Abstract
In 1986, Mosmann and Coffman identified 2 subsets of activated CD4 T cells, Th1 and Th2 cells, which differed from each other in their pattern of cytokine production and their functions. Our understanding of the importance of the distinct differentiated forms of CD4 T cells and of the mechanisms through which they achieve their differentiated state has greatly expanded over the past 2 decades. Today at least 4 distinct CD4 T-cell subsets have been shown to exist, Th1, Th2, Th17, and iTreg cells. Here we summarize much of what is known about the 4 subsets, including the history of their discovery, their unique cytokine products and related functions, their distinctive expression of cell surface receptors and their characteristic transcription factors, the regulation of their fate determination, and the consequences of their abnormal activation.
Introduction
CD4 T cells play a central role in immune protection. They do so through their capacity to help B cells make antibodies, to induce macrophages to develop enhanced microbicidal activity, to recruit neutrophils, eosinophils, and basophils to sites of infection and inflammation, and, through their production of cytokines and chemokines, to orchestrate the full panoply of immune responses. Beginning with the groundbreaking work of Mossman and Coffman in 19861 showing that long-term CD4 T-cell lines could be subdivided into 2 groups, those that made IFNγ as their signature cytokine and those that produced IL-4, it has been realized that CD4 T cells are not a unitary set of cells but represent a series of distinct cell populations with different functions
While some of these CD4 T-cell populations are actually distinct lineages of cells already distinguished from one another when they emerge from the thymus, such as “natural” regulatory T (nTreg) cells2,3 and natural killer T cells (NKT cells),4 several represent alternative patterns of differentiation of naive CD4 T cells. It is to the description of these cells, their functions, their patterns of differentiation, the sets of genes they express, and the consequences of abnormalities in them that this review is devoted.
Naive conventional CD4 T cells have open to them 4 (and possibly more) distinct fates that are determined by the pattern of signals they receive during their initial interaction with antigen. These 4 populations are Th1, Th2, Th17, and induced regulatory T (iTreg) cells. Mossman and Coffman recognized the Th1 and Th2 phenotypes among the set of long-term T-cell lines that they studied and the early history of this field was devoted to understanding these 2 cell populations, with Th1 cells being regarded as critical for immunity to intracellular microorganisms and Th2 cells for immunity to many extracellular pathogens, including helminths.5,6
Abnormal activation of Th1 cells was seen as the critical event in most organ-specific autoimmune diseases while Th2 cells were responsible for allergic inflammatory diseases and asthma. Th17 cells have been recognized much more recently but there is now a growing body of work indicating not only that these cells exist but that they play a critical function in protection against microbial challenges, particularly extracellular bacteria and fungi.7 Further, some of the autoimmune responses formally attributed to Th1 cells, such as experimental autoimmune encephalomyelitis (EAE), collagen induced arthritis (CIA), and some forms of inflammatory bowel disease (IBD), have now been shown to be mediated, at least in part, by Th17 cells. iTreg cells are also now well established as an inducible cell population that phenotypically resembles nTreg cells, although distinguishing the function of iTreg cells from that of nTreg cells and, particularly, the relative importance of the 2 Treg populations in humans and experimental animals has been difficult. In this review, we will deal with the function of Treg cells as a group except where we explicitly speak of iTreg cells. There are also other regulatory CD4 T cells including Th3 and TR1 cells. Th3 cells are transforming growth factor β (TGF-β)–producing cells induced by oral tolerance.8 Most of them are likely inducible regulatory T cells that express Foxp3.9 Whether or not there are TGFβ-producing Foxp3− CD4 T cells is unclear. TR1 cells are IL-10 producing cells.10 Because all the CD4 T-cell sets including Th1, Th2, Th17 as well as Treg cells are capable of producing IL-10 under certain circumstances,11-13 TR1 cells may not be a distinct lineage but rather may represent a certain state of each existing lineage. Finally, there may well be other sets of conventional CD4 T cells and even among the more conventional sets, important differences exist, such as the detailed pattern of cytokines that they produce.
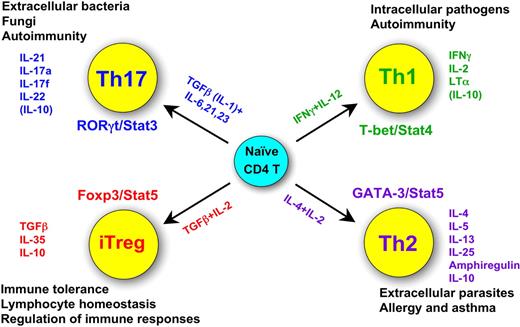
Figure 1 summarizes much of what we know about the major sets of CD4 T cells, including their unique products, the characteristic transcription factors and cytokines critical for their fate determination and some of their functions. Each of these topics will be discussed in some depth in the subsequent sections of this review.
Summary of the 4 CD4 T helper cell fates: their functions, their unique products, their characteristic transcription factors, and cytokines critical for their fate determination.
Summary of the 4 CD4 T helper cell fates: their functions, their unique products, their characteristic transcription factors, and cytokines critical for their fate determination.
A little history
Initially, immunologists believed that there were fundamentally 2 types of immune responses that require the action of CD4 T cells. One was antibody-mediated and the other cell-mediated. However, there was very little progress in this area until the early 1980s, when T-cell cloning technology was developed, many cytokines were discovered and cloned, and assays for them became available.
Tim Mosmman and Bob Coffman recognized that mature CD4 T cells could be subdivided into 2 distinct populations with different sets of products and that this would endow them with unique functions.1 Kim Bottomly was also working on this subject; she and her colleagues subdivided CD4 T-cell lines based on functional criteria, distinguishing inflammatory and helper CD4 T cells, with the latter being IL-4 producers.14
The translation of the differences observed in long-term CD4 T-cell lines to the behavior of normal CD4 T cells, first in vitro and then in vivo, constitutes the beginning of the Th field as a biologic subject. The earliest description of in vitro differentiation was reported in 1990 by our group and that of Susan Swain, demonstrating first that naive CD4 T cells failed to make IL-4 (or most other effector cytokines) and that these cells could be induced to develop into vigorous IL-4 producers if they were stimulated both with T-cell receptor ligands and IL-4, itself.15,16 Within 2 to 3 days after the initiation of culture, the stimulated cells acquire the capacity to produce IL-4. It was subsequently shown that this in vitro differentiation requires a signaling pathway that includes the IL-4 receptor, the signal transducer and activator of transcription (Stat) 6 and the DNA-binding factor GATA-3.17,18 As we will discuss later, this is far from the whole story, but “it gets us off to the races.” We note in passing that in our original 1990 paper, we found that IL-2 was also necessary for cells to acquire IL-4–producing capacity, although that was largely overlooked and didn't come back for serious analysis for more than a decade.19
Three years later, Ken Murphy, Anne O'Garra, and their colleagues showed that naive CD4 T cells could acquire the capacity to produce IFNγ in vitro.20 They stimulated T-cell receptor transgenic naive CD4 T cells and antigen-presenting cells with cognate antigen and heat-killed Listeria monocytogenes organisms; the heat-killed Listeria caused cells in the culture to produce IL-12, which was critical for Th1 differentiation in this system.
At first, it appeared that there was a fundamental dichotomy between the logic of differentiation process for Th1 and Th2 cells, with a CD4 T-cell endogenous product, IL-4, playing a major positive feedback role in Th2 differentiation and an exogenous product, IL-12, probably mainly from dendritic cells, playing the major inductive role for Th1 cells. However, with time and attention, the logic of the differentiation processes appears to be much closer than initially appreciated. Neutralizing IFNγ strikingly diminishes Th1 differentiation; IL-12 appears to induce some IFNγ production which then acts to up-regulate the key transcription factor T-bet21,22 and leads to much more IFNγ production, showing a positive feedback loop for Th1 cells as well.
Immunologists attributed many autoimmune diseases, including multiple sclerosis, rheumatoid arthritis, and their experimental models, to the action of Th1 cells. However, they were puzzled by the paradoxical finding that neutralizing or knocking out IL-12 and IFNγ had different effects on the induction of experimental autoimmune encephalomyelitis (EAE), a mouse model for multiple sclerosis. IL-12 p40 knockout mice are resistant to EAE induction whereas IFNγ knockout mice are more sensitive. The discovery of IL-23, which consisted of IL-12p40 paired with a distinctive chain (p19), led to a reassessment of the relative contributions of IL-12 and IL-23 in EAE induction.23 Indeed, it is IL-23, not IL-12, that plays the major role in inducing EAE. Due to the linkage between IL-23 and the expression of IL-17, a new Th lineage, Th17, was soon identified.24,25 Th17 cells are different from classical Th1/Th2 cells based on the following evidence: Th17 cells do not produce the “classical” Th1/Th2 cytokines; Th17 cells express low levels of T-bet and GATA-3; and the Th1/Th2 signature cytokines, IL-4 and IFNγ, suppress Th17 cell differentiation.24,25
In 2006, Stockinger, Weaver, Kuchroo, and their colleagues each showed that Th17 cells could be induced in vitro from naive mouse CD4 T cells by stimulation through their T-cell receptor (TCR) in the presence of IL-6 and TGF-β.26-28 RORγt was identified as the master regulator gene for Th17 cells.29 More work has revealed that the role of TGF-β in human cells may not be central to Th17 differentiation but that IL-1 has an important role.30,31 However, very recently, 3 groups independently reported that TGF-β was also critical for human Th17 cell differentiation.32-34 The discrepancy between these reports and previous studies may be explained by the potentially different purity of the naive T-cell population each group prepared because a small contamination with effector/memory cells may suppress de novo Th17 cell differentiation. In addition, in the earlier studies, the amount of TGF-β added to the culture and/or present in the serum is much higher than the amount required for Th17 differentiation and high levels of TGF-β inhibit Th17 cell differentiation and favor iTreg differentiation.
IL-21 produced by Th17 cells, induced in the course of Th17 differentiation,35-37 fulfills the role of the powerful positive feedback stimulant, reinforcing the Th17 induction process and showing that Th17 development has the logic similar to that of Th1 and Th2 cells.
The Treg “revolution” has been one of the defining themes of modern immunology but reaching an understanding of how these cells differentiate has been complex. In 1995, Sakaguchi and his colleagues discovered that regulatory T cells express CD25.38 Transfer of CD4 T cells that had been depleted of the CD25+ population into congenitally athymic mice induced autoimmune diseases while transfer of intact populations of CD4 T cells did not. In 2001, the autoimmune Scurfy mice and a human immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) patient were found to have mutations in Foxp3.39-41 In 2003, Foxp3 was reported as the master transcriptional regulator for nTreg cells.42,43
Weiner and colleagues had reported in 1994 that oral tolerance regimens induced TGF-β–producing CD4 T regulatory cells.8 This cell population was designated Th3 cells. In 2003, Chen et al reported that TGF-β can convert Foxp3− naive CD4 T cells into Foxp3+ CD4 T cells, that is iTreg cells.44 It is now clear that activated naive CD4 T cells stimulated by TGF-β in the absence of proinflammatory cytokines develop into iTreg cells. The positive feedback factor here is TGF-β itself, although there is still much uncertainty as to the relative biologic importance of nTreg and iTreg cells, particularly in humans.
Converting the Th paradigm from in vitro to in vivo situations initially met with much resistance but with time it became clear that memory and memory/effector T cells from normal priming events do display polarization in their cytokine-producing capacity, in their functions and in the range of cell surface molecules they express. Indeed, the recent description of the selective deficit in development of Th17 cells in patients with hyper-IgE syndrome (HIES or Job syndrome) strikingly validates this concept.45 HIES patients have a genetically determined inability to signal through Stat3, due to dominant negative mutations in the SH2 domain or the DNA-binding domain of this molecule.45-47 In humans and mice, the 3 major inducers and/or sustainers of Th17 differentiation, IL-6, IL-21 and IL-23, each use Stat3 for signal transduction. Indeed, the principal difficulties HIES patients face, recurrent staphylococcal and fungal infections, are precisely those observed in mice that cannot develop Th17 cells, strikingly validating the importance of the CD4 T-cell differentiation concept and indicating that lessons are learned, although not always perfectly, by studying experimental animals.
Th cells: cytokine produced and functions
Th cells play critical roles in orchestrating the adaptive immune responses. They exert such functions mainly through secreting cytokines and chemokines that activate and/or recruit target cells.
Th1 cells mediate immune responses against intracellular pathogens.5,6 In humans, they play a particularly important role in resistance to mycobacterial infections. Th1 cells are also responsible for the induction of some autoimmune diseases. Their principal cytokine products are IFNγ, lymphotoxin α (LTα), and IL-2. IFNγ produced by Th1 cells is important in activating macrophages to increase their microbicidal activity.48 LTα has been implicated as a marker for the disease progression in multiple sclerosis patients.49 LTα-deficient mice are resistant to EAE.50 IL-2 production is important for CD4 T-cell memory. IFNγ+IL-2+ cells are regarded as precursors of the Th1 memory cells.51 IL-2 stimulation of CD8 cells during their priming phase is critical for CD8 memory formation.52
Th2 cells mediate host defense against extracellular parasites including helminths.5,6 They are important in the induction and persistence of asthma and other allergic diseases. Th2 cells produce IL-4, IL-5, IL-9, IL-10, IL-13, IL-25, and amphiregulin. IL-4 is the positive feedback cytokine for Th2 cell differentiation15,16 and is the major mediator of IgE class switching in B cells.53 IgE binds to FcϵRI on basophils and mast cells and, when interacting with a multivalent ligand, cross-links FcϵRI, leading to the secretion of active mediators such as histamine and serotonin and to the production of several cytokines including IL-4, IL-13, and tumor necrosis factor α (TNF-α).
IL-5 plays a critical role in recruiting eosinophils.54 In addition to its effect on mast cells and lymphocytes, IL-9 induces mucin production in epithelial cells during allergic reactions.55 IL-10, produced by Th2 cells, suppresses Th1 cell proliferation.56 IL-10 can also suppress dendritic cell function.57 IL-13 is the effector cytokine in the expulsion of helminths and in the induction of airway hypersensitivity.58,59 Amphiregulin is a member of the epidermal growth factor (EGF) family. It induces epithelial cell proliferation. In the absence of amphiregulin, the expulsion of the nematode Trichuris muris is delayed.60 Amphiregulin may also be important for the induction of airway hypersensitivity.
IL-25 (also known as IL-17E) is also a Th2 cytokine.61,62 IL-25, signaling through IL-17RB, enhances the production of IL-4, IL-5, and IL-13 by a unique c-kit+FcϵRI− nonlymphocyte population.63 Interestingly, IL-25 is also produced by lung epithelial cells in response to allergens.55 Thus, IL-25 serves as an initiation factor as well as an amplification factor for Th2 responses. IL-25 can induce the production of chemokines including RANTES (CCL5) and eotaxin (CCL11) that recruit eosinophils.
Th17 cells mediate immune responses against extracellular bacteria and fungi.7 They are responsible for, or participate in, the induction of many organ-specific autoimmune diseases. Th17 cells produce IL-17a, IL-17f, IL-21, and IL-22. IL-17a was originally cloned as CTLA-8 and is homologous to a Herpesvirus saimiri gene. It was renamed IL-17 when its receptor was cloned.64 IL-17a and IL-17f are genetically linked and presumably under the control of the same locus control region (LCR). Thus, IL-17a and IL-17f are often coexpressed at the single cell level although there are also IL-17a- and IL-17f-single producing cells, suggesting the regulation of IL-17a and IL-17f expression in Th17 cells mirrors that of IL-4 and IL-13 in Th2 cells (see below). IL-17a and IL-17f both use the IL-17RA chain for their signaling, implying that they have similar functions, although IL-17a binds to IL-17RA with much higher affinity.65 IL-17a can induce many inflammatory cytokines, IL-6 as well as chemokines such as IL-8 (also known as CXCL8), and thus has an important role in inducing inflammatory responses.64 Both IL-17a and IL-17f recruit and activate neutrophils during immune responses against extracellular bacteria and fungi. IL-21 made by Th17 cells is a stimulatory factor for Th17 differentiation and serves as the positive feedback amplifier,35-37 as does IFNγ for Th1 and IL-4 for Th2 cells. IL-21 also acts on CD8 T cells, B cells, natural killer (NK) cells, and dendritic cells.66 IL-22 is produced by Th17 cells through IL-6– or IL-23–mediated Stat3 activation67 ; TGF-β inhibits IL-22 expression.13 The aryl hydrocarbon receptor (AHR), a receptor for dioxin, is highly expressed in Th17 cells and plays an important role in the expression of IL-22.68 IL-22 mediates IL-23–induced acanthosis and dermal inflammation.67 IL-22 also protects hepatocytes during acute liver inflammation.69 Strikingly, IL-22 mediates host defense against bacterial pathogens such as Klebsiella pneumoniae70 and Citrobacter rodentium.71 However, these functions may largely depend upon IL-23 stimulation of innate cells to produce IL-22 rather than on the action of Th17 cells.71
Treg cells play a critical role in maintaining self-tolerance as well as in regulating immune responses.2 Increasing Treg numbers and/or enhancing their suppressive function may be beneficial for treating autoimmune diseases and for preventing allograft rejection. Indeed, Treg cells stimulated in vitro with alloantigen prevent both acute and chronic allograft rejection in mice.72 On the other hand, depletion of Treg cells and/or inhibition of their function could enhance immunity against tumors and chronic infectious agents. Treg cells exert their suppressive functions through several mechanisms, some of which require cell-cell contact.3 The molecular basis of suppression in some cases is through their production of cytokines, including TGF-β, IL-10, and IL-35. TGF-β produced by Treg cells may also result in the induction of iTreg cells from naive CD4 T cells. Although TGF-β is not absolutely required for suppression in some settings, particularly in vitro, it is very important in mediating suppression in several circumstances in vivo.73,74 IL-10 production is critical for Treg-mediated prevention and cure of inflammatory bowel disease.75,76 Specific deletion of IL-10 in Treg cells by Foxp3-Cre results in the development of spontaneous colitis and enhanced lung inflammation.77 IL-10 also plays an important role in limiting the severity of EAE at later stages. During Leishmania infection, Treg IL-10 production in the lesion maintains a homeostasis between the host and the pathogen, allowing a low level of pathogen persistence and a consequent continued stimulation of protective immunity.78 IL-35, which consists of EBI3, a chain shared with IL-27, and IL-12 p35, is produced by Treg cells and contributes to suppressive activity.79
CD4 T cells other than Th2 and Treg cells can also produce IL-10. IL-10 production by Th1 or Th17 cells may play an important role in limiting their own effector function.11-13 IL-10, IL-27, and TGFβ can induce IL-10 production.10,13,80 Interestingly, Foxp3-deleted “Treg cells,” judged by expression of GFP encoded by a Foxp3null locus, produce high levels of IL-10, suggesting that IL-10 production in Treg cells is independent of Foxp3.81 The originally described TR1 cells (IL-10–producing regulatory T cells) may include many different types of cells that are capable of producing IL-10. Thus, IL-10 production by all CD4 T cells serves as a negative regulatory mechanism for limiting the immune responses to prevent host tissue damage.
Expression of cytokine and chemokine receptors by Th cells
Th1 cells
IL-12Rβ2 expression is induced by TCR activation and then maintained by IL-12 as well as by IFNγ stimulation.82-84 IL-12Rβ1 is constitutively expressed on naive CD4 T cells and its expression is further increased in Th1 cells through an IRF1-dependent mechanism.85 Up-regulation of the IL-12R complex conveys IL-12 hyperresponsiveness to activated cells. IL-18Rα is also up-regulated during Th1 differentiation. Although IL-18 is not involved in the differentiation of Th1 cells, it can synergize with IL-12 in inducing IFNγ, implying that IL-18 plays an important role in Th1 responses.86,87 Although chemokine receptor expression and differentiated Th phenotype are not strictly coordinate, some receptors, such as CXCR388,89 and CCR5,90 show a striking preferential expression on Th1 cells.
Th2 cells
IL-4Rα is up-regulated by IL-4 during Th2 differentiation. However, other γc cytokines may also induce IL-4Rα. CD25 (IL-2Rα) expression is higher in Th2 cells than in Th1 cells, possibly due to the action of c-Maf.91 Such higher expression of CD25 may confer hyperresponsiveness to IL-2. The most important cell surface marker for Th2 cells is T1/ST2 (IL-33Rα).92 T1/ST2, also known as IL-1R like 1, belongs to the IL-1R superfamily, which includes IL-1R and IL-18Rα. The function of IL-33Rα on Th2 cells may mirror the function of IL-18Rα on Th1 cells. Among the chemokine receptors, CCR3,93 CCR4,88,89 CCR8,94 and CRTh295 tend to be expressed on Th2 cells.
Th17 cells
Th17 cells express high levels of IL-23R.27,31,37 In addition, Th17 cells express substantial amounts of IL-1R1 and of IL-18Rα. The function of IL-18Rα on Th17 cells is unclear while IL-1R1 appears critical for IL-17 production; mice deficient in IL-1R1 are resistant to EAE, which is correlated with reduced IL-17 production.96 This is also consistent with a requirement for IL-1 in induction of human Th17 cells. Surprisingly, there has been little study of the expression of TGFβR on various Th cells. Among the chemokine receptors, human Th17 cells coexpress CCR6 and CCR4.97
Treg cells
The majority of the nTreg cells express CD25.2 Although all activated T cells express CD25, Treg cells express the highest levels of CD25 and do so constitutively, whereas expression by conventional CD4 T cells is transient and lower. The high level of expression of CD25, IL-2Rα, on Treg cells suggests the importance of IL-2 for these cells. Treg cells also express CTLA-4, GITR, and Folr4. However, these markers are only useful for distinguishing Treg cells from naive conventional CD4 T cells because each can be induced by activation of conventional T cells. Treg cells, especially in human, express little or no IL-7Rα. The absence of IL-7Rα in combination with high levels of CD25 provides an approach to identifying Treg cells and separating them from other cells.98 An interesting subset of Treg cells, those that express CD103,99 also known as alpha E integrin, is mainly found in the gut or at sites of inflammation. Most iTreg cells induced in vitro express CD103.
Transcription factors critical for each T helper lineage
Transcription factors including members of the nuclear factor of activated T cell (NFAT), NF-κB, and activator protein-1 (AP-1) families are critically involved in cytokine production upon TCR and/or cytokine stimulation. Presumably, those factors are also important during the process of T helper differentiation. However, they are not the factors directly determining T helper lineage fates and are usually expressed in all lineages. Below, we will focus on the transcription factors that either are specifically expressed, or function differently, in each of the lineages.
Transcription factors for Th1 differentiation
T-bet,21 the Th1 master regulator, is up-regulated during Th1 differentiation. Stat1, the major transducer of IFNγ signaling, plays a critical role in the IFNγ-mediated induction of T-bet.22 Overexpression of T-bet in Th2 cells induces them to produce IFNγ and inhibits their production of IL-4. T-bet−/− cells have severe defects in Th1 cell differentiation. T-bet−/− mice spontaneously develop asthma-like diseases.100
However, T-bet−/− Th1 cells still produce some IFNγ. Eomesodermin (Eomes),101 another T-box family member critical for IFNγ production in CD8 T cells, is up-regulated during Th1 differentiation, suggesting that it may also be involved in IFNγ production by CD4 T cells. Indeed, IL-21 treatment of Th1 cells partially inhibits IFNγ production, correlating with suppression of Eomes but not T-bet.102
Stat4, an IL-12 signal transducer, is important for amplifying Th1 responses.103,104 In addition, Stat4 can directly induce IFNγ-production in activated CD4 T cells, which can initiate the positive feedback loop in which IFNγ, acting through T-bet, induces more IFNγ. IL-12/Stat4, together with an NF-κB inducer, can cause IFNγ production independent of TCR stimulation. This is best illustrated by the capacity of IL-12 and IL-18, whose receptor is expressed on Th1, but not Th2, cells to induce IFNγ production by Th1 cells in a cyclosporine A–independent matter.86,87
Runx3,105,106 a transcriptional repressor important for silencing CD4 during CD8 T-cell development, is also up-regulated in Th1 cells. Overexpression of Runx3 in Th2 cells induces IFNγ production independent of T-bet (our unpublished data). Runx3-deficient cells produce less IFNγ than wild type Th1 cells.106
Hlx, a transcription factor induced by T-bet, interacts with T-bet and enhances T-bet-mediated IFNγ production.107
Transcription factors for Th2 differentiation
Stat6, activated by IL-4, is the major signal transducer in IL-4–mediated Th2 differentiation.108-110 Stat6-deficient cells fail to develop IL-4–producing capacity in vitro; in vivo, Th2 responses independent of Stat6 activation can be obtained.111-113 In vitro, Stat6 activation is necessary and sufficient for inducing high expression levels of the Th2 master regulator gene, GATA-3.114,115
Overexpression of GATA-3 in Th1 cells induces IL-4 production116 and in the absence of GATA-3, Th2 differentiation is totally abolished in vitro and in vivo.117,118 Even in fully differentiated Th2 cells, deleting GATA-3 completely blocks the subsequent production of IL-5 and IL-13,117 although it has only a modest effect on IL-4 production, consistent with the presence of GATA-3-binding sites in the promoters of IL-5 and IL-13 but not in the IL-4 promoter.
There are 2 Stat5 family members, Stat5a and Stat5b.119 They are important for cytokine-driven cell proliferation and cell survival. IL-2 potently stimulates Stat5 activation. Th2 cell differentiation requires strong Stat5 signaling.19,120 Thus, Stat5a single knockout cells have profound defects in Th2 cell differentiation both in vitro and in vivo despite the presence and activation of Stat5b. Stat5 has been shown to directly bind to DNase I hypersensitive sites (HSII and HSIII) in the second intron of the Il4 locus.120
c-Maf, which is selectively up-regulated in Th2 cells, also enhances IL-4 production but does not play a role in the production of other Th2 cytokines.121 IRF-4 expression is required for Th2 cell differentiation.122,123 IRF-4–deficient cells produce much less IL-4, but this defect can be rescued by overexpression of GATA-3, suggesting that IRF-4 up-regulates GATA-3.122
Transcription factors for Th17 differentiation
RORγt is important in Th17 cell differentiation.29 Overexpressing RORγt induces IL-17 production, whereas RORγt-deficient cells produce very little IL-17. Indeed, RORγt-deficient mice are partially resistant to EAE.
Another related nuclear receptor, RORα, is also up-regulated in Th17 cells.126 Although RORα deletion has minimal effect on IL-17 production, deficiency in both RORγt and RORα completely abolished IL-17 production.
Stat3, the major signal transducer for IL-6, IL-21 and IL-23, is indispensable for IL-17 production and deletion of Stat3 results in the loss of IL-17 producing cells.127-129 Stat3 is also responsible for the induction of IL-23R.
Interferon regulatory factor–4 (IRF4) has been recently reported to be critical for Th17 cell differentiation.130 IRF4−/− T cells fail to produce any IL-17. EAE cannot be induced in IRF4−/− mice. IRF4 appears to play a role in RORγt expression but not in Foxp3 induction.
Transcription factors for Treg differentiation
As noted above, most patients with IPEX and Scurfy mice have FOXP3/Foxp3 mutations, which result in loss of functional Treg cells. Overexpression of Foxp3 in conventional T cells converts them to a Treg phenotype and endows them with anergy and suppressive activity.42 TGF-β induces Foxp3 expression.44 Continuous expression of Foxp3 is critical for maintaining the suppressive activity of Treg cells.131 Diminishing the degree of Foxp3 expression may convert Treg cells to Th2 like cells, implying a close relationship of the Th2 and Treg lineages.132 Stat5 activation by IL-2, important for Th2 differentiation, is also required for Treg development.133 Stat5 may contribute to Foxp3 induction through binding to its promoter.134,135
T helper differentiation
Th1 cell differentiation
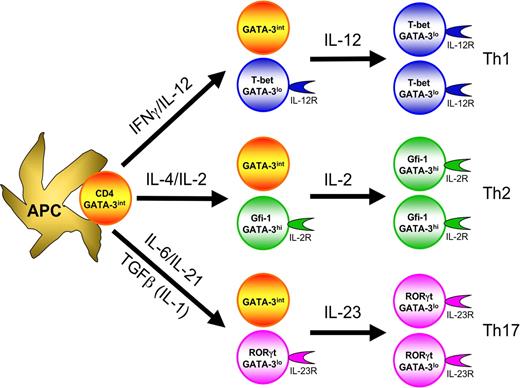
In the initiation of Th1 responses, antigen-presenting cells (APCs), particularly activated dendritic cells, stimulate naive CD4 T cells possessing cognate T-cell receptors. APCs that produce large amounts of IL-12 as a result of their activation136 (eg through either a combination of TLR3, TLR4, TLR7, TLR8, TLR9, and TLR11 stimulation or a single TLR activation in the presence of type I IFNs, IFNγ, or CD40L-mediated signaling) promote Th1 cell differentiation by acting on both NK cells and T cells. IL-12 activates NK cells to produce IFNγ, which in turn activates Stat1 in the responding CD4 T cells, up-regulating their T-bet expression. T-bet, in turn, induces T-cell IFNγ production and up-regulates IL-12Rβ2. Then, the IL-12Rβ2-expressing T cells, with high levels of T-bet, can be selected by IL-12, which is produced by APCs (Figure 2). IL-12, through activation of Stat4, induces IFNγ production and sustains expression of IL-12Rβ2. Thus, collaboration between IFNγ and IL-12 induces full Th1 differentiation.137
T-cell differentiation involves instructive differentiation as well as selective expansion of differentiated cells. The cytokines critical for the differentiation of each lineage instruct activated CD4 T cells to express their master transcription factors, T-bet for Th1, GATA-3 for Th2 and RORγt for Th17, as well as other lineage specific factors, IL-12R for Th1, Gfi-1 for Th2 and IL-23R for Th17. In many instances, only a portion of cells expresses the indicated transcription factors and adopts the differentiated phenotype. Such differentiated cells express the factors that determine responsiveness to particular cytokines, IL-12 for Th1, IL-2 for Th2 and IL-23 for Th17 cells, thus leading to selective expansion of those differentiated cells.
T-cell differentiation involves instructive differentiation as well as selective expansion of differentiated cells. The cytokines critical for the differentiation of each lineage instruct activated CD4 T cells to express their master transcription factors, T-bet for Th1, GATA-3 for Th2 and RORγt for Th17, as well as other lineage specific factors, IL-12R for Th1, Gfi-1 for Th2 and IL-23R for Th17. In many instances, only a portion of cells expresses the indicated transcription factors and adopts the differentiated phenotype. Such differentiated cells express the factors that determine responsiveness to particular cytokines, IL-12 for Th1, IL-2 for Th2 and IL-23 for Th17 cells, thus leading to selective expansion of those differentiated cells.
At later stages of Th1 differentiation, IL-18Rα is also up-regulated. IL-18Rα up-regulation requires IL-12/Stat4 signaling and is further increased by IFNγ. IL-12 and IL-18 jointly induce IFNγ production by Th1 cells in the absence of TCR stimulation. Such antigen-independent cytokine production is probably important for amplifying Th1 responses by recruiting other preexisting Th1 cells.
Th2 differentiation
Both IL-4 and IL-2 are required for Th2 differentiation (Figure 3) in vitro.15,19 IL-4 can be provided exogenously, in which case IL-4–mediated Stat6 activation induces GATA-3 expression. If exogenous IL-4 is not provided, naive CD4 T cells can produce limited amounts of IL-4, as a result of TCR-mediated Gata3 transcription and IL-2 mediated Stat5 activation.138 Such endogenous IL-4 production only occurs when cells receive low strength signals. The endogenous IL-4 then acts like exogenous IL-4 to up-regulate GATA-3 expression. GATA-3 has been reported to induce it own expression,139 probably when it has reached a threshold level. The IL-4/Stat6 pathway also induces expression of Gfi-1, a transcriptional repressor, which plays an important role in selecting GATA-3high cells to grow, providing a selective component in the Th2 development pathway124,125 (Figure 2). GATA-3 binds to regions of the Il4/Il13 loci including DNaseI hypersensitive site Va and CNS-1 sites (see “Epigenetic changes in Th differentiation”); however, GATA-3 alone is not sufficient to induce IL-4 production. IL-2–mediated activation of Stat5 plays a critical role in inducing/maintaining accessibility at the second intron HSII and HSIII DNase I hypersensitive sites of the Il4 locus.120 Indeed, Stat5 is bound to these 2 sites in Th2 but not Th1 cells. The collaboration of Stat5 and GATA-3 accounts for full Th2 differentiation in vitro.140
Th2 differentiation driven by low concentration of peptide stimulation in vitro consists of an IL-4–independent initiation phase and an IL-4–dependent amplification phase. (A) TCR stimulation by low concentration of peptide induces IL-4–independent GATA-3 expression and IL-2–mediated Stat5 activation. (B) GATA-3 binds to CNS-1 and VA whereas activated Stat5 binds to HSII and HSIII of Il4 locus. Both are critical for TCR-mediated IL-4 production at the initial phase of Th2 cell differentiation. (C) IL-4 produced by T cells can further induce GATA-3 expression through Stat6 activation. GATA-3 also regulates itself once it reaches a certain threshold. Thus, IL-4–mediated GATA-3 expression together with IL-2–mediated Stat5 activation drives full Th2 differentiation. (D) High levels of GATA-3 and activated Stat5 play critical roles in inducing large amount of IL-4 production.
Th2 differentiation driven by low concentration of peptide stimulation in vitro consists of an IL-4–independent initiation phase and an IL-4–dependent amplification phase. (A) TCR stimulation by low concentration of peptide induces IL-4–independent GATA-3 expression and IL-2–mediated Stat5 activation. (B) GATA-3 binds to CNS-1 and VA whereas activated Stat5 binds to HSII and HSIII of Il4 locus. Both are critical for TCR-mediated IL-4 production at the initial phase of Th2 cell differentiation. (C) IL-4 produced by T cells can further induce GATA-3 expression through Stat6 activation. GATA-3 also regulates itself once it reaches a certain threshold. Thus, IL-4–mediated GATA-3 expression together with IL-2–mediated Stat5 activation drives full Th2 differentiation. (D) High levels of GATA-3 and activated Stat5 play critical roles in inducing large amount of IL-4 production.
Accumulating in vivo studies indicate that IL-4 is not essential for Th2 differentiation in some settings, particularly for primary Th2 responses to Nippostrongylus brasiliensis and Schistosoma mansoni infection.111-113 The absence of IL-4 abolishes IgE switching in B cells in these infections, but Th2 cell differentiation is retained, at least partially. On the other hand, in vivo Th2 responses are completely dependent on GATA-3,117 suggesting that there is an IL-4–independent pathway for GATA-3 induction in vivo. It has been suggested that IL-4 can be induced by Notch signaling.141 However, Notch's role in IL-4–independent in vivo Th2 responses is still debatable. IL-4–independent Th2 responses in vivo may reflect hyperactivation of Stat5 by cytokines like IL-2, IL-7 or TSLP, because only limited amounts of GATA-3 are needed for Th2 differentiation when Stat5 is overexpressed.120 In fact, GATA-3 expression levels in in vivo–primed Th2 cells are substantially lower than those of in vitro–primed Th2 cells.
Th17 differentiation
TGFβ is critical for Th17 cell differentiation.26-28,32-34 TGFβ1-deficient mice are devoid of Th17 cells. More importantly, T cell– specific deletion of TGFβ1 blocks differentiation of Th17 cells during EAE induction and such mice are resistant to EAE.74 IL-6 is produced by the cells of the innate immune system that have been activated through TLR signaling. In the presence of IL-6, TGFβ induces Th17 differentiation,26-28 production of IL-21 and expression of IL-23R and RORγt. IL-21 can replace IL-6 in inducing RORγt and IL-17 expression.35-37 Thus, IL-21 could serve as an amplification cytokine for Th17 differentiation. The importance of IL-21 during in vivo Th17 differentiation in different models needs to be further studied. IL-23, initially proposed as the differentiation factor for Th17 cells, fails to induce Th17 differentiation from naive mouse CD4 T cells but is critical for Th17 cell survival and/or for maintaining their function (Figure 2). Therefore, Th17 cell differentiation consists of 3 stages: a differentiation stage, based on TGFβ and IL-6; an amplification stage, mediated by IL-21; and a stabilization stage due to IL-23. Importantly, all 3 cytokines, IL-6, IL-21, and IL-23, activate Stat3.
Treg cell differentiation
TGFβ also plays a major role in iTreg differentiation44 and is important for nTreg development.142 Deleting TGFβ from Treg cells results in diminished suppressive function and poor survival in vivo.74,143 In the absence of proinflammatory cytokines, TGFβ induces iTreg differentiation from naive mouse CD4 T cells.26 TGFβ activates Smad3 while TCR stimulation induces NFAT activation. Smad3 and NFAT collaborate in remodeling the Foxp3 enhancer region and promote Foxp3 expression.144 IL-2–mediated Stat5 activation is also required for the induction of Foxp3 expression.133,135,145 Both TGFβ and IL-2 are required for the survival and function of Treg cells even after they have differentiated.
Cross-regulation of T-helper differentiation
As described, Th differentiation involves positive feedback by cytokines. The differentiation process also actively involves cross-inhibition of other lineage fates. Mutual suppression between IFNγ and IL-4 signaling was the take-off point for studies of cross-regulation.5,6 TGFβ was also found to suppress both Th1 and Th2 differentiation,146 and both IL-4 and IFNγ inhibit Th17 differentiation.24,25
The cross-regulation of Th cell differentiation by cytokines may be partly explained by interaction of master genes. T-bet suppresses GATA-3 function by direct binding of the factors.147 Although it has not been studied carefully, such interactions may also be important for IL-4–mediated suppression of Th1 development. TGFβ induces RORγt expression in both Th17 and Treg cells, whereas Foxp3 is only found in Treg cells.148 Despite RORγt expression, Treg cells do not produce IL-17. The suppression of RORγt function in Treg cells is explained by the direct protein-protein binding between it and Foxp3. In addition, a low concentration of TGFβ can induce RORγt expression, whereas Foxp3 induction requires high concentrations of TGFβ. Thus, the amount of TGFβ as well as the presence or absence of proinflammatory cytokines determines the balance of RORγt and Foxp3 expression and thus whether the Th17 or the Treg fate is adopted. Besides direct interaction between lineage-specific transcription factors, competition for DNA binding has also been reported. Stat5 may compete with Stat3 for binding to the promoter of Il17, with the consequence that IL-17 production is suppressed.129
Another level of cross-regulation is through transcriptional regulation of critical factors. GATA-3 has been reported to down-regulate Stat4.149 Strong Stat5 activation inhibits T-bet expression.120 On the other hand, T-bet can suppress GATA-3 expression.84
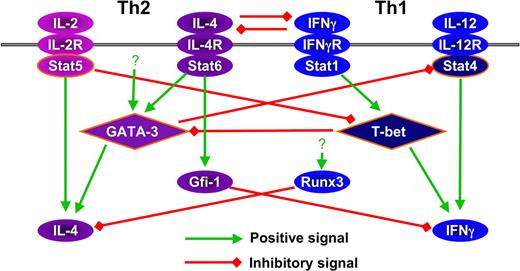
Finally, cross-regulation occurs at levels of cytokine transcription. Foxp3 suppresses IL-2 through its binding to NFAT150 as well as to Runx1.151 Runx3 inhibits IL-4 production through binding to the HSIV region of the Il4 locus.105 GATA-3 deficiency results in spontaneous IFNγ production, independent of IL-12 and IFNγ.117 Gfi-1, which acts to favor Th2 cell growth, suppresses both IFNγ125 and IL-17 production (our unpublished data). The factors expressed in Th17 cells that are responsible for suppressing cytokine production of other lineages are unknown. Interestingly, interchromosomal interaction occurs between Ifng and Il4 in naive T cells152 ; this may prove of importance in cross-regulation. The cross-regulation between Th1 and Th2 factors are shown in Figure 4.
Cross regulation among the factors that are involved in Th1 and Th2 differentiation.
Cross regulation among the factors that are involved in Th1 and Th2 differentiation.
Epigenetic changes in Th differentiation
As with all processes of differentiation, whole sets of genes are activated or repressed during the transition of naive CD4 T cells to Th1, Th2, Th17, and iTreg cells, and these differentiated states are associated with heritable changes in the conformation of key genes. Indeed, new technologies now being brought to bear will give a fuller assessment of the degree of genome-wide epigenetic modification than could previously be achieved. Zhao and his colleagues153 are pioneers in the analysis of genome-wide patterns of histone modification that are critical for regulation of gene expression in the 4 major types of Th cells.
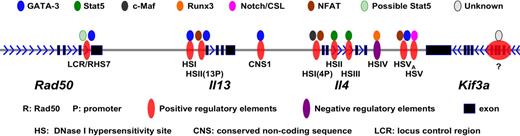
Much work has been done on how the accessibility of signature cytokine genes for each of the differentiated cell types is modified in the course of differentiation. Of these, most is known about Il4 and its congener Il13 and it is on these that we will concentrate (see Figure 5 for detailed regulatory elements and their binding to transcription factors). The Il4 and Il13 genes are closely linked on human chromosome 5q31 and the syntenic region on mouse chromosome 11 as part of a larger genetic assemblage that includes Il3, Csf2, Irf1, Il5, Rad50, and Kif3a.
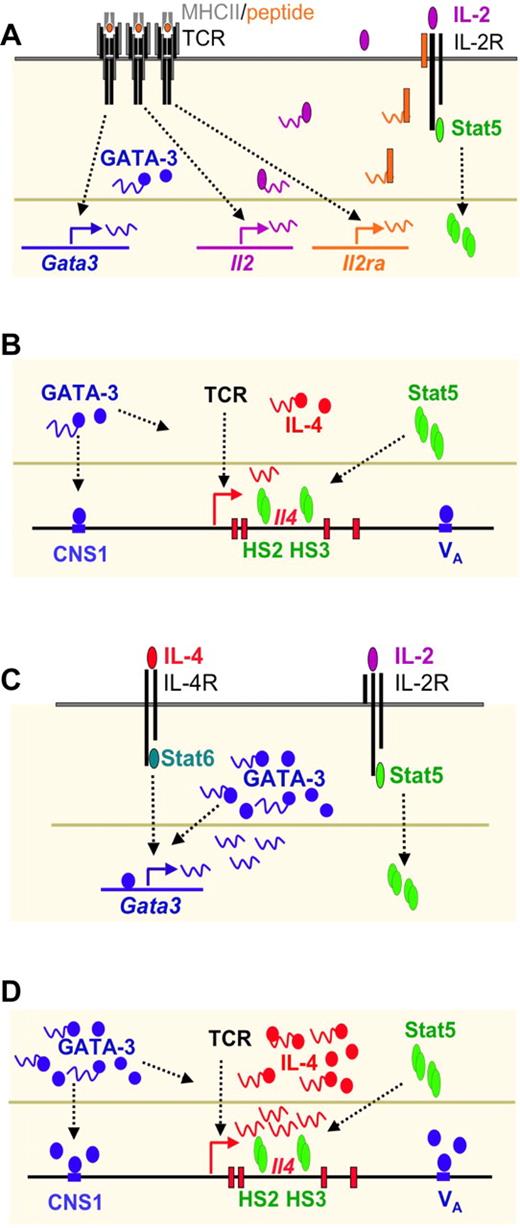
Positive and negative regulatory elements within Il4/Il13 loci and their binding to transcription factors.
Positive and negative regulatory elements within Il4/Il13 loci and their binding to transcription factors.
An LCR for Il4-Il13 has been identified that lies in a 25 Kb region at the 3′ end of Rad50, approximately 20 Kb and 40 Kb 5′ of Il13 and Il4, respectively.154 The LCR was defined by using a bacterial artificial chromosome (BAC) containing Il4 and Il13 and showing that transgenic mice expressing this BAC displayed copy number–dependent, position-independent expression of the cytokine genes. By carrying out a set of deletions, Flavell and his colleagues showed that the region in Rad50 described above contained the LCR. This Th2 LCR is both necessary and sufficient for locus control activity directed toward the neighboring Il4 and Il13 genes. In cells such as fibroblasts, which do not transcribe Th2 cytokines, the Il4, Il13, and Il5 genes form a minimal core interacting structure. In naive T cells as well as in Th1 and Th2 cells, the LCR is recruited into this interacting structure. In contrast to naive and Th1 cells, one particular site within the LCR (RHS7)155 becomes DNase I hypersensitive and is demethylated154,156 within 48 hours of the initiation of Th2 differentiation. It is known that deleting this portion of the LCR diminishes but does not abolish IL-4 production in Th2 cells. Precisely how the LCR regulates the accessibility and transcription of Il4 and Il13 is not certain. Although GATA-3 binds to RHS7, it is not sufficient to induce the activation of LCR. The demethylation of RHS7 during Th2 differentiation seems to be partially dependent on IL-2/Stat5 signaling.156 It is possible that GATA-3 and Stat5 besides directly regulating Il4 gene also collaborate in regulating the LCR.
Analysis of Il4 in Th1 and Th2 cells revealed a series of notable differences in DNase I hypersensitivity. Among a series of sites, a set within an Il4-Il13 intergenic region (conserved noncoding sequence 1 or CNS1),157,158 2 in the second intron of Il4159 and 2 3′ of the Il4 coding region (HSV and HSVa, associated with CNS2)160 appear particularly important. The CNS-1 and HSVa regions were shown by chromatin immunoprecipitation studies to have bound GATA-3 in Th2 but not Th1 cells and 2 DNase I hypersensitivity sites (those within the Il4 second intron, designated HSII and HSIII) to have bound Stat5a. It has been shown that overexpressing either GATA-3 or constitutively active Stat5a in cells stimulated under Th1-inducing conditions allows the cells to produce IL-4.120 The Stat5a effect does not occur in cells that are genetically deficient in GATA-3117 and anti–IL-2 diminishes the capacity of GATA-3 overexpression to allow IL-4 production.19 Thus, it would appear that GATA-3 and Stat5, the former induced by TCR and/or IL-4/Stat6 stimulation and the latter by IL-2, bind to sequences in the Il4 locus and lead to accessibility, as measured either by patterns of histone modification or restriction enzyme accessibility.
In addition to genetic regions that enhance IL-4 expression, there is a region in the 3′ portion of Il4, HSIV, acted on by Runx3, that represses IL-4 transcription.105 Runx3 is expressed at substantially higher levels in Th1 than Th2 cells.106 This is one of several examples of cross-inhibition between the differentiated Th cells as discussed above.
Much still remains to be established as to how the distinctive patterns of gene accessibility are initially induced and how they are maintained but the detailed analysis of the Il4 region and the ease of achieving alternative patterns of Th differentiation indicate that Il4 and the other key cytokine genes can provide insight into mechanisms of gene regulation in immune cells.
One very striking property of some of the cytokine genes, most notably Il4 and Il13, is that they are often expressed monoallelically. This monoallelic expression can be explained by probabilistic determination of transcription such that each Il4 (or Il13) allele has a given probability of expression in Th2 cells that is determined by its pattern of gene accessibility.161 Because these probabilities are often relatively low, many (but not all) cells express only one of the 2 alleles during any one stimulation period. We have suggested that probabilistic regulation of transcription may provide a selective advantage because of the biology of cytokine-producing cells and the functions they mediate. A particular example is IL-4's control of immunoglobulin class switching to IgE. Switching requires a direct interaction between antigen-specific T cells and B cells, with the formation of an immunologic synapse. IL-4 mainly acts across short distances so the IL-4–producing T cells can only stimulate their interacting B cells to switch. We argue that regulating the proportion of Th2 cells that make IL-4 through probabilistic transcription (with monoallelism as the consequence) would provide finer control over the switching process than trying to regulate the amount of IL-4 each CD4 T cell makes.
Immunologic abnormalities resulting from mutations or polymorphisms in the pathways of Th differentiation
One of the most telling pieces of evidence regarding the importance of the various differentiated cell types is the consequence of their absence or abnormalities in their development in humans. We presented in “A little history” the consequences of dominant negative mutations in STAT3, which were the failure of human CD4 T cells to develop into Th17 cells.45 This failure can explain a principal abnormality suffered by individuals with HIES, susceptibility to staphylococcal and fungal infections. This established both the key role of “Stat3 users” in human Th17 differentiation and the central role of Th17 cells in protection against certain types of infections.
A second striking example of a human mutation causing an impact on one of the key T-cell subsets is the effect of disabling mutations in FOXP3,41 which lead to the human IPEX syndrome. IPEX is the acronym for immunodysregulation, polyendocrinopathy, and enteropathy, X-linked. The key elements of IPEX are the appearance early in life of intractable diarrhea, eczema, hemolytic anemia, diabetes mellitus, or thyroid autoimmunity. In the initial description, there were exaggerated responses to viral infections. Remarkably, affected infants often display type I diabetes within the first days after birth. This constellation of events appears to be accounted for by the inability of affected individuals to develop nTreg or iTreg cells. The mouse genetic equivalent, the Scurfy mouse, also demonstrates a serious autoimmune disease resulting in death between 16 and 25 days of age. The immunopathology of Scurfy mice has a substantial Th2 component. Chatila and colleagues have proposed designating the human disorder X-linked autoimmunity-allergic dysregulation syndrome (XLAAD) because of a Th2 bias in the response of affected humans.162 Here again, the impact of the human mutation illustrates the critical role Treg cells play in controlling autoimmune/immunopathologic responses by conventional T cells and validates the importance of Foxp3 in the induction and/or function of these cells. It further argues that in the absence of Treg cells there is a greater likelihood of Th2 differentiation. Interestingly, mutations in IL2RA (encoding CD25, IL-2Rα), which is constitutively expressed on most Treg cells, results in an IPEX-like syndrome.163
Individuals with haploinsufficiency of GATA3 develop the hypoparathyroidism, sensorineural deafness, and renal dysplasia (HDR) syndrome.164 An analysis of these patients revealed that their levels of Th2 cells and the capacity of their naive CD4 T cells to develop into Th2 cells in vitro is diminished as was their serum concentration of IgG4, switching to which is dependent upon IL-4.165 Pykäläinen and colleagues have reported that polymorphisms in GATA3 in Finnish populations are associated with elevated IgE levels and greater susceptibility to asthma.166 Polymorphisms have also been shown to exist in TBX21 (the gene that encodes T-bet); some are associated with enhanced incidence of asthma and airway hyperresponsiveness.167 The former results imply that hyperactivity of GATA-3 favors Th2 differentiation and the latter that diminished activity of T-bet relieves the restraint on Th2 differentiation normally exerted by T-bet or other proteins in the Th1 differentiation pathway.
A mutation from glutamine to arginine at position 576 in the cytoplasmic domain of the IL-4Rα is common among the patients with elevated IgE and severe atopic dermatitis.168 However, this single mutation by itself does not affect IL-4–mediated CD23 induction.169 Another IL-4Rα variant Ile50Val is also associated with atopic asthma and has a dominant effect on Stat6 activation and IgE production.170,171 Mutations in IL12RB1 (the gene that encodes IL-12Rβ1) and IL12B (encoding IL-12 p40) are associated with increased susceptibility to mycobacterial and salmonella infection172,173 and, in one instance, to infection with Nocardia.174 IL-12 and IL-23 both use p40 as a constituent and their receptors both use IL-12Rβ1. Because IL-12 plays an important role in inducing Th1 differentiation and IL-23 is important in sustaining the Th17 phenotype, such mutations could diminish levels of either or both Th1 and Th17 cells. Mutations in IFNG or IFNGR1 in humans are associated with increased susceptibility to intracellular infections.175-177 This suggests that the major abnormality in individuals with mutations in IL12RB1 or IL12B is in the development of Th1 cells rather than Th17 cells. Furthermore, IL23R mutation is associated with inflammatory bowel diseases including Crohn disease.178
Minegishi and colleagues have reported an unusual form of HIES that is associated with mutations in TYK2, encoding Tyk2, a member of the Jak family of protein tyrosine kinases.179 Tyk2 plays a role in signaling by type I IFN, IL-6, IL-10, IL-12, and IL-23. While the cellular defects in this individual are not completely clear, the results are consistent with diminished development of Th1 and Th17 cells and enhanced development of Th2 cells.
Closing remarks
CD4 T cells represent a remarkable cell population. They are central to protection against a wide range of pathogens and do so through the adoption of a series of distinct differentiated states, each evolved under the pressure of a particular set of pathogens. The process through which the naive cells differentiate into these distinct states shows several similar features. TCR engagement is essential. A major product of the differentiated cells is a principal stimulant, providing a potent positive feedback that can enforce the development of a high degree of polarization. The Jak/Stat pathways and a specific Stat in association with one of 4 master regulators, T-bet, GATA-3, RORγt, and Foxp3, are essential for the differentiation process. In a real sense, the study of this process has illuminated how central cytokines are to the mounting of effective immune responses and, through the commonalties in their pathway of differentiation, support the assertion that cytokine biology is more than a collection of isolated facts but rather involves a set of principles in which knowledge about any of the pathways points the way to a deeper understanding of the others. The analysis of the effects of mutations in key players in the differentiation process has also provided a much deeper understanding of the true biologic function of this set of cells that are so central to the mounting of effective and regulated immune responses.
Acknowledgments
We thank Dr Hidehiro Yamane for insightful discussions in preparation of this review. We apologize to those authors whose related work is not appropriately cited because of the limitations of the space and/or our knowledge.
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health, under project Z01 AI000493-22-LI.
National Institutes of Health
Authorship
Contribution: J.Z. and W.E.P. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jinfang Zhu or William E. Paul, Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, 9000 Rockville Pike, Building 10, Room 11N311, Bethesda, MD 20892; e-mail: jfzhu@niaid.nih.gov or wpaul@niaid.nih.gov.