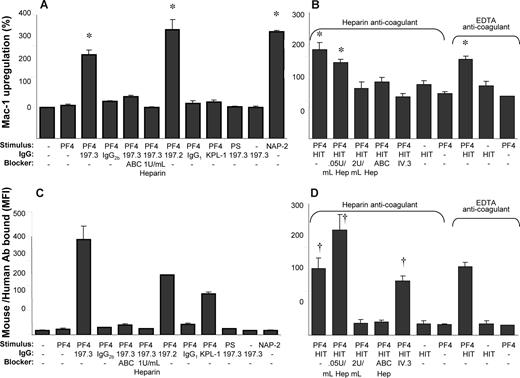
PF4 has been shown to bind chondroitin sulfates on neutrophils7 and platelets,12 and thus we aimed to determine whether this molecule contributes to leukocyte activation (Figure 4). Here, we observed that both 1 U/mL chondroitinase ABC and heparin blocked neutrophil activation induced by costimulation as measured by cell surface Mac-1 expression (Figure 4A). These treatments also inhibited PF4-antibody complex binding to neutrophils (Figure 4C). Isotype nonbinding (IgG1 and IgG2b) and binding control mAb (anti–CD162/KPL-1, IgG1) did not support Mac-1 up-regulation. Further, substitution of PF4 with cationic peptide protamine sulfate (PS), a reagent used clinically to neutralize the anticoagulation effects of heparin, failed both to activate human neutrophils and to promote binding of 197.3 to leukocytes. Finally, costimulation with immune complex (PF4 + 197.3/197.2) resulted in Mac-1 up-regulation that was comparable to the effects of the potent neutrophil agonist NAP-2 (20 nM). Overall, these observations are consistent with a role for chondroitin sulfates in cell activation, and the higher affinity of PF4 for heparin compared with the chondroitin sulfates. Studies with PS confirm that our experimental observations are due to the specific nature of PF4-neutrophil interactions and not due to the electrostatic properties of PF4, a highly basic protein.
Costimulation with both anti-PF4 mAb and HIT patient IgG results in Mac-1 up-regulation that is blocked by chondroitinase ABC and heparin. The ability of 1 U/mL chondroitinase ABC (ABC), heparin, and 10 μg/mL anti-CD32/FcγRII mAb (IV.3) to block Mac-1 up-regulation following 9-minute costimulation with 2 μM hPF4, and either 10 μg/mL anti-PF4 mAbs (197.3 and 197.2; A,C) or HIT patient IgG (B,D) was examined. Whereas 10 μg/mL nonbinding isotype-matched Abs (IgG2b and IgG1) were used in some runs, anti–PSGL-1/CD162 mAb KPL-1 served as an isotype-matched, neutrophil-binding control Ab for studies performed with 197.2. Protamine sulfate (PS; 25 μg/mL) was added in lieu of human PF4 in other controls. (A,B) Mac-1 up-regulation (%). (C,D) Total antibody binding to neutrophils (MFI) determined using Alexa 488 goat anti–mouse F(ab′)2 IgG (C) and Alexa 488 goat anti–human F(ab′)2 IgG (D). *P < .05, with respect to all negative controls (no stimulus, hPF4 or 197.3 alone, hPF4 plus IgG1 or IgG2b isotype controls, hPF4 plus KPL-1, and PS plus 197.3) and blocker treatments (ABC, ≥ 1U/mL heparin, IV.3). †P < .05, with respect to all other treatments except that HIT + PF4 is not different from HIT + PF4 + IV.3. In panels B and D, whereas blood was drawn in heparin in some runs, EDTA was used as anticoagulant in other runs. Low doses of unfractionated heparin (0.05 U/mL) enhance PF4-HIT complex binding to neutrophils. Error bars represent SEM (n ≥ 3).
Costimulation with both anti-PF4 mAb and HIT patient IgG results in Mac-1 up-regulation that is blocked by chondroitinase ABC and heparin. The ability of 1 U/mL chondroitinase ABC (ABC), heparin, and 10 μg/mL anti-CD32/FcγRII mAb (IV.3) to block Mac-1 up-regulation following 9-minute costimulation with 2 μM hPF4, and either 10 μg/mL anti-PF4 mAbs (197.3 and 197.2; A,C) or HIT patient IgG (B,D) was examined. Whereas 10 μg/mL nonbinding isotype-matched Abs (IgG2b and IgG1) were used in some runs, anti–PSGL-1/CD162 mAb KPL-1 served as an isotype-matched, neutrophil-binding control Ab for studies performed with 197.2. Protamine sulfate (PS; 25 μg/mL) was added in lieu of human PF4 in other controls. (A,B) Mac-1 up-regulation (%). (C,D) Total antibody binding to neutrophils (MFI) determined using Alexa 488 goat anti–mouse F(ab′)2 IgG (C) and Alexa 488 goat anti–human F(ab′)2 IgG (D). *P < .05, with respect to all negative controls (no stimulus, hPF4 or 197.3 alone, hPF4 plus IgG1 or IgG2b isotype controls, hPF4 plus KPL-1, and PS plus 197.3) and blocker treatments (ABC, ≥ 1U/mL heparin, IV.3). †P < .05, with respect to all other treatments except that HIT + PF4 is not different from HIT + PF4 + IV.3. In panels B and D, whereas blood was drawn in heparin in some runs, EDTA was used as anticoagulant in other runs. Low doses of unfractionated heparin (0.05 U/mL) enhance PF4-HIT complex binding to neutrophils. Error bars represent SEM (n ≥ 3).
We next tested whether similar observations are made with IgG purified from HIT patients as with the mAbs 197.2 and 197.3 (Figure 4B,D). Here, we confirmed that addition of PF4 with HIT IgG induced cell activation, even in the absence of exogenous heparin and when EDTA was used as anticoagulant during venipuncture. Immune-complex binding to leukocytes and cell activation could be blocked by high heparin doses (2 U/mL) and chondroitinase ABC. Indeed, although 0.05 U/mL heparin (low concentration) did enhance immune-complex binding to neutrophils, the level of cell activation was comparable in this case to that in the absence of heparin. Together, these results suggest a key role for leukocyte surface chondroitin sulfates in regulating cell activation.
Studies with recombinant PF4 confirm observations made with human platelet protein
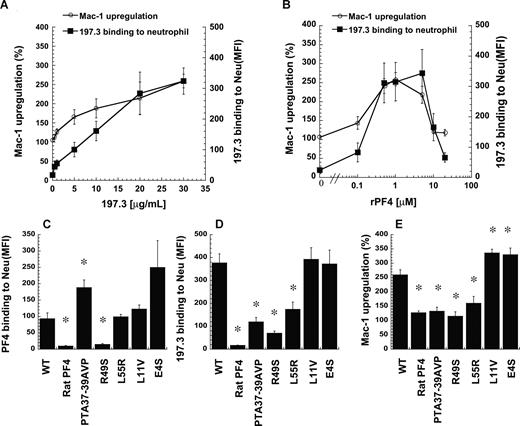
The purity of PF4 obtained from human platelets is sometimes cited as a reason for differences in results reported by diverse investigators.25 Thus, we conducted additional studies to confirm our findings using recombinant human PF4 (rPF4) from E coli. Dose-response studies were performed with neutrophils stimulated either with fixed rPF4 concentrations (1 μM) and various 197.3 concentrations (0-30 μg/mL; Figure 5A) or fixed 197.3 concentration (30 μg/mL) and various rPF4 concentrations (0-20 μM; Figure 5B). Results obtained were similar to Figure 1, with Mac-1 levels increasing with the extent of 197.3 binding to leukocytes in Figure 5A, and cell activation peaking at an intermediate PF4 dose (1-5 μM) in Figure 5B. In Figure 5B, increasing PF4 concentration beyond a critical level likely reduced the extent of immune-complex formation since it saturated the available 197.3 and reduced multimer complex formation. Significant and reproducible Mac-1 up-regulation was observed at 1 to 2 μM rPF4 and 10 to 30 μg/mL 197.3.
Recombinant human PF4 and 197.3 costimulation induce neutrophil Mac-1 up-regulation. (A) In costimulation studies, 197.3 dose was varied from 0.5 to 30 μg/mL in the presence of 1 μM wild-type (WT) recombinant PF4 (rPF4). Mac-1 up-regulation (left axis) and 197.3 binding to neutrophil (right axis) at 9 minutes after stimulation were measured. Mac-1 expression increased with 197.3 binding to cells. (B) In costimulation studies where rPF4 dose was varied in the presence of constant levels of 197.3 (30 μg/mL), Mac-1 up-regulation was maximum at 1 μM. For studies in panels C-E, 5 PF4 mutants were generated based on the primary sequence of rat-PF4. Costimulation experiments were performed with 1 μM PF4 (recombinant or rat source) and 30 μg/mL 197.3 for 9 minutes. Fixed samples were used to quantify (C) PF4 binding to neutrophils using polyclonal rabbit anti–human PF4 Ab followed by Alexa 488 goat anti–rabbit IgG, (D) 197.3 binding to neutrophil using Alexa 488 goat anti–mouse F(ab′)2 IgG, and (E) Mac-1 up-regulation using PE-labeled anti–Mac-1 mAb. rPF4 binding to neutrophils was abrogated by R49S mutation. Binding of 197.3 was partially dependent on residues 37 to 39. *Significantly different from WT rPF4 run (P < .05). Error bars represent SEM (n = 3).
Recombinant human PF4 and 197.3 costimulation induce neutrophil Mac-1 up-regulation. (A) In costimulation studies, 197.3 dose was varied from 0.5 to 30 μg/mL in the presence of 1 μM wild-type (WT) recombinant PF4 (rPF4). Mac-1 up-regulation (left axis) and 197.3 binding to neutrophil (right axis) at 9 minutes after stimulation were measured. Mac-1 expression increased with 197.3 binding to cells. (B) In costimulation studies where rPF4 dose was varied in the presence of constant levels of 197.3 (30 μg/mL), Mac-1 up-regulation was maximum at 1 μM. For studies in panels C-E, 5 PF4 mutants were generated based on the primary sequence of rat-PF4. Costimulation experiments were performed with 1 μM PF4 (recombinant or rat source) and 30 μg/mL 197.3 for 9 minutes. Fixed samples were used to quantify (C) PF4 binding to neutrophils using polyclonal rabbit anti–human PF4 Ab followed by Alexa 488 goat anti–rabbit IgG, (D) 197.3 binding to neutrophil using Alexa 488 goat anti–mouse F(ab′)2 IgG, and (E) Mac-1 up-regulation using PE-labeled anti–Mac-1 mAb. rPF4 binding to neutrophils was abrogated by R49S mutation. Binding of 197.3 was partially dependent on residues 37 to 39. *Significantly different from WT rPF4 run (P < .05). Error bars represent SEM (n = 3).
Selected rPF4 mutants were generated to determine the critical residues that participate in cell activation. Specifically, the goal was to begin defining the epitope recognized by mAbs used in this project and the residues that are critical for PF4-neutrophil interaction. Here, the ability of rPF4 mutants to bind neutrophils was quantified by assessing anti-PF4 polyclonal Ab binding to neutrophils (Figure 5C). Additional studies were performed to quantify mutation effects on 197.3 binding to cells (Figure 5D) and cell surface Mac-1 expression (Figure 5E). All mutants were designed based on the sequence of rat PF4, a protein sharing 74% homology with human PF4 and having equivalent basic properties.26 Rat PF4 itself did not bind human cells (Figure 5C), although independent experiments verified that the polyclonal Ab used in these assays cross-reacts between the 2 species. Consequently, neither 197.3 binding (Figure 5D) nor Mac-1 up-regulation (Figure 5E) was observed upon addition of rat PF4. In studies with mutants, although PTA37–39AVP and L55R bound neutrophils at similar or higher levels than the wild-type molecule, these reagents did not bind 197.3 efficiently. Thus, 197.3 binding to PF4 is regulated by these C-terminal residues. Due to the reduced 197.3 binding, Mac-1 up-regulation was also reduced. Mutation R49S abrogated PF4 binding to leukocytes and subsequent activation, suggesting that this amino acid may be a critical moiety distinguishing human and rat protein. Since R49S bound heparin during the purification step described in “Methods,” the results also suggest that arginine 49 may be more important for chondroitin binding compared with heparin recognition. Two N-terminal mutants tested (L11V and E4S) resulted in either similar or 30% higher Mac-1 up-regulation compared with wild-type rPF4 in the presence of 197.3. In control studies, none of the mutants induced Mac-1 up-regulation or neutrophil shape change in the absence of 197.3.
Overall, the studies confirm (1) observations made with isolated human PF4 with recombinant protein, (2) the dose dependence of the feature peaking at intermediate PF4 dose, and (3) the importance of selected PF4 C-terminus residues in regulating PF4 binding to neutrophil chondroitin sulfates, antibody binding, and cell activation.
Neutrophil adhesion function is augmented by immune complex
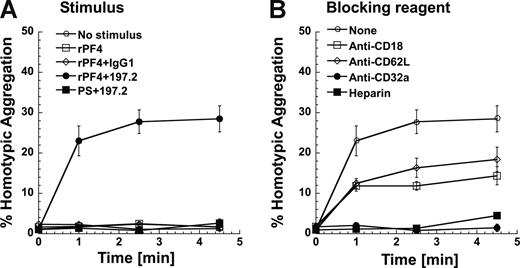
To establish a link between Mac-1 expression and leukocyte adhesion, functional studies that assess neutrophil homotypic aggregation were performed under fluid shear using a cone-plate viscometer (Figure 6). Unstimulated neutrophils did not aggregate since their Mac-1 is in a low-affinity state (Figure 6A). Costimulation with rPF4 and 197.2 led to 30% of the neutrophils forming homotypic aggregates. Addition of rPF4 alone did not lead to homotypic aggregation. Cell aggregation was also not observed in costimulation runs where 197.2 was replaced with an isotype control Ab, or when rPF4 was replaced with PS.
Costimulation induces neutrophil homotypic adhesion under shear. Neutrophils (2 × 106/mL) were preincubated with or without inhibiting reagents at room temperature for 3 minutes. Neutrophils were then stimulated with rPF4 (1 μM) or protamine sulfate (PS; 25 μg/mL) in the presence or absence of 10 μg/mL 197.2 or IgG1 isotype control at 37°C for 5 minutes prior to shear application at 1077/s. Mac-1 is up-regulated in this 5-minute interval. t = 0 designates time of shear application in this figure. (A) Kinetics of neutrophil homotypic aggregation. (B) Neutrophil homotypic aggregation inhibited by 1 U/mL heparin and 10 μg/mL of function blocking mAbs directed against CD32a (IV.3), L-selectin/CD62L (DREG56), or β2-integrin/CD18 (IB4). Error bars represent SEM (P ≥ 3).
Costimulation induces neutrophil homotypic adhesion under shear. Neutrophils (2 × 106/mL) were preincubated with or without inhibiting reagents at room temperature for 3 minutes. Neutrophils were then stimulated with rPF4 (1 μM) or protamine sulfate (PS; 25 μg/mL) in the presence or absence of 10 μg/mL 197.2 or IgG1 isotype control at 37°C for 5 minutes prior to shear application at 1077/s. Mac-1 is up-regulated in this 5-minute interval. t = 0 designates time of shear application in this figure. (A) Kinetics of neutrophil homotypic aggregation. (B) Neutrophil homotypic aggregation inhibited by 1 U/mL heparin and 10 μg/mL of function blocking mAbs directed against CD32a (IV.3), L-selectin/CD62L (DREG56), or β2-integrin/CD18 (IB4). Error bars represent SEM (P ≥ 3).
Cell adhesion was partially blocked (∼ 50%) by anti–L-selectin and anti-CD18 antibodies, and completely abrogated by anti-CD32 antibody and heparin (Figure 6B). These data are consistent with our earlier publication23 that shows that homotypic neutrophil aggregation following chemotactic stimulation is mediated by L-selectin and CD18 integrins. Similar levels of homotypic aggregation and function blocking data were observed in studies where rPF4 was replaced by human platelet PF4 in these runs (data not shown). Overall, Mac-1 up-regulation following costimulation is accompanied by increased molecular affinity/avidity that results in enhanced cell adhesion.
Discussion
We report that neutrophil activation can be induced by immune complexes that contain PF4 and anti-PF4 mAb. The dose of PF4 used in this study is physiological and it mimics previous investigations of platelet activation. Further, cell activation observed is robust since in addition to profound changes in neutrophil shape observed using flow cytometry there is a 2.5- to 3-fold increase in cell surface CD11b/Mac-1 expression, and degranulation of secondary and tertiary granules. Both PF4 purified from human platelets and recombinant PF4 from E coli induced similar neutrophil activation and adhesion behavior, in the presence of anti-PF4 antibodies (197.2 and 197.3) and HIT patient IgG. Thus, the underlying observations are not complicated by artifacts of protein purification or antibody specificity.
Role for chondroitin sulfate and CD32a
The mechanism of immune complex–mediated cell activation is similar to that which has been reported previously for platelets4,10-12,27,28 (Figure 7). Similar to these previous studies, Mac-1 up-regulation and anti-PF4 antibody binding to neutrophils displayed an optimum in a narrow PF4 dose range. Two principal players in this process are cell surface chondroitin sulfates and FcγRIIa/CD32a.
Potential role of activated neutrophils based on our and other published data. Region left of the dashed line shows the situation prior to heparin infusion. Here, PF4 is bound to the surface of vascular cells. Heparin infusion (right of the dashed line) results in PF4-heparin complexes in blood and immune response. Platelets and neutrophils may be activated via 3 modes based on results in this paper and other published data4,11,12 : PF4–anti-PF4 complexes formed on platelet/neutrophil surface chondroitin sulfates may activate either the same cell (mode A) or an adjacent cell (mode B) via CD32a. In addition, PF4–heparin–anti-PF4 immune complexes formed in solution may ligate CD32a and trigger cell activation in both cells (mode C). Platelet activation results in release of α-granule content including PF4, amplification of cell activation processes, and thrombocytopenia. Neutrophil activation results in degranulation of secondary and tertiary granules and up-regulation of cell surface Mac-1. Newly expressed P-selectin on platelets and functionally active Mac-1 on neutrophils likely contribute to new adhesive interactions in the vasculature. These may include neutrophil-platelet adhesion, enhanced neutrophil-endothelial interactions, or neutrophil-neutrophil secondary interactions that amplify leukocyte-endothelial cell-binding events.
Potential role of activated neutrophils based on our and other published data. Region left of the dashed line shows the situation prior to heparin infusion. Here, PF4 is bound to the surface of vascular cells. Heparin infusion (right of the dashed line) results in PF4-heparin complexes in blood and immune response. Platelets and neutrophils may be activated via 3 modes based on results in this paper and other published data4,11,12 : PF4–anti-PF4 complexes formed on platelet/neutrophil surface chondroitin sulfates may activate either the same cell (mode A) or an adjacent cell (mode B) via CD32a. In addition, PF4–heparin–anti-PF4 immune complexes formed in solution may ligate CD32a and trigger cell activation in both cells (mode C). Platelet activation results in release of α-granule content including PF4, amplification of cell activation processes, and thrombocytopenia. Neutrophil activation results in degranulation of secondary and tertiary granules and up-regulation of cell surface Mac-1. Newly expressed P-selectin on platelets and functionally active Mac-1 on neutrophils likely contribute to new adhesive interactions in the vasculature. These may include neutrophil-platelet adhesion, enhanced neutrophil-endothelial interactions, or neutrophil-neutrophil secondary interactions that amplify leukocyte-endothelial cell-binding events.
In support of the role of neutrophil surface chondroitin sulfates, we observed that both heparin and chondroitinase ABC block cell activation by preventing immune-complex binding to cells. Such treatments consequently prevented neutrophil activation and adhesion. Chondroitin sulfates in these assays have 2 potential roles: (1) they may enhance the binding of PF4-heparin complexes to neutrophil CD32a by enhancing rebinding effects,29 or (2) the binding of PF4 to chondroitin sulfate may enhance the affinity of PF4 for the HIT antibody, and this in turn may enhance immune-complex formation. In experiments with HIT patient IgG, consistent with published literature, we observed that addition of 0.05 U/mL heparin enhanced antibody binding to neutrophils, whereas higher doses (2 U/mL) had an inhibitory effect. Cell activation was observed even in the absence of heparin when EDTA was used as anticoagulant, and this suggests that HIT IgG can recognize PF4 that is bound to chondroitin sulfates on the cell surface. These observations are consistent with a recent report where a mouse mAb (KKO) mimicking human HIT antibodies has been shown to bind PF4 engaged on platelet chondroitin sulfate. Such binding induces platelet activation even in the absence of heparin.12 It is necessary to note that our observations do not rule out the possibility that heparin may be necessary for cell activation in the case of other HIT patient antibodies. The possible contribution of heparin-binding proteins expressed on neutrophils to cell activation is also not addressed in this study.
Following the formation of the immune complex, activation is mediated through ligation and possible cross-linking of neutrophil Fc receptor CD32a. In support of this, cell activation is completely blocked by the mAb IV.3.30 Microscopy investigations reveal clusters of CD32 on the cell surface when PF4–anti-PF4 immune complexes are formed. Such cell activation likely proceeds via activation of the immunoreceptor tyrosine–based activation motif (ITAM) and downstream pathways described elsewhere.31
Activation of neutrophils via the PF4 anti-PF4 immune complex mechanism may enhance leukocyte-platelet adhesion and leukocyte interactions with other vascular components, such as the endothelial cells that have been shown to up-regulate various selectins and immunoglobulins in response to HIT patient sera14 (Figure 7). Indeed, there is a very rich literature that demonstrates that leukocyte activation and adhesion play a principal role in a variety of inflammatory processes (Luster et al32 and references cited therein).
Partial mapping of Abs and PF4-binding epitopes
Although homologous to human PF4, PF4 from rats did not induce neutrophil activation. Mutants of human PF4 generated based on the primary sequence of rats revealed 2 notable findings. First, mutation of Pro-Thr-Ala (37-39) augmented PF4 binding to neutrophils while it decreased the binding of 197.3 to these cells. ELISA studies also show that addition of heparin enhances 197.3-PF4 interactions (Figure S1). In this regard, Poncz and coworkers (Li et al33 ) suggest that HIT patient IgG recognize at least 1 of 2 distinct antigenic sites on PF4. Site 1 consists of amino acids DLQ7-9 and P34, whereas site 2 includes P37. A mouse mAb described by these investigators called KKO mimics HIT antibody since it binds proline 37. In this context, our observation that 197.3 binding depends on the Pro-Thr-Ala (37-39) residues suggests that the binding epitope of this reagent is either close to or overlaps with HIT patient Ab and the KKO-binding site. Second, mutations at arginine 49 completely inhibited mutant binding to neutrophil, suggesting that this is a critical residue that may participate in chondroitin-neutrophil recognition. This residue may also be a key in distinguishing between the activities of human and rat PF4.
Neutrophil activation and adhesion
Costimulation resulted in neutrophil adhesion that was blocked by anti-CD32a and heparin. Blocking CD18 was only partially effective. This is unlike our previous observations where neutrophil adhesion stimulated by either formyl peptide, IL-8 (interleukin-8, CXCL8), or NAP-2 could be completely blocked by anti-CD18 mAb.23,34 The partial efficacy of CD18 blockers in this study may be due to the high electrostatic charge of PF4 and/or the strength and multivalency of immune-complex interactions with CD32a on one cell and chondroitin sulfates on another. Such strong interactions may allow immune complexes to form bridges stabilizing neutrophil aggregates. In agreement with this, in our microscopy experiments, we observed instances where neutrophil doublets had immune complexes located between them. These complexes have the appearance of stabilizing cell doublets. Other subtle distinctions between previous cell-adhesion studies23,34 and the current observations with regard to the kinetics and extent of cell adhesion can be explained based on differences in the rates of CD18-integrin up-regulation and activation in the 2 systems. In this context, all integrins are simultaneously and maximally activated upon stimulation with chemokines, whereas immune complex–mediated CD18-integrin activation proceeds gradually over a period of minutes.
Complex features regulating leukocyte function
A potential role for immune complex–mediated neutrophil activation during HIT is suggested in this paper based on studies with isolated neutrophils (main text) and whole blood assays (Figure S5). This proposition is supported by previous reports that demonstrate that HIT patient sera have enhanced soluble levels of L-selectin,35 and that they promote neutrophil activation and neutrophil-platelet adhesion.17
More detailed studies are warranted in the future to examine the exact role of neutrophils, and to quantify their contribution to disease relative to monocytes and platelets. In this context, it is important to note that platelet degranulation results in the release of numerous other chemokines in addition to PF4. Examples include IL-8 and connective tissue activating peptide-III (CTAP-III), with the latter being the precursor of NAP-2 (CXCL7 variant).36,37 IL-8 and NAP-2 bind CXC receptors on neutrophils and are potent activators of CD18-integrin function. Further, recent evidence demonstrates that PF4 binds numerous chemokines and/or influences their functions, including IL-8,38 tumor necrosis factor-α (TNF-α),25 and RANTES.39 In the case of RANTES, PF4 forms heterotypic complexes with this chemokine, and such complexes promote monocyte arrest on the endothelium. In addition, activated platelets express high levels of P-selectin on their surface and they release microparticles that bind neutrophils with high affinity.40,41 P-selectin–mediated leukocyte-platelet adhesion17,42 and neutrophil activation43 can play an additional role in disease context. Further studies that measure the levels of neutrophil/leukocyte activation in HIT patients, soluble indicators of leukocyte activation including serum levels of soluble L-selectin (sL-selectin) in patients with thrombotic episodes, and detailed investigations of leukocyte composition in HIT-associated vascular thrombi may reveal the true contribution of leukocytes to this disease.
Overall, we present a novel mechanism of neutrophil activation that may be important in the context of thrombotic and inflammatory complications that are associated with HIT.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dr Wade Sigurdson for helping us acquire the confocal microscopy images. This work was supported by National Institutes of Health (NIH, Bethesda, MD) grants HL63014 and HL77258
National Institutes of Health
Authorship
Contribution: Z.X. and K.M.D. performed experiments; G.P.V. designed research, contributed vital reagents, and performed experiments; S.N. performed experiments, designed research, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sriram Neelamegham, 906 Furnas Hall, State University of New York, Buffalo, NY 14260; e-mail: neel@eng.buffalo.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal