Abstract
The chemokine receptor CCR5 is implicated in the pathogenesis of various inflammatory diseases, such as multiple sclerosis (MS), atherosclerosis, transplant rejection, and autoimmunity. In previous studies, we have shown that MS lesions are characterized by enhanced expression of transcription factors associated with stress responses, ie, IRF-1, NF-κB, and CREB-1, which modulate expression of both classes of major histocompatibility complex (MHC) molecules. The expression of MHC-I and MHC-II molecules greatly overlaps with the expression of CCR5 in MS lesions. Therefore, we investigated whether these factors are also involved in the transcriptional regulation of CCR5. Using in vitro assays, we determined that neither IRF-1 nor NF-κB is involved in the activation of the CCR5 promoter. This is corroborated by the finding that these factors are not involved in the induction of endogenous CCR5 transcription in various cell types. In contrast, we show that CCR5 expression is regulated by the cAMP/CREB pathway and that interference in this pathway affects endogenous CCR5 transcription. From this, we conclude that the cAMP/CREB pathway is involved in the regulation of CCR5 transcription and that, given the ubiquitous nature of CREB-1 protein expression, additional regulatory mechanisms must contribute to cell type-specific expression of CCR5.
Introduction
Chemokines are a family of cytokines that mediate the migration of leukocytes toward sites of inflammation on activation through specific chemokine receptors on their cell surface. The CC chemokine receptor 5 (CCR5) regulates trafficking and effector functions of memory/effector T lymphocytes, macrophages, and immature dendritic cells (DCs). Interactions between chemokines and chemokine receptors are promiscuous, ie, most chemokines activate more than one receptor and most chemokine receptors can bind several chemokines.1 It is well established that CCR5 binds the chemokines CCL5 (RANTES), CCL3 (MIP-1α), and CCL4 (MIP-1β) and regulates differentiation and anatomic distribution of leukocytes to meet with local requirements for an adequate immune response against pathogens. Because of its important immune regulatory role, CCR5 is also implicated in the pathogenesis of various inflammatory diseases, such as atherosclerosis, transplant rejection and autoimmunity, and neurodegenerative diseases.2-6 In addition, CCR5 also serves as a coreceptor for viral entry of HIV-1.7,8 CCR5 is mainly expressed on a subset of T lymphocytes, monocytes, macrophages, DCs, and microglia
Multiple sclerosis (MS) is a demyelinating disease with inflammatory aspects that are mediated by infiltrating leukocytes and resident cells, resulting in extensive inflammation and demyelination of the central nervous system (CNS).9 Enhanced expression of CCR5 has been noted in MS-affected CNS tissue compared with normal-appearing white matter of patients and with control brain tissue of non-MS patients.10,11 In particular, increased expression of CCR5 has been detected on reactive microglia near the edges of active demyelinating MS lesions and on phagocytic (foamy) macrophages present inside these lesions.10,11 In addition, expression of CCR5 has been found on reactive astrocytes in several MS cases.10
In earlier studies, we have found that expression of major histocompatibility complex (MHC)-II and MHC-I is enhanced in MS lesions.12 This enhanced expression is mainly found on activated microglia and foamy macrophages. We have shown that this enhanced expression is the result of a concomitant increase in the expression of transcription factors controlling expression of both classes of MHC genes. These include MHC-specific transcription factors and general factors, such as IRF-1, NF-κB, and CREB-1.12,13 The fact that the latter group can be activated by a variety of stresses, including viral and bacterial infection, inflammation, and tissue damage,14-16 suggests that a general state of cell activation is present in MS lesions.
Because the expression of CCR5 on CNS resident cell resembles to a great extent the expression of both classes of MHC molecules found in MS lesions, we investigated whether IRF-1, NF-κB, and CREB-1 are also involved in the regulation of CCR5 transcription in various cell types.
Using established human monocytic and glioma cell lines, cultured monocyte-derived DCs, primary astrocytes and microglia, and primary T lymphocytes, we determined that neither IRF-1 nor NF-κB is involved in the activation of CCR5 promoters or in the induction of endogenous CCR5 transcription. In contrast, we found that the CREB pathway regulates the activity of the CCR5 promoter. Considering the ubiquitous nature of CREB-1, these findings suggest that additional genetic or epigenetic mechanisms contribute to the cell-type specific transcriptional regulation of CCR5.
Methods
Cell culture
The cell lines U251 (VUMC, Amsterdam, The Netherlands), THP-1 and Tera-2 (ATCC, Manassas, VA) were cultured in Iscove modified Dulbecco medium (Lonza Verviers, Verviers, Belgium) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum (FCS; Greiner, Alphen a/d Rijn, The Netherlands), 100 IU/mL streptomycin, and 100 IU/mL penicillin.
Transcription factor binding site search
Potential transcription factor binding sites were identified using the TFSEARCH program (http://www.cbrc.jp/research/db/TFSEARCH.html), which searches the TRANSFAC database.17,18 Cutoff was set at 85% of the consensus TF binding site.
Preparation of nuclear extracts and electrophoretic mobility shift assay
Nuclear extracts were prepared, and electrophoretic mobility shift assays (EMSAs) were performed as previously described.19 Briefly, 2 μL nuclear extracts was incubated with 2 ng [33P]-labeled double strand (ds-) DNA probe for 30 minutes on ice. Samples were run on a 6% polyacrylamide gel in 0.25× Tris-borate/ethylenediaminetetraacetic acid buffer at 200 V for 150 minutes. The ds-oligonucleotides used as probe are depicted in Table 1.
ds-oligonucleotide sequences used for EMSA
| EMSA probe . | Oligonucleotide sequence* . | Location† . |
|---|---|---|
| ISRE β2m21 | 5′-TAAGAAAAGGAAACTGAAAACG-3′ | |
| CCR5-ISRE-1 | 5′-CTCCGCATGGTGAAAGTAAGAACC-3′ | −4026 |
| CCR5-ISRE-2 | 5′-GCAATTAGCTTTACCTTTTCAGCTTCT-3′ | −3367 |
| CCR5-ISRE-3 | 5′-GGACTGCTGAAAGAGTAACTAAGAGTT-3′ | −3255 |
| κB β2m21 | 5′-ACGGGAAAGTCCCTC-3′ | |
| CCR5-κB-1 | 5′-GAACAGAGTGAAAATCCCCACTAAGA-3′ | −2650 |
| CCR5-κB-2 | 5′-CTTACTGTTGAAAAGCCCTGTGATCT-3′ | −2340 |
| CCR5-κB-3 | 5′-ATCCAGTGAGAAAAGCCCGTAAATAA-3′ | −2155 |
| CRE consensus27 | 5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′ | |
| CCR5-CRE-1 | 5′-AACACAAAAGTGGAGTAACGCACA-3′ | −4369 |
| CCR5-CRE-2 | 5′-CAGGTCTAGCACGTCATTTAACAG-3′ | −4226 |
| CCR5-CRE-3 | 5′-TATCTTGCCGAGGTCACAAAGCAA-3′ | −4172 |
| CCR5-CRE-B1 | 5′-GATTGGGGGCACGTAATTTTGCTG-3′ | −3028 |
| CCR5-CRE-B2 | 5′-AGCCAAGGTCACGGAAGCCCAGAG-3′ | −2950 |
| CCR5-CRE-B3 | 5′-AGATTTTCAGATGTCACCAACCGC-3′ | −2866 |
| CCR5-CRE-B4 | 5′-CCATATACTTATGTCATGTGGAAA-3′ | −2788 |
| CCR5-CRE-B5 | 5′-GGTTAATGTGAAGTCCAGGATCCC-3′ | −2445 |
| CCR5-CRE-B6 | 5′-TGGGCTTTTGACTAGATGAATGTA-3′ | −2267 |
| CCR5-CRE-B7 | 5′-TAGTGGGATGAGCAGAGAACAAAA-3′ | −2187 |
| CCR5-CRE-B8 | 5′-GCTTATTTTAAGCTCAACTTAAAA-3′ | −2102 |
| CCR5-CRE-B9 | 5′-TCTAGCTCTGATATCCTTTATTCT-3′ | −2235 |
| CCR5-CRE-B10 | 5′-CGTAAATAAACCTTCAGACCAGAG-3′ | −2138 |
| CCR5-CRE-B11 | 5′-ATTCTTTTCGCCTTCAATACACTT-3′ | −2061 |
| CCR5-CRE-B12 | 5′-ACTCCACCCTCCTTCAAAAGAAAC-3′ | −2028 |
| CCR5-CRE-B13 | 5′-TGATTTGCACAGCTCATCTGGCCA-3′ | −1972 |
| EMSA probe . | Oligonucleotide sequence* . | Location† . |
|---|---|---|
| ISRE β2m21 | 5′-TAAGAAAAGGAAACTGAAAACG-3′ | |
| CCR5-ISRE-1 | 5′-CTCCGCATGGTGAAAGTAAGAACC-3′ | −4026 |
| CCR5-ISRE-2 | 5′-GCAATTAGCTTTACCTTTTCAGCTTCT-3′ | −3367 |
| CCR5-ISRE-3 | 5′-GGACTGCTGAAAGAGTAACTAAGAGTT-3′ | −3255 |
| κB β2m21 | 5′-ACGGGAAAGTCCCTC-3′ | |
| CCR5-κB-1 | 5′-GAACAGAGTGAAAATCCCCACTAAGA-3′ | −2650 |
| CCR5-κB-2 | 5′-CTTACTGTTGAAAAGCCCTGTGATCT-3′ | −2340 |
| CCR5-κB-3 | 5′-ATCCAGTGAGAAAAGCCCGTAAATAA-3′ | −2155 |
| CRE consensus27 | 5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′ | |
| CCR5-CRE-1 | 5′-AACACAAAAGTGGAGTAACGCACA-3′ | −4369 |
| CCR5-CRE-2 | 5′-CAGGTCTAGCACGTCATTTAACAG-3′ | −4226 |
| CCR5-CRE-3 | 5′-TATCTTGCCGAGGTCACAAAGCAA-3′ | −4172 |
| CCR5-CRE-B1 | 5′-GATTGGGGGCACGTAATTTTGCTG-3′ | −3028 |
| CCR5-CRE-B2 | 5′-AGCCAAGGTCACGGAAGCCCAGAG-3′ | −2950 |
| CCR5-CRE-B3 | 5′-AGATTTTCAGATGTCACCAACCGC-3′ | −2866 |
| CCR5-CRE-B4 | 5′-CCATATACTTATGTCATGTGGAAA-3′ | −2788 |
| CCR5-CRE-B5 | 5′-GGTTAATGTGAAGTCCAGGATCCC-3′ | −2445 |
| CCR5-CRE-B6 | 5′-TGGGCTTTTGACTAGATGAATGTA-3′ | −2267 |
| CCR5-CRE-B7 | 5′-TAGTGGGATGAGCAGAGAACAAAA-3′ | −2187 |
| CCR5-CRE-B8 | 5′-GCTTATTTTAAGCTCAACTTAAAA-3′ | −2102 |
| CCR5-CRE-B9 | 5′-TCTAGCTCTGATATCCTTTATTCT-3′ | −2235 |
| CCR5-CRE-B10 | 5′-CGTAAATAAACCTTCAGACCAGAG-3′ | −2138 |
| CCR5-CRE-B11 | 5′-ATTCTTTTCGCCTTCAATACACTT-3′ | −2061 |
| CCR5-CRE-B12 | 5′-ACTCCACCCTCCTTCAAAAGAAAC-3′ | −2028 |
| CCR5-CRE-B13 | 5′-TGATTTGCACAGCTCATCTGGCCA-3′ | −1972 |
Potential protein binding sites are underlined.
For competition assays, nuclear extracts were incubated with an unlabeled consensus ds-oligonucleotide (Table 1) for 30 minutes before incubation with the labeled ds-oligonucleotides. For supershift assays, after oligonucleotide/nuclear extract incubation, 1 μg antibody was added and incubation was continued for an additional 60 minutes on ice. The antibodies used (all Santa Cruz Biotechnology, Santa Cruz, CA) are depicted in Table 2.
Antibodies used for EMSA supershift and ChIP assays
| Antibody name . | Protein reactivity . | Catalog no.* . | Application† . |
|---|---|---|---|
| IRF-1 (C-20) | IRF-1 | sc-497 | E/C |
| IRF-2 (C-19) | IRF-2 | sc-498 | E |
| IRF-4 (M-17) | IRF-4 | sc-6059 | E |
| ICSBP (C-19) | IRF-8 | sc-6058 | E |
| NF-κB p50 (NLS) | NF-κB p50 | sc-114 | E |
| NF-κB p65 (A) | NF-κB p65 (RelA) | sc-109 | E/C |
| RelB (C-19) | RelB p68 | sc-226 | E |
| cRel (C) | c-Rel p75 | sc-71 | E |
| CREB-1 (24H4B) | CREB-1 p43 | sc-271 | E |
| ATF-1 (C41–5.1) | ATF-1 p35 | sc-243 | E |
| ATF-1 (25C10G; ATF/CREB) | ATF-1, CREB-1, CREM-1 | sc-270 | E |
| CREB-1 | CREB-1 | Boss | C |
| acetyl-histone H3 | Acetylated Lys9 and 14 | up-06–599 | C |
| RNA Pol II (N-20) | RNA Polymerase II | sc-899 | C |
| Antibody name . | Protein reactivity . | Catalog no.* . | Application† . |
|---|---|---|---|
| IRF-1 (C-20) | IRF-1 | sc-497 | E/C |
| IRF-2 (C-19) | IRF-2 | sc-498 | E |
| IRF-4 (M-17) | IRF-4 | sc-6059 | E |
| ICSBP (C-19) | IRF-8 | sc-6058 | E |
| NF-κB p50 (NLS) | NF-κB p50 | sc-114 | E |
| NF-κB p65 (A) | NF-κB p65 (RelA) | sc-109 | E/C |
| RelB (C-19) | RelB p68 | sc-226 | E |
| cRel (C) | c-Rel p75 | sc-71 | E |
| CREB-1 (24H4B) | CREB-1 p43 | sc-271 | E |
| ATF-1 (C41–5.1) | ATF-1 p35 | sc-243 | E |
| ATF-1 (25C10G; ATF/CREB) | ATF-1, CREB-1, CREM-1 | sc-270 | E |
| CREB-1 | CREB-1 | Boss | C |
| acetyl-histone H3 | Acetylated Lys9 and 14 | up-06–599 | C |
| RNA Pol II (N-20) | RNA Polymerase II | sc-899 | C |
sc indicates from Santa Cruz Biotechnology; Boss, gift of Dr J. M. Boss, Emory University School of Medicine, Atlanta, GA; and up, from Upstate Biotechnology, Lake Placid, NY.
E indicates EMSA supershift; and C, ChIP.
Transient transfection
CCR5 promoter constructs were a kind gift from Prof S. K. Ahuja (University of Texas Health Science Center at San Antonio). Tera-2 cells were transfected with the CCR5 upstream and downstream promoter-luciferase reporter constructs shown in Figure 1 20 or the pGL3-Basic luciferase reporter plasmid (Promega, Madison, WI), in combination with the actin driven Renilla pGL3 reporter construct (pRL, Promega) as an internal control. pGL3-B250 and pGL3-β2m have been described previously.19,21
Cells were transfected in triplicate with 1 μg of promoter construct and 0.1 μg of actin-pRL construct, using the calcium phosphate coprecipitation method.22 For cytokine induction experiments, cells were treated with 500 U/mL interferon-γ (IFN-γ; Boehringer Ingelheim, Ingelheim, Germany) or 10 ng/mL tumor necrosis factor-α (TNF-α; BioSource International, Camarillo, CA) for 24 hours after transfection. For transcription factor induction experiments, cells were cotransfected with 0.5 μg of the previously described23 pRc/RSV expression vectors of IRF-1, CREB-1, CBP, p300, P/CAF, ATF-1, or ICER. Cells were harvested 48 hours after transfection, and luciferase activity was measured using the dual-luciferase reporter assay system (Promega).
Primary cell culture and stimulation
Peripheral blood mononuclear cells (PBMCs) were isolated from blood of normal healthy donors using a Ficoll gradient (Pharmacy, Leiden University Medical Center, Leiden, The Netherlands). To obtain a high number of T lymphocytes, PBMCs were propagated with 10 μg/mL phytohemagglutinin and irradiated allogeneic PBMCs (3000 cGy) in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% human serum, 10 U/mL interleukin-2, 100 U/mL streptomycin, 100 U/mL penicillin, and 2 mM l-glutamine. Activated T lymphocytes were treated with 20 μM forskolin (Calbiochem, San Diego, CA) for 6 hours, after which RNA was isolated and chromatin was prepared for chromatin immunoprecipitation (ChIP).
To obtain DCs, monocytes derived from freshly isolated PBMCs were cultured in RPMI 1640 medium with regular supplements and stimulated with a combination of 800 U/mL recombinant human granulocyte-macrophage colony-stimulating factor and 500 U/mL IL-4 (both BioSource International) for 7 days. Subsequently, the obtained cells were stimulated with 100 ng/mL lipopolysaccharide (LPS; Sigma-Aldrich, Steinheim, Germany) or 500 U/mL IFN-γ for 24 hours each, or 20 μM forskolin for 6 hours, after which RNA was isolated.
Human brain tissue was obtained by rapid autopsy according to standardized procedures under the management of the Netherlands Brain Bank (Amsterdam, The Netherlands). All patients or their next of kin had given written consent for autopsy and use of brain tissue for research purposes. Astrocytes and microglia were isolated and cultured as described previously.24 Astrocytes were stimulated with LPS (100 ng/mL) or IFN-γ (500 U/mL) each for 8 hours before RNA isolation. Microglia were stimulated with TNF-α (10 ng/mL) or IFN-γ (500 U/mL) each for 8 hours, followed by RNA isolation.
RNA isolation and reverse transcriptase polymerase chain reaction
Total RNA was isolated using the RNA-Bee extraction method (Tel-Test, Friendswood, TX). cDNA was synthesized from 4 μg of each RNA sample using avian myeloblastosis virus reverse transcriptase (Promega). CCR5, NF-κB p50, HLA-DRA, CIITA promoter IV isoform (CIITA-pIV), ICER, CREB-1, and GAPDH products were amplified by polymerase chain reaction (PCR) reaction. Primer sequences and conditions used are depicted in Table 3. PCR products were separated and visualized on an ethidium bromide-stained agarose gel and densitrometrically analyzed using Tina 2.09f software (Raytes Isotopenmeβgeräte, Straubentardt, Germany).
RT-PCR primer sequences and conditions
| PCR product . | Primer sequence (5′-3′) . | Tann . | No. of cycles . | MgCl2 concentration . |
|---|---|---|---|---|
| CCR5 | F-CTGAGACATCCGTTCCCCTA | 60°C | 30 | 3 mM |
| R-GCTCTTCAGCCTTTTGCAGT | ||||
| NF-κB p50 | F-GAACTCCTCCATTGTGGAACC | 62°C | 32 | 1.5 mM |
| R-CCCGGAGATTTGCTGTCATG | ||||
| HLA-DRA | F-GGCCATAAGTGGAGTCCC | 55°C | 30 | 3 mM |
| R-CTATACTCCGATCACCAA | ||||
| CIITA-PIV | F-AGCTGGCGGGAGGGAGAGGCCACC | 60°C | 35 | 1.75 mM |
| R-CATACTGGTCCAGTTCCGCGATATTGG | ||||
| ICER | F-CAGATCCGAGCTCCTACTGC | 60°C | 27 | 3 mM |
| R-CAACTCGGCTCTCCAGACAT | ||||
| CREB-1 | F-AACCAGCAGAGTGGAGATGCAGCT | 60°C | 30 | 4 mM |
| R-CTGTAGGAAGGCCTCCTTGAAAGA | ||||
| GAPDH | F-GGTCGGAGTCAACGGATTTG | 60°C | 22 | 1.5 mM |
| R-ATGAGCCCCAGCCTTCTCCAT |
| PCR product . | Primer sequence (5′-3′) . | Tann . | No. of cycles . | MgCl2 concentration . |
|---|---|---|---|---|
| CCR5 | F-CTGAGACATCCGTTCCCCTA | 60°C | 30 | 3 mM |
| R-GCTCTTCAGCCTTTTGCAGT | ||||
| NF-κB p50 | F-GAACTCCTCCATTGTGGAACC | 62°C | 32 | 1.5 mM |
| R-CCCGGAGATTTGCTGTCATG | ||||
| HLA-DRA | F-GGCCATAAGTGGAGTCCC | 55°C | 30 | 3 mM |
| R-CTATACTCCGATCACCAA | ||||
| CIITA-PIV | F-AGCTGGCGGGAGGGAGAGGCCACC | 60°C | 35 | 1.75 mM |
| R-CATACTGGTCCAGTTCCGCGATATTGG | ||||
| ICER | F-CAGATCCGAGCTCCTACTGC | 60°C | 27 | 3 mM |
| R-CAACTCGGCTCTCCAGACAT | ||||
| CREB-1 | F-AACCAGCAGAGTGGAGATGCAGCT | 60°C | 30 | 4 mM |
| R-CTGTAGGAAGGCCTCCTTGAAAGA | ||||
| GAPDH | F-GGTCGGAGTCAACGGATTTG | 60°C | 22 | 1.5 mM |
| R-ATGAGCCCCAGCCTTCTCCAT |
Tann indicates annealing temperature.
Chromatin immunoprecipitation
ChIP was perfomed as described earlier.25 Shortly after cross-linking DNA and DNA-binding proteins with formaldehyde, cells were harvested and lysed sequentially in cell lysis buffer and nuclear lysis buffer at a concentration of 5 × 106 cells/mL. After sonication into fragments with an average length of 0.5 to 3 kb using a Branson 250 sonifier (Boom, Meppel, The Netherlands), chromatin was precleared using preblocked protein A agarose beads (Upstate Biotechnology, Charlottesville, VA); 5 μg of antibody (Table 2) was added to 2 mL of precleared diluted sonicated chromatin (equivalent to 106 cells) and allowed to bind overnight, after which immune complexes were precipitated with 140 μL preblocked protein A agarose beads (25%). After extensive washing and elution from the beads, immune-precipitated chromatin complexes were de–cross-linked and proteins were digested. After phenol/chloroform extraction and precipitation, the obtained DNA was resuspended in 60 μL distilled H2O, of which 5 μL was used per 25 μL PCR reaction. Quantification of the immune-precipitated chromatin was performed on an iQ-Cycler system (Bio-Rad, Hercules, CA) using the iQ SYBR Green Supermix and the following primers: CCR5 ChIP sense: 5′-TGTGGGCTTTTGACTAGATGA-3′ and CCR5 ChIP antisense: 5′-TAGGGGAACGGATGTCTCAG-3′ (respectively, nt −2577 to −2555 and −1951 to −1932 from ORF), GAPDH ChIP sense: 5′-TACTAGCGGTTTTACGGGCG-3′ and GAPDH ChIP antisense: 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′ (respectively, nt −145 to −126 and nt +20 to −3 from transcription start site).
Results
Location of potential binding sites in the CCR5 promoter region
The organization and promoter usage of the CCR5 gene has been elucidated previously by Mummidi et al using luciferase-reporter constructs containing CCR5 regulatory regions (Figure 1).20,26 Two distinct functional promoter regions for the CCR5 gene were identified: a downstream promoter region, designated P1, and an upstream promoter region, designated P2. Using TFSearch analysis, we evaluated the CCR5 promoter regions for potential interferon-stimulated response elements (ISREs), which bind interferon regulatory factors (IRFs), κB elements, which bind nuclear factor κB (NF-κB), and cAMP-response elements (CRE elements), which can bind the common activator of transcription CREB-1 (cAMP-response element [CRE]–binding protein) and its family members.
Gene and promoter organization of human CC chemokine receptor CCR5. Schematic representation of the CCR5 gene and its promoters. Exons (1-3) are depicted by boxes. Both promoter regions, the downstream promoter P1 and the upstream promoter P2, are indicated above the scheme. Locations of the identified putative transcription factor binding sites are depicted above. Underneath, promoter-luciferase-reporter constructs used in this study are shown. Two sets of constructs were used: the upstream promoter constructs CCR5-pA1 through -pA4 and the downstream promoter constructs CCR5-pB1 and -pB3 through -pB5. Nucleotide positions for the exons and the promoter constructs are depicted relative to the ORF in exon 3. Numbering of nucleotides and exons is according to the CCR5 numbering system proposed at the 1999 CCR5-AIDS symposium.26,34
Gene and promoter organization of human CC chemokine receptor CCR5. Schematic representation of the CCR5 gene and its promoters. Exons (1-3) are depicted by boxes. Both promoter regions, the downstream promoter P1 and the upstream promoter P2, are indicated above the scheme. Locations of the identified putative transcription factor binding sites are depicted above. Underneath, promoter-luciferase-reporter constructs used in this study are shown. Two sets of constructs were used: the upstream promoter constructs CCR5-pA1 through -pA4 and the downstream promoter constructs CCR5-pB1 and -pB3 through -pB5. Nucleotide positions for the exons and the promoter constructs are depicted relative to the ORF in exon 3. Numbering of nucleotides and exons is according to the CCR5 numbering system proposed at the 1999 CCR5-AIDS symposium.26,34
Figure 1 shows the location of the potential binding sites for these transcription factors (Table 1). The potential NF-κB binding sites (CCR5-κB-1 through κB-3) are situated only in the downstream promoter region, whereas the putative CRE sites (CCR5-CRE-1 through CRE-3) are all located in the upstream promoter region. One of the identified potential IRF-binding sites, CCR5-ISRE-1, is located in the upstream promoter region, whereas the other ISREs, CCR5-ISRE-2 and ISRE-3, are located in the region in which the upstream promoter and the downstream promoter overlap each other.
Protein binding to the putative transcription factor binding sites in the CCR5 promoter region
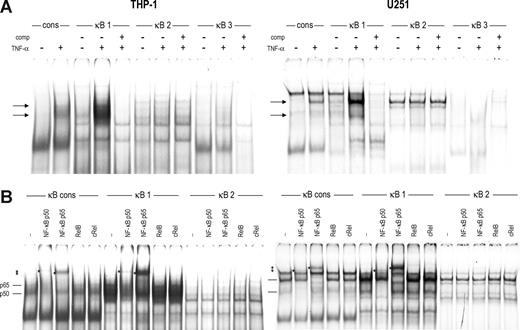
We tested the capacity of the identified regulatory sites in the promoter regions of the CCR5 gene to bind specific transcription factors in vitro by EMSA. Using extracts of THP-1 and U251 cells stimulated with TNF-α, which induces NF-κB activity, we studied in vitro complex formation at the κB sites present in the CCR5 downstream promoter. We could detect constitutive protein binding to the CCR5-κB-1 and CCR5-κB-2 probes but not to the CCR5-κB-3 probe using nuclear extracts of both cell types. Similar to binding to the κB element of the β2m promoter,21 the amount of protein binding to the CCR5-κB-1 was dramatically increased after TNF-α stimulation (Figure 2A). Binding of this TNF-α–induced factor to the CCR5-κB-1 was specific for the κB sequence in the oligonucleotide probe because it could be competed away with the β2m-κB probe. In contrast, protein binding to CCR5-κB2 proved to be nonspecific.
Transcription factor binding to the κB sites in the CCR5 downstream promoter. EMSA showing binding of complexes to the κB sites of CCR5 (κB-1 through -3) and the κB site of human β2m as a consensus probe (cons) using nuclear extracts of THP-1 cells (left panels) or U251 cells (right panels). (A) Cells were left untreated or stimulated with TNF-α (10 ng/mL) for 2 hours. EMSA analysis revealed formation of 2 complexes to CCR5-κB-1 on TNF-α treatment (→). CCR-5-kB-2 showed nonspecific complex binding constitutively, whereas CCR-5-κB-3 did not generate any significant binding, either constitutive or on TNF-α treatment. (B) Using specific antibodies, the proteins binding to CCR5-κB-1 on TNF-α treatment were identified as NF-κB p65 and p50. Binding of these factors to CCR5-κB-2 could not be detected. *Supershifted complexes. Shown are representatives of 2 independent experiments.
Transcription factor binding to the κB sites in the CCR5 downstream promoter. EMSA showing binding of complexes to the κB sites of CCR5 (κB-1 through -3) and the κB site of human β2m as a consensus probe (cons) using nuclear extracts of THP-1 cells (left panels) or U251 cells (right panels). (A) Cells were left untreated or stimulated with TNF-α (10 ng/mL) for 2 hours. EMSA analysis revealed formation of 2 complexes to CCR5-κB-1 on TNF-α treatment (→). CCR-5-kB-2 showed nonspecific complex binding constitutively, whereas CCR-5-κB-3 did not generate any significant binding, either constitutive or on TNF-α treatment. (B) Using specific antibodies, the proteins binding to CCR5-κB-1 on TNF-α treatment were identified as NF-κB p65 and p50. Binding of these factors to CCR5-κB-2 could not be detected. *Supershifted complexes. Shown are representatives of 2 independent experiments.
By supershift analysis with antibodies directed against several members of the NF-κB family, we identified the factors binding to CCR5-kB-1 on TNF-α stimulation as the subunits p50 and p65 of NF-κB in both THP-1 and U251 cells (Figure 2B). In addition, we could identify the factor that constitutively binds to the CCR5-κB-1 site in THP-1 cells as p50 and as p65 in U251 cells. In contrast, the complex found with CCR5-κB-2 could not be shifted with any of the antibodies, confirming that this binding is nonspecific.
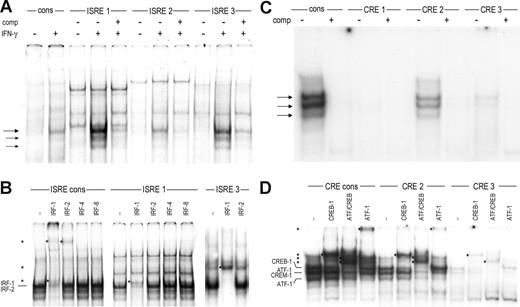
Using nuclear extracts of U251 cells stimulated with IFN-γ, we next found that IFN-γ stimulation induced binding of 1 main and 2 minor protein complexes to CCR5-ISRE-1 and CCR5-ISRE-3 and, to a lesser extent, to CCR5-ISRE-2 (Figure 3A). By competition analysis, these IFN-γ-induced complexes were shown to be specific for the ISRE sequence in the probes. In THP-1 cells, we could detect only very weak binding to CCR5-ISRE-1 and CCR5-ISRE-3 (not shown).
Transcription factor binding to the ISREs and CRE sites in the CCR5 promoter. (A) EMSA showing binding of complexes to the ISREs of CCR5 (ISRE-1 through ISRE-3) and the ISRE of human β2m as a consensus probe (cons) using nuclear extracts of U251 cells either unstimulated or stimulated with 500 U/mL of IFN-γ for 2 hours. IFN-γ stimulation induced formation of 1 major ( ) and 2 minor (→) complexes to CCR5-ISRE-1 and CCR5-ISRE-3 and weak binding of these complexes to CCR5-ISRE-2, specific for the ISRE in the probes. (B) Using specific antibodies, the proteins binding to the β2m-ISRE on IFN-γ treatment were identified as IRF-1 and IRF-2, whereas the complexes binding to ISRE-1 and ISRE-3 contained only IRF-1. (C) EMSA showing binding of complexes to the CRE sites located in the CCR5 upstream promoter (CRE-1 through CRE-3) and a CRE consensus probe (cons) using nuclear extracts of U251 cells. Binding of 3 complexes (
) and 2 minor (→) complexes to CCR5-ISRE-1 and CCR5-ISRE-3 and weak binding of these complexes to CCR5-ISRE-2, specific for the ISRE in the probes. (B) Using specific antibodies, the proteins binding to the β2m-ISRE on IFN-γ treatment were identified as IRF-1 and IRF-2, whereas the complexes binding to ISRE-1 and ISRE-3 contained only IRF-1. (C) EMSA showing binding of complexes to the CRE sites located in the CCR5 upstream promoter (CRE-1 through CRE-3) and a CRE consensus probe (cons) using nuclear extracts of U251 cells. Binding of 3 complexes ( ) to CCR5-CRE-2 and, to a lesser extent, CCR5-CRE-3, specific for the CRE in the probes, was detected. (D) Using specific antibodies, the proteins binding to CCR5-CRE-2 and CCR5-CRE-3 were identified as CREB-1 and ATF-1. In addition, the CCR5-CRE-2 binding complex was revealed to also contain CREM-1. *Supershifted complexes. Shown are representatives of 2 independent experiments.
) to CCR5-CRE-2 and, to a lesser extent, CCR5-CRE-3, specific for the CRE in the probes, was detected. (D) Using specific antibodies, the proteins binding to CCR5-CRE-2 and CCR5-CRE-3 were identified as CREB-1 and ATF-1. In addition, the CCR5-CRE-2 binding complex was revealed to also contain CREM-1. *Supershifted complexes. Shown are representatives of 2 independent experiments.
Transcription factor binding to the ISREs and CRE sites in the CCR5 promoter. (A) EMSA showing binding of complexes to the ISREs of CCR5 (ISRE-1 through ISRE-3) and the ISRE of human β2m as a consensus probe (cons) using nuclear extracts of U251 cells either unstimulated or stimulated with 500 U/mL of IFN-γ for 2 hours. IFN-γ stimulation induced formation of 1 major ( ) and 2 minor (→) complexes to CCR5-ISRE-1 and CCR5-ISRE-3 and weak binding of these complexes to CCR5-ISRE-2, specific for the ISRE in the probes. (B) Using specific antibodies, the proteins binding to the β2m-ISRE on IFN-γ treatment were identified as IRF-1 and IRF-2, whereas the complexes binding to ISRE-1 and ISRE-3 contained only IRF-1. (C) EMSA showing binding of complexes to the CRE sites located in the CCR5 upstream promoter (CRE-1 through CRE-3) and a CRE consensus probe (cons) using nuclear extracts of U251 cells. Binding of 3 complexes (
) and 2 minor (→) complexes to CCR5-ISRE-1 and CCR5-ISRE-3 and weak binding of these complexes to CCR5-ISRE-2, specific for the ISRE in the probes. (B) Using specific antibodies, the proteins binding to the β2m-ISRE on IFN-γ treatment were identified as IRF-1 and IRF-2, whereas the complexes binding to ISRE-1 and ISRE-3 contained only IRF-1. (C) EMSA showing binding of complexes to the CRE sites located in the CCR5 upstream promoter (CRE-1 through CRE-3) and a CRE consensus probe (cons) using nuclear extracts of U251 cells. Binding of 3 complexes ( ) to CCR5-CRE-2 and, to a lesser extent, CCR5-CRE-3, specific for the CRE in the probes, was detected. (D) Using specific antibodies, the proteins binding to CCR5-CRE-2 and CCR5-CRE-3 were identified as CREB-1 and ATF-1. In addition, the CCR5-CRE-2 binding complex was revealed to also contain CREM-1. *Supershifted complexes. Shown are representatives of 2 independent experiments.
) to CCR5-CRE-2 and, to a lesser extent, CCR5-CRE-3, specific for the CRE in the probes, was detected. (D) Using specific antibodies, the proteins binding to CCR5-CRE-2 and CCR5-CRE-3 were identified as CREB-1 and ATF-1. In addition, the CCR5-CRE-2 binding complex was revealed to also contain CREM-1. *Supershifted complexes. Shown are representatives of 2 independent experiments.
IRF-1 is the main IRF induced by IFN-γ in U251 and THP-1 cells.21 In addition, U251 cells constitutively express IRF-2, whereas THP-1 cells constitutively express both IRF-2 and the lymphoid/myeloid-specific factors IRF-4 and IRF-821 (data not shown). Supershift analysis revealed binding of IRF-1 to CCR5-ISRE-1 and CCR5-ISRE-3 in U251 cells, whereas IRF-2, present in these cells, as determined by its binding to the β2m ISRE, did not bind to the CCR5 ISREs (Figure 3B). Furthermore, CCR5-ISRE-1 and CCR5-ISRE-3 failed to bind IRF-4 and IRF-8 expressed in THP-1 nuclear extracts, whereas these factors did interact with the β2m-ISRE21 (data not shown). These findings indicate that only IRF-1 binds to CCR5-ISRE-1 and CCR5-ISRE-3 after IFN-γ stimulation.
Finally, we detected constitutive binding of 3 complexes in U251 and THP-1 (data not shown) cells to the CCR5-CRE-2 and -CRE-3, although binding to the latter was very weak (Figure 3C). Binding to both CCR5-CRE-sites was specific for the CRE region in the probe, as determined by competition with a CRE consensus probe.27
The CREB family consists of 3 members: CREB-1, activating transcription factor 1 (ATF-1) and cAMP-responsive element modulator (CREM), which all bind to CRE sequences as homodimers or heterodimers.28 Using antibodies directed against either CREB-1 or ATF-1 and an antibody that recognizes CREB-1, ATF-1, and CREM-1, supershift patterns revealed that both CREB-1 and ATF-1 bind to CCR5-CRE-2 as well as CCR5-CRE-3 (Figure 3D). In addition, the intermediate complex formed at CCR5-CRE-2 was identified as CREM-1 binding because it was only shifted by the antibody directed against multiple members of the CREB family (ATF/CREB), whereas the upper and lower complexes were shifted by both this antibody and the antibodies against CREB-1 and ATF-1, respectively (Figure 3D).
Taken together, these analyses reveal that the CCR5 promoter contains at least 2 ISREs, one κB site and one CRE site, that can be bound in vitro by their respective transcription factors that are present in THP-1 or U251 cells, either constitutively or induced by IFN-γ or TNF-α, respectively.
Transactivation capacity of the putative regulatory sites
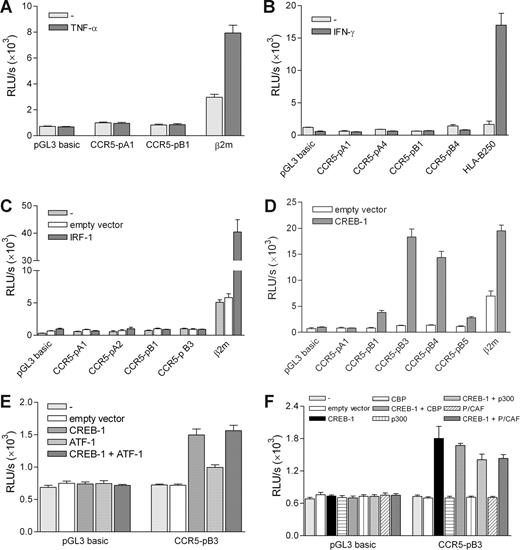
As previously described,21,23 we used the cytokine-responsive teratocarcinoma cell line Tera-2 to test the functionality of the binding sites displaying in vitro binding of transcription factors in luciferase-reporter assays using various promoter constructs generated by Mummidi et al20 (Figure 1).
Compared with U251 and THP-1 cells (not shown), the CCR5 promoter constructs displayed low constitutive activity in Tera-2 cells (Figure 4). We first investigated whether we could activate the CCR5-κB-1 element containing CCR5 downstream promoter by TNF-α treatment. Although a promoter construct containing the TNF-α-responsive β2m promoter21 did display enhanced activity on TNF-α treatment, the full-length CCR5 downstream promoter construct (CCR5-pB1) could not be activated by TNF-α (Figure 4A). This indicates that the putative CCR5-κB-1 site found in the CCR5 promoter is not functional, despite in vitro NF-κB protein binding. In addition, activity of the CCR5-pA1 upstream promoter construct could not be induced by TNF-α treatment (Figure 4A), which excludes the presence of functional nonconsensus NF-κB binding sites in the CCR5 promoter region. These data are confirmed by the observation that the CCR5-κB-1–containing promoter constructs could not be activated by cotransfecting expression vectors of NF-κB p50 and p65 (not shown).
Transactivation capacity of the regulatory sites in the CCR5 promoter. Transient transfection of promoter constructs in Tera-2 cells. (A) Transient transfection of the full-length CCR5 upstream (CCR5-pA1) and downstream (CCR5-pB1) promoter constructs. Cells were left untreated or treated with TNF-α (10 ng/mL) for 24 hours, leading to activation of only the β2m promoter. (B) Transient transfection of full-length and truncated upstream (CCR5-pA1 and CCR5-pA4) and downstream (CCR5-pB1 and CCR5-pB4) promoter constructs and the HLA-B250 promoter construct, as a positive control. Cells were left untreated or treated with IFN-γ (500 U/mL) for 24 hours, leading to HLA-B250 promoter activity only. (C) Transient transfection of CCR5 promoter constructs and a β2m promoter construct as a positive control with an IRF-1 expression vector or empty control vector, showing lack of CCR5 promoter transactivation by IRF-1. (D) Transient transfection of CCR5 promoter constructs with a CREB-1 expression vector, showing transactivation of the downstream promoter constructs by CREB-1. (E) Cotransfection of the CCR5 downstream promoter construct CCR5-pB3 with CREB-1 and ATF-1 expression vectors, revealing low transactivation activity of ATF-1 and absent enhancement of CREB-1 induced promoter activity. (F) Cotransfection of the CCR5 downstream promoter construct CCR5-pB3 with CREB-1 and CBP, p300, and P/CAF expression vectors, revealing lack of coinduction of the CCR5 promoter by CBP, p300, and P/CAF. Depicted are relative light units (RLU) per second obtained for luciferase activity and normalized with Renilla luciferase activity. Shown are means plus or minus SEM of 3 independent experiments (A,C,E,F) or a representative of 3 independent experiments (B,D) performed in triplicate.
Transactivation capacity of the regulatory sites in the CCR5 promoter. Transient transfection of promoter constructs in Tera-2 cells. (A) Transient transfection of the full-length CCR5 upstream (CCR5-pA1) and downstream (CCR5-pB1) promoter constructs. Cells were left untreated or treated with TNF-α (10 ng/mL) for 24 hours, leading to activation of only the β2m promoter. (B) Transient transfection of full-length and truncated upstream (CCR5-pA1 and CCR5-pA4) and downstream (CCR5-pB1 and CCR5-pB4) promoter constructs and the HLA-B250 promoter construct, as a positive control. Cells were left untreated or treated with IFN-γ (500 U/mL) for 24 hours, leading to HLA-B250 promoter activity only. (C) Transient transfection of CCR5 promoter constructs and a β2m promoter construct as a positive control with an IRF-1 expression vector or empty control vector, showing lack of CCR5 promoter transactivation by IRF-1. (D) Transient transfection of CCR5 promoter constructs with a CREB-1 expression vector, showing transactivation of the downstream promoter constructs by CREB-1. (E) Cotransfection of the CCR5 downstream promoter construct CCR5-pB3 with CREB-1 and ATF-1 expression vectors, revealing low transactivation activity of ATF-1 and absent enhancement of CREB-1 induced promoter activity. (F) Cotransfection of the CCR5 downstream promoter construct CCR5-pB3 with CREB-1 and CBP, p300, and P/CAF expression vectors, revealing lack of coinduction of the CCR5 promoter by CBP, p300, and P/CAF. Depicted are relative light units (RLU) per second obtained for luciferase activity and normalized with Renilla luciferase activity. Shown are means plus or minus SEM of 3 independent experiments (A,C,E,F) or a representative of 3 independent experiments (B,D) performed in triplicate.
Likewise, IFN-γ treatment did not activate the CCR5-pA1 upstream promoter construct containing both CCR5-ISRE-1 and CCR5-ISRE-3 or the CCR5-pB1 downstream promoter construct containing CCR5-ISRE-3, activity of which was comparable to the activity of the truncated constructs (CCR5-pA4 and CCR5-pB4) lacking these sites, and the control plasmid. In contrast, the IFN-γ–responsive HLA-B promoter19 did show activity on IFN-γ treatment (Figure 4B). In addition, cotransfection of an expression vector of IRF-1 did not lead to activation of the 2 CCR5 upstream promoter constructs containing both CCR5-ISREs (CCR5-pA1 and CCR5-pA2) or the CCR5-ISRE-3–containing downstream construct (CCR5-pB1; Figure 4C). Again, activity did not rise above activity of a construct lacking CCR5-ISREs (CCR5-pB3) or the control plasmid, whereas the IFN-γ inducible β2m promoter,21 was activated by IRF-1 (Figure 4C). This demonstrates that, like the CCR5-κB-1 site, the putative CCR5-ISREs in the CCR5 promoter are not functional, although they do bind IRF-1 in vitro. Furthermore, these analyses exclude the presence of nonconsensus ISREs in the full-length promoter-reporter constructs.
Finally, we tested whether CREB-1 is able to transactivate the CCR5 upstream promoter, which contains the CRE-2 and CRE-3 sites. Cotransfection with a CREB-1 expression vector did not lead to activation of the full-length upstream promoter construct (Figure 4D, CCR5-pA1). Furthermore, CREB-1 cotransfection was not able to activate any of the truncated upstream promoter constructs, CCR5-pA2 through −pA4, either (not shown). Surprisingly, however, CREB-1 did activate the full-length downstream promoter construct CCR5-pB1 and, to an even higher extent, the 2 truncated downstream promoter constructs CCR5-pB3 and CCR5-pB4, activation of which reached a level comparable with CREB-1–induced activity of the β2m promoter construct (Figure 4D). Further truncation of the promoter region resulted in loss of transactivation by CREB-1 (Figure 4D, CCR5-pB5). These findings imply that, whereas the CRE sites found in the upstream promoter are not functional, the downstream promoter region spanning −2333 to −2128 must contain at least 1 functional CREB-1 binding site.
Using the construct that displayed maximal CREB-1 transactivation, CCR5-pB3, we found that, in addition to CREB-1, ATF-1 also induced CCR5 promoter activity (Figure 4E). However, its transactivation capacity was considerably lower than that of CREB-1, and ATF-1 could not further enhance CREB-1-induced transactivation. Subsequently, we also investigated whether the coactivators CBP (CREB-binding protein), p300, and P/CAF (p300/CREB-associated factor)28,29 would modulate CREB-1–mediated activation of CCR5. Cotransfection of the CREB-responsive CCR5-pB3 construct with CREB-1, and these coactivators revealed that CBP, p300, and P/CAF did not enhance CREB-1 induction of CCR5 promoter activity (Figure 4F), indicating that exogenous expression of CREB-1 is sufficient to activate the CCR5 promoter.
Taken together, these data suggest that the CREB pathway is involved in the transactivation of the CCR5 promoter region, whereas the IRF and the NF-κB pathways are not.
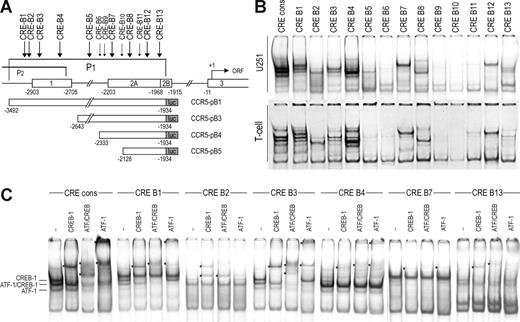
Protein binding to CRE sites in the downstream promoter
In view of these findings, we analyzed the CCR5 downstream promoter region more closely for potential CRE sites. Figure 5A depicts the position of putative CRE sites identified in the downstream promoter region (designated CCR5-CRE-B1 through -CRE-B13). Using EMSA, we observed protein/DNA complex formation with several of the CCR5-CRE-B sites with nuclear extracts of both U251 and primary activated T lymphocytes, which are known to express CCR5 (Figure 5B), which we further analyzed by supershift analysis. CCR5-CRE-B1 through CRE-B4 are located in the region that is included in the downstream CCR5 promoter construct CCR5-pB1. Of these sites, CCR5-CRE-B1 and CRE-B3 displayed clear binding of CREB-1 and weaker binding of ATF-1 (Figure 5C). We detected weak binding of CREB-1 to CCR5-CRE-B2 and CREB-1 and ATF-1 to CRE-B4. However, this region is also included in the upstream promoter constructs (CCR5-pA1 through pA4), which are not responsive to CREB-1 (Figures 1 and 5A).
Transcription factor binding to the CRE sites in the CCR5 downstream promoter. (A) Scheme depicting the CCR5 gene and the downstream promoter region (P1). Locations of the identified putative CRE sites in the downstream promoter regions are depicted above. Sites at which significant protein binding was detected are indicated by large arrows, whereas sites that did not generate protein binding are indicated by small arrows. Underneath, the downstream promoter-luciferase-reporter constructs are shown. (B) EMSA showing binding of complexes to the CRE sites in the CCR5 downstream promoter region (CRE-B1 through -B13) and a CRE consensus probe (cons) using nuclear extracts of U251 cells (top panels) and primary T lymphocytes (bottom panels). Protein/DNA complexes are formed with probe CRE-B1 through -B5, -B7, -B8, -B12, and -B13. (C) Using specific antibodies, the complexes binding to CCR5-CRE-B1, -B3, -B4, and -B7 were shown to contain both CREB-1 and ATF-1, whereas the complexes formed with CCR5-CRE-B2, and CRE-B13 contained only CREB-1. *Supershifted complexes. Shown are representatives of 2 independent experiments. Empty lanes were deleted from the images.
Transcription factor binding to the CRE sites in the CCR5 downstream promoter. (A) Scheme depicting the CCR5 gene and the downstream promoter region (P1). Locations of the identified putative CRE sites in the downstream promoter regions are depicted above. Sites at which significant protein binding was detected are indicated by large arrows, whereas sites that did not generate protein binding are indicated by small arrows. Underneath, the downstream promoter-luciferase-reporter constructs are shown. (B) EMSA showing binding of complexes to the CRE sites in the CCR5 downstream promoter region (CRE-B1 through -B13) and a CRE consensus probe (cons) using nuclear extracts of U251 cells (top panels) and primary T lymphocytes (bottom panels). Protein/DNA complexes are formed with probe CRE-B1 through -B5, -B7, -B8, -B12, and -B13. (C) Using specific antibodies, the complexes binding to CCR5-CRE-B1, -B3, -B4, and -B7 were shown to contain both CREB-1 and ATF-1, whereas the complexes formed with CCR5-CRE-B2, and CRE-B13 contained only CREB-1. *Supershifted complexes. Shown are representatives of 2 independent experiments. Empty lanes were deleted from the images.
In addition to these sites, several other sites displayed protein binding in EMSA analysis. Of these sites, CCR5-CRE-B13 is located in the region that is included in all CREB-responsive downstream promoter constructs, whereas CCR5-CRE-B7 is present only in CCR5-pB1 through CCR5-pB4 (Figure 5A). CCR5-CRE-B7 displayed rather weak binding of both CREB-1 and ATF-1, whereas we only found weak binding of CREB-1 to CCR5-CRE-B13 (Figure 5C). In addition, we found very weak binding of CREB-1 and ATF-1 to CCR5-CRE-B5, located in promoter constructs CCR5-pB1 and CCR5-pB3, and hardly detectable binding of CREB-1 to CCR5-CRE-B8 and CCR5-CRE-B12 (not shown), located in all downstream promoter constructs. Together with the data presented in Figure 4D, this suggests that the effect of CREB-1 on CCR5 promoter activation is mediated by CRE-B7.
Involvement of the IRF and NF-κB pathways in endogenous CCR5 expression
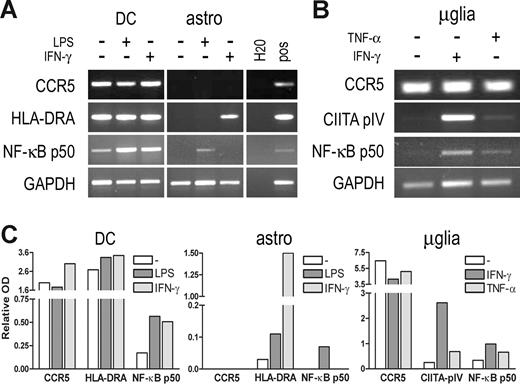
Next, we evaluated whether stimulation of the investigated signaling pathways could lead to transcription activation or modulation of CCR5 expression in primary human monocyte-derived DCs and in primary cells originating from the CNS. First, we observed that neither LPS (a potent inducer of NF-κB activation) nor IFN-γ enhanced the constitutive transcription of CCR5 in DCs significantly, as determined by RT-PCR (Figure 6A,C). Likewise, LPS and IFN-γ did not modulate the constitutive transcription of HLA-DRA, whereas both stimuli did enhance expression of the p50 subunit of NF-κB in these cells.
Induction of CCR5 transcription by inflammatory stimuli. (A) mRNA expression levels of CCR5, HLA-DRA, and NF-κB p50 in human monocyte-derived DCs (DC) and human primary astrocytes (astro) after stimulation with LPS (100 ng/mL) or IFN-γ (500 U/mL) for 24 (DC) or 8 (astro) hours, as determined by RT-PCR. CCR5 expression is not enhanced in DCs nor induced in astrocytes by either LPS or IFN-γ treatment, whereas NF-κB p50 expression is enhanced in DCs by either treatment, and HLA-DR and NF-κB p50 expression is induced in astrocytes by IFN-γ and TNF-α, respectively. (B) mRNA expression levels of CCR5, CIITA-PIV, and NF-κB p50 in human primary microglia (μglia) after stimulation with TNF-α (10 ng/mL) or IFN-γ (500 U/mL) for 8 hours, as determined by RT-PCR. IFN-γ or TNF-α does not enhance CCR5 expression in microglia, whereas expression of CIITA-PIV and NF-κB p50 is enhanced by either treatment. (C) Quantification of the PCR products. Relative optical densities (ODs) corrected for GAPDH expression are shown.
Induction of CCR5 transcription by inflammatory stimuli. (A) mRNA expression levels of CCR5, HLA-DRA, and NF-κB p50 in human monocyte-derived DCs (DC) and human primary astrocytes (astro) after stimulation with LPS (100 ng/mL) or IFN-γ (500 U/mL) for 24 (DC) or 8 (astro) hours, as determined by RT-PCR. CCR5 expression is not enhanced in DCs nor induced in astrocytes by either LPS or IFN-γ treatment, whereas NF-κB p50 expression is enhanced in DCs by either treatment, and HLA-DR and NF-κB p50 expression is induced in astrocytes by IFN-γ and TNF-α, respectively. (B) mRNA expression levels of CCR5, CIITA-PIV, and NF-κB p50 in human primary microglia (μglia) after stimulation with TNF-α (10 ng/mL) or IFN-γ (500 U/mL) for 8 hours, as determined by RT-PCR. IFN-γ or TNF-α does not enhance CCR5 expression in microglia, whereas expression of CIITA-PIV and NF-κB p50 is enhanced by either treatment. (C) Quantification of the PCR products. Relative optical densities (ODs) corrected for GAPDH expression are shown.
Subsequently, we investigated CCR5 transcription in cultured human astrocytes and microglia stimulated with LPS, TNF-α, or IFN-γ. It has been shown that astrocytes may express CCR5 protein ex vivo.30 However, as proposed before, this expression seems to be lost during culture because we could not detect constitutive CCR5 mRNA expression in cultured astrocytes (Figure 6A,C), whereas primary cultured microglia did express CCR5 constitutively (Figure 6B,C). In astrocytes, LPS stimulation did not induce CCR5 expression, whereas NF-κB p50 expression was induced by LPS treatment. Likewise, IFN-γ treatment did not induce CCR5 expression in astrocytes, whereas HLA-DRA expression was induced (Figure 6A,C). Consistent with the results obtained with DCs, neither IFN-γ nor TNF-α treatment resulted in enhanced CCR5 expression in microglia, whereas these treatments did enhance expression of the inducible Class II Transactivator (CIITA) PIV isoform or induce NF-κB p50 expression (Figure 6B,C). Similarly, we were not able to induce CCR5 mRNA expression by either IFN-γ or TNF-α treatment in U251 or THP-1 cells (not shown).
Involvement of the CREB pathway in endogenous CCR5 expression
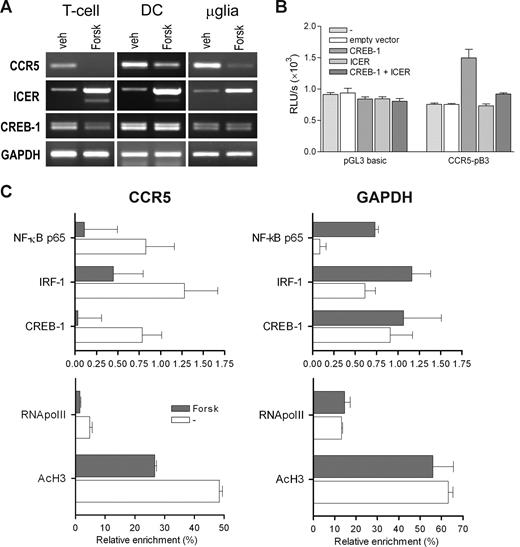
Finally, we investigated whether the CREB pathway is involved in endogenous expression of CCR5 as found in activated T lymphocytes, DCs, and microglia. Therefore, we treated these cells with forskolin, an agent that induces the CREM isoform inducible cAMP early repressor (ICER) in various cell types, including T lymphocytes.31,32 ICER competes with CREB for DNA binding and acts as a repressor of CREB-mediated transcription.31 We found that CCR5 mRNA expression was greatly reduced in T lymphocytes after forskolin treatment, whereas expression of ICER was markedly increased (Figure 7A). In these cells, CREB-1 expression was slightly reduced by forskolin treatment. In addition, forskolin treatment greatly reduced CCR5 expression in monocyte-derived DCs and microglia, concomitant with increased expression of ICER (Figure 7A). Interestingly, no change in CREB-1 expression was observed in these cells. To confirm that ICER affects CCR5 promoter activity, we cotransfected the CREB-responsive CCR5 promoter construct CCR5-pB3 with CREB-1 and ICER expression constructs. Indeed, we found that ICER strongly inhibited CREB-induced transactivation of the CCR5 promoter (Figure 7B).
Inhibition of CCR5 transcription and CCR5 promoter activation by forskolin and ICER. (A) mRNA expression levels of CCR5, ICER, and CREB in T lymphocytes, DCs, and microglia treated with forskolin (Forsk; 20 μM) for 6 hours, as determined by RT-PCR. Forskolin treatment reduces amounts of CCR5 transcript in T lymphocytes. Shown are representatives of at least 2 independent experiments. (B) Transient transfection of the CREB-responsive CCR5 downstream promoter construct CCR5-pB3 with CREB-1 and ICER expression vectors, showing inhibition of CREB-1-induced CCR5 promoter activity by ICER. Depicted are relative light units (RLU) per second obtained for luciferase activity and normalized with Renilla luciferase activity expressed as means plus or minus SEM. Shown is a representative of 3 independent experiments performed in triplicate. (C) Chromatin immunoprecipitation (ChIP) analysis of CREB-1, NF-κB, and IRF-1 binding, and acetylated histone H3 (AcH3) and RNA polymerase II (RNApolII) binding to the CCR5 and GAPDH promoter regions in T lymphocytes treated with forskolin (Forsk; 20 μM) for 6 hours. Protein binding is depicted as percentage enrichment, relative to input chromatin and corrected for background binding. Shown are means plus or minus SEM of 2 PCR reactions of a representative of 3 independent experiments.
Inhibition of CCR5 transcription and CCR5 promoter activation by forskolin and ICER. (A) mRNA expression levels of CCR5, ICER, and CREB in T lymphocytes, DCs, and microglia treated with forskolin (Forsk; 20 μM) for 6 hours, as determined by RT-PCR. Forskolin treatment reduces amounts of CCR5 transcript in T lymphocytes. Shown are representatives of at least 2 independent experiments. (B) Transient transfection of the CREB-responsive CCR5 downstream promoter construct CCR5-pB3 with CREB-1 and ICER expression vectors, showing inhibition of CREB-1-induced CCR5 promoter activity by ICER. Depicted are relative light units (RLU) per second obtained for luciferase activity and normalized with Renilla luciferase activity expressed as means plus or minus SEM. Shown is a representative of 3 independent experiments performed in triplicate. (C) Chromatin immunoprecipitation (ChIP) analysis of CREB-1, NF-κB, and IRF-1 binding, and acetylated histone H3 (AcH3) and RNA polymerase II (RNApolII) binding to the CCR5 and GAPDH promoter regions in T lymphocytes treated with forskolin (Forsk; 20 μM) for 6 hours. Protein binding is depicted as percentage enrichment, relative to input chromatin and corrected for background binding. Shown are means plus or minus SEM of 2 PCR reactions of a representative of 3 independent experiments.
In addition to this, we investigated whether forskolin treatment altered in vivo CREB binding to the CCR5 downstream promoter in T lymphocytes by ChIP. This analysis revealed that the presence of CREB-1 in CCR5 promoter chromatin was indeed abolished by forskolin treatment, whereas the presence of CREB-1 in GAPDH chromatin was not altered (Figure 7C). Interestingly, we could also detect binding of NF-κB p65 and IRF-1 to CCR5 promoter chromatin, which was also decreased by forskolin treatment. In addition, forskolin treatment led to a marked reduction in RNA polymerase II recruitment to CCR5 promoter chromatin and a reduction in the amount of acetylated histone H3 (Ac-H3) in CCR5 promoter chromatin, whereas these alterations were not observed with the GAPDH promoter chromatin (Figure 7C bottom panels).
These findings substantiate that of the signaling pathways investigated: only the cAMP/CREB pathway contributes to CCR5 transcriptional activation.
Discussion
In the current study, we have evaluated the contribution of IRF-1, NF-κB, and CREB-1 to the transcriptional regulation of the chemokine receptor CCR5. Our results indicate that only CREB-1 is involved in the transcriptional regulation of CCR5 and that CCR5 expression is neither induced nor modulated by IRF-1 and NF-κB.
Although it has been previously suggested that NF-κB could up-regulate CCR5 in T lymphocytes,33 our data indicate that the identified NF-κB binding sites do not seem to play a role in the transcriptional regulation of CCR5 expression in the cell types investigated in this study. This is illustrated by the lack of CCR5 promoter activation by TNF-α and NF-κB and the failure of LPS and TNF-α to induce or enhance endogenous CCR5 transcription.
Likewise, we show a lack of inducibility of CCR5 transcription by IFN-γ in several cell types, which is corroborated by the failure of IFN-γ and IRF-1 to activate the promoter constructs containing CCR5-ISRE-1 and -3, that displayed in vitro IRF-1 binding. The lack of CCR5 promoter activation by IFN-γ and IRF-1 does not seem to result from the presence of an upstream inhibitory region,32,34 because the truncated constructs, which did include CCR5-ISRE-3, were not responsive to IFN-γ or IRF-1 either. In addition, it seems unlikely that binding of an inhibitory IRF causes the unresponsiveness of the ISRE-containing promoter constructs. Previously, we have shown that IRF-2 inhibits the transactivation of the β2m promoter by IRF-1.21 However, whereas we did find binding of IRF-2 to the ISRE of β2m in U251 cells, we could detect no binding of IRF family members other than IRF-1 to CCR5-ISRE-1 and CCR5-ISRE-3 (Figure 3B).
In contrast to the lack of CCR5 transcriptional regulation through IFN-γ– and TNF-α–induced activating pathways, our findings indicate that CREB-1 is involved in the transcriptional regulation of CCR5. Of the identified CRE sites in the downstream promoter, CCR5-CRE-B1 and CCR5-CRE-B3 showed the most prominent binding of CREB-1/ATF-1. However, these sites are located in the region that is also included in the upstream promoter constructs, which are not responsive to CREB-1. Therefore, it is reasonable to assume that these sites do not contribute to the transactivation of the downstream promoter by CREB-1. In contrast, CCR5-CRE-B7, which displayed weaker binding of CREB-1/ATF-1 than the upstream CRE sites, is located in the region that is most responsive to CREB-1 and therefore most likely contributes to the transactivation of the CCR5 promoter by CREB-1.
Several polymorphisms in the CCR5 regulatory region are known, some of which have been suggested to be associated to some degree with altered cell surface expression of CCR5 and/or altered rates of HIV-1 disease progression.26,34 However, these polymorphisms are not located in any of the downstream CRE sites and therefore do not seem to be involved in the transactivation of the CCR5 promoter by CREB-1. It does seem, however, that CCR5 promoter polymorphisms and the alternative usage of the CCR5 upstream and downstream promoters can affect translational efficiency of CCR5 transcripts.35
The CRE site in the upstream promoter region, CCR5-CRE-2, proved also not to contribute to CCR5 transcription despite binding of CREB-1/ATF-1 (Figure 3D). However, this site also seems to bind CREM-1. It is known that, depending on the isoform obtained from alternative splicing, CREM-1 can act as either an activator or repressor of transcription.28,36 For example, ICER is an inducible CREM isoform that lacks a transactivation domain and therefore is a natural antagonists of CREB-1, repressing its transactivation function.31 Therefore, it could be argued that the CREM-isoform binding to CCR5-CRE-2 might represent a repressive CREM isoform or ICER, inhibiting activation of the CCR5 upstream promoter by CREB-1. However, EMSA analysis revealed considerable binding of both CREB-1 and ATF-1, which suggests that the CREM binding to CCR5-CRE2 is not able to compete with the binding of CREB-1 and ATF-1.
The fact that the upstream promoter constructs are not responsive to CREB-1, whereas the downstream promoter constructs are, might suggest a repressive function of the upstream promoter region. This hypothesis is underscored by the fact that the longest downstream promoter construct (CCR5-pB1), including part of the upstream region (Figure 1), is less responsive to CREB-1 than the truncated downstream promoter constructs CCR5-pB3 and -pB4 (Figure 4D). Therefore, it seems that this upstream region indeed suppresses CREB-1-mediated activation of the downstream promoter. These findings corroborate those of others that mapped a repressive element, corresponding to the region upstream of −2750, affecting the CCR5 promoter.33,35,37 This region is included in CCR5-pB1 and not in CCR5-pB3 and -pB4 (Figure 1).
Many genes contain consensus sites for CREB-1 binding and, as such, the transcription factor CREB-1 has been implicated in a wide variety of cellular processes.38 In the CNS CREB-1 plays an important role in various neuronal processes, such as neuronal development, neuroprotection, and disease, and various signaling pathways, including neurotrophin-mediated signaling, result in enhanced CREB-1 expression.39 In effect, CREB-1 plays an important role in the transcriptional control of antiapoptotic factors such as Bcl-2 after neurotrophin signaling.40 Furthermore, CREB-1 has also been implicated in axonal regeneration in the injured CNS.41 Previously, we have detected enhanced expression of CREB-1 in MS-affected microglia, which could reflect a stress signaling–induced up-regulation of CREB-1 transcription.12 In addition, a concomitant increase in CCR5 expression on MS-affected microglia has also been documented.10,11 The fact that we now have shown that CREB-1 is a bona fide inducer of CCR5 promoter activation is in line with these observations.
In conclusion, we show that CREB-1 is an important inducer of CCR5 expression. Interestingly, whereas CREB-1 is ubiquitously expressed and its expression can be induced by various stimuli, expression of CCR5 in healthy subjects is confined mainly to T lymphocytes, DCs, macrophages, monocytes, and microglia. This specificity of CCR5 expression could be the result of differences in signal transduction pathways that lead to differential phosphorylation and additional modifications of CREB-1, altering its transcriptional activation state.28,29 In addition, differential expression of CCR5 could be the result of cell-specific splicing of the CREM gene, giving rise to either activating or repressive CREM isoforms that could affect CREB-1 transactivation of the CCR5 promoter.36 However, these mechanisms are also common and therefore not likely to account for the cell-specific expression of CCR5. Thus, these data suggest the existence of additional regulatory constraints imposed on CCR5, which control tissue-specific expression. These could include DNA methylation of the CCR5 promoter region and/or association of specific histone modifications with this region, leading to a repressive chromatin status. Alterations in these regulatory mechanisms might also explain the aberrant expression of CCR5 in MS and several other inflammatory diseases.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
CCR5 promoter constructs were a kind gift of Prof S. K. Ahuja (University of Texas Health Science Center at San Antonio). Human monocyte-derived DCs were kindly provided by Dr T. B. H. Geijtenbeek (VU University Medical Center, Amsterdam, The Netherlands). The authors thank Prof F. Koning for critically reading the manuscript.
This work was supported by grants MS 00-407 and MS 04-543 from the Dutch MS Research Foundation (Voorschoten, The Netherlands).
Authorship
Contribution: H.F.K. performed experiments and wrote the manuscript; P.J.B. performed experiments; L.J.M. and E.S.v.H. provided the cultured primary glial cells; P.v.d.V. supervised the glial cell work; and P.J.v.d.E. had overall supervision and is responsible for the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter J. van den Elsen, Division of Molecular Biology, Department of Immunohematology and Blood Transfusion, Building 1, E3-Q, Leiden University Medical Center, Leiden, The Netherlands; e-mail: pjvdelsen@lumc.nl.