Abstract
Familial Mediterranean fever (FMF) is an autoinflammatory disease caused by mutations in MEFV, which encodes a 781–amino acid protein denoted pyrin. We have previously shown that pyrin regulates caspase-1 activation and IL-1β production through interaction of its N-terminal PYD motif with the ASC adapter protein, and also modulates IL-1β production by interaction of its C-terminal B30.2 domain with the catalytic domains of caspase-1. We now asked whether pyrin might itself be a caspase-1 substrate, and found that pyrin is cleaved by caspase-1 at Asp330, a site remote from the B30.2 domain. Pyrin variants harboring FMF-associated B30.2 mutations were cleaved more efficiently than wild-type pyrin. The N-terminal cleaved fragment interacted with the p65 subunit of NF-κB and with IκB-α through its 15-aa bZIP basic domain and adjacent sequences, respectively, and translocated to the nucleus. The interaction of the N-terminal fragment with p65 enhanced entrance of p65 into the nucleus. The interaction of N-terminal pyrin with IκB-α induced calpain-mediated degradation of IκB-α, thus potentiating NF-κB activation. Absolute and relative quantities of cleaved pyrin and IκB-α degradation products were substantially increased in leukocytes from FMF patients compared with healthy controls. Our data support a new pyrin/caspase-1 pathway for NF-κB activation.
Introduction
Familial Mediterranean fever (FMF, MIM249100) is a recessively inherited systemic autoinflammatory disorder characterized by short recurrent bouts of fever and serosal, synovial, or cutaneous inflammation, sometimes leading to AA amyloidosis. The gene responsible for FMF (designated MEFV for Mediterranean fever) was identified by positional cloning.1,2 The protein product of MEFV, denoted pyrin (or marenostrin), is a 781-aa protein expressed in neutrophils, eosinophils, monocytes, dendritic cells, and synovial fibroblasts.3,4 Nearly all of the disease-associated mutations are missense substitutions and most of the major mutations are clustered in the C-terminal B30.2 domain. Carrier frequencies as high as 1:3 have been observed in the Middle East.5 MEFV appears to play a pivotal role in the regulation of the systemic inflammatory response, as evidenced by the fact that high levels of MEFV expression have been correlated with adverse outcomes in critically ill children with multiple organ dysfunction syndrome.6
When it was first identified, pyrin was hypothesized to be a transcription factor, largely because of its homology with other known transcription factors and the computational deduction of 2 overlapping nuclear targeting signals.1 Subsequent transfection experiments have placed full-length pyrin in the cytoplasm, associated with the cytoskeleton.7,8 Native pyrin is predominantly nuclear in granulocytes, dendritic cells, and synovial fibroblasts, whereas it is cytoplasmic in monocytes.4
Another important clue to the function of pyrin came from the recognition that the N-terminal approximately 90 amino acids of pyrin define a motif, variously called the PYRIN domain,9 PYD,10 PAAD,11 or DAPIN,12 that has been found in several regulators of apoptosis and inflammation. Pyrin interacts with an adapter protein denoted apoptosis-associated speck-like protein with a caspase-recruitment domain (ASC)13 through homotypic interaction of their respective N-terminal PYD domains.14 ASC has been shown to oligomerize and mediate the proteolytic activation of caspase-1 in macromolecular complexes denoted inflammasomes.15,16 Pyrin modulates caspase-1 and IL-1β activation in part through its interactions with ASC. Studies of mice expressing a C-terminal truncation of pyrin and functional analyses of human pyrin demonstrate an inhibitory role17-19 under some experimental conditions. However, human pyrin may potentiate IL-1β production under other conditions.20-22
Pyrin may also have a role in the regulation of NF-κB activation in conjunction with ASC, as has been shown for several other PYD-containing proteins.23-30 In transfection studies, coexpression of pyrin with ASC has been shown to have positive,31,32 negative,33,34 or no regulatory effects20 on ASC-dependent NF-κB activation. Factors determining the effect of pyrin on NF-κB activation—whether dependent on or independent of ASC—remain unclear.
In the present paper, we explore a novel mechanism by which pyrin might be at the crossroads between caspase-1 activation and NF-κB signaling. The current line of investigation derives from recent observations that the C-terminal B30.2 domain of pyrin binds to the catalytic domains of caspase-1 and inhibits enzyme activity.18,19 We therefore hypothesized that, if pyrin binds directly to caspase-1, it might also be a substrate for caspase-1–mediated cleavage. Indeed, we found that caspase-1 cleaves pyrin at Asp330, producing a 330-residue N-terminal fragment that enhances ASC-independent NF-κB activation. Comparing the susceptibility of FMF-associated B30.2 pyrin mutants to cleavage with wild-type (WT) pyrin, we found increased cleavage in the mutants, suggesting another possible basis for the FMF autoinflammatory phenotype. Moreover, we found that the absolute and relative quantities of cleaved pyrin are substantially increased in peripheral blood mononuclear cells (PBMCs) from FMF patients compared with healthy controls. These data identify a new pyrin/caspase-1 pathway for NF-κB activation, and suggest a molecular basis for selection of pyrin mutants in humans.
Methods
Cleavage analysis of pyrin
All human samples were obtained with informed consent in accordance with the Declaration of Helsinki under a protocol approved by the Institutional Review Board of the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS, Bethesda, MD). For in vitro cleavage analysis, in vitro–translated 35S-labeled WT pyrin, which was produced by the TNT coupled transcription/translation kit (Promega, Madison, WI), was incubated with recombinant human caspase-1 (Calbiochem, San Diego, CA) at 37°C and subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography. WT and mutant pyrin proteins were produced from transfected PT67 cells (Clontech, Mountain View, CA). The cell lysates were incubated with 20 U recombinant human caspase-1 for 10 minutes at room temperature (RT), and analyzed by Western blotting.
For in vivo cleavage analysis, WT and mutant pyrin were cotransfected into PT67 cells with caspase-1. Cos-7 cells were cotransfected with WT pyrin, caspase-1, and IL-1β, and treated with various amounts of z-WEHD-fmk, a caspase-1 inhibitor (R&D Systems, Minneapolis, MN). After 24 hours, equal amounts of total protein were subjected to Western blot, and cell culture supernatants from Cos-7 cells were analyzed by IL-1β enzyme-linked immunosorbent assay (ELISA; R&D Systems).
To identify the cleavage site, PT67 cells were cotransfected with myc-tagged B30.2 domain–deleted pyrin (NBC-myc) and caspase-1. The C-terminal cleaved fragment was purified by immunoprecipitation (IP) using protein A–conjugated antimyc antibody (Pierce, Rockford, IL). Bound proteins were eluted and separated by SDS-PAGE followed by Western blot or Coomassie blue staining. The band corresponding to the C-terminal cleaved fragment was excised from the Coomassie blue–stained PVDF membrane, and subjected to N-terminal Edman sequencing.
Cleavage of endogenous pyrin
PBMCs were isolated by Ficoll-Hypaque centrifugation of freshly drawn peripheral venous blood from healthy controls and FMF patients in remission on colchicine therapy. All FMF patients met Tel-Hashomer clinical criteria for this disorder, and had given informed consent to participate in this study (National Institutes of Health [NIH, Bethesda, MD] protocol 94-AR-0105, approved by the National Institute of Diabetes and Digestive and Kidney Diseases [NIDDK, Bethesda, MD]/NIAMS Intramural Institutional Review Board). The patients' genotypes were M694V/M694V, V726A/K695R, M694I/M694I, M694V/M680I, M694V/V726A, M694V/−, M694V/−, M694V/V726A, V726A/R761H, V726A/−, V726A-E148Q/E148Q, V726A-E148Q/−, V726A-E148Q/V726A, M694V/E148Q, M694V/M680I, and V726A/V726A, representing the spectrum of common FMF genotypes. A total of 12 μg protein was subjected to Western blotting with a polyclonal antipyrin Ab that was developed against the N-terminal 374 aa's of pyrin.18 The intensity of uncleaved and cleaved bands was compared by densitometry (Bio-Rad GS800 Scanner, Quantity One software; Bio-Rad, Hercules, CA). Means and standard errors were calculated and compared using the Student t statistic.
Co-IP, Western blots, and GST pull-down assays
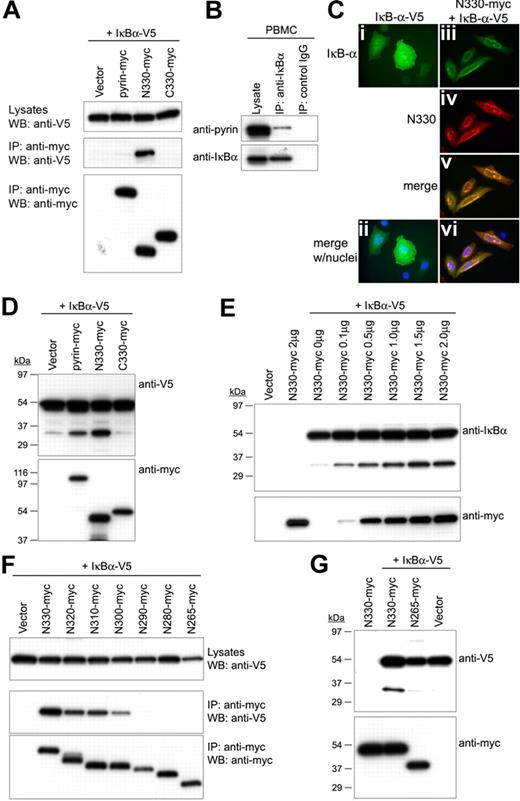
PBMCs from healthy donors were lysed with M-PER mammalian protein extraction buffer (Pierce) and incubated for 16 hours at 4°C with agarose-conjugated anti-p65 Ab, anti–IκB-α Ab, or control IgG (Santa Cruz Biotechnology, Santa Cruz, CA). After washing the beads 4× with the same lysis buffer, bound proteins were eluted with 2 × SDS sample buffer. Co-IP of N330 and endogenous p65 were also performed from U937 cells stably expressing N330, which were established as in a previous study.17 For in vivo glutathione-S-transferase (GST) pull down and co-IP, PT67 cells were cotransfected with each of the myc- or GST-tagged pyrin variants with myc- or V5-tagged p50, p65, and IκB-α; and each of the myc-tagged 10-aa deleted N-terminal fragments from aa 330 to aa 266, with V5-tagged p50, p65, and IκB-α. After 24 hours, cell lysates were incubated with glutathione Sepharose 4B (GE Healthcare, Piscataway, NJ) for GST pull down or agarose-conjugated antimyc Ab (Clontech) for co-IP at 4°C. After 16 hours, the beads were washed (4×) and eluted with 2 × SDS sample buffer, and subjected to Western blotting.
Immunofluorescence analysis
HeLa cells were cultured in 2-well chamber slides. Myc-tagged full-length pyrin, N330, or C330 was transiently transfected or cotransfected with V5-tagged p65 or IκB-α using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). After 24 hours, cells were fixed with 4% paraformaldehyde in PBS, and stained with AlexaFluor 488–conjugated anti-V5 Ab and AlexaFluor 568–conjugated anti-myc Ab. Cells were counterstained with DAPI, and visualized on a Leica DMR microscope (Heidelberg, Germany) with a HCX PL APO 63×/1.32-0.6 oil-immersion objective lens. The images were acquired using Magnafire software (Olympus America, Center Valley, PA) and were processed using Adobe Photoshop CS2 (Adobe Systems, San Jose, CA).
NF-κB DNA-binding activity assays
Electromobility shift assay (EMSA) was used to measure NF-κB DNA-binding activity. From the transfected HeLa cells, nuclear extracts were produced using NE-PER nuclear and cytoplasmic extraction reagents (Pierce). Nuclear extracts (4 μg) were incubated with a 32P-end–labeled double-strand consensus NF-κB oligonucleotide (Promega) probe. For competition assays, a 50× molar excess of unlabeled NF-κB consensus or mutant oligonucleotide (Santa Cruz Biotechnology) was added. DNA-protein complexes were separated by nondenaturing PAGE, and analyzed by autoradiography.
IκB-α cleavage analysis
HeLa cells were cotransfected with myc-tagged pyrin variants and V5-tagged IκB-α. Cells were also treated with an increasing amount of calpain inhibitors (Calbiochem), ubiquitination inhibitor (Calbiochem), or colchicine (Sigma-Aldrich, St Louis, MO). Total protein (20 μg) was subjected to Western blotting with anti-V5 or anti–IκB-α Abs (Cell Signaling Technology, Danvers, MA). PBMCs from FMF patients and healthy controls were lysed immediately after purification or after 24 hours' treatment with IFN-γ, and subjected to Western blotting.
Computational analysis of subcellular localization
The potential subcellular localization of the N330 pyrin fragment was computationally analyzed with the following programs: WolF-PSORT (National Institute of Advanced Science and Technology, Computational Biology Research Center, Tsukuba, Japan; http://wolfpsort.org), PSORT II (Human Genome Center, Institute for Medical Science, University of Tokyo, Tokyo, Japan; http://psort.nibb.ac.jp/form2.html), SubLoc v1.0 (Virginia Bioinformatics Institute, Virginia Polytechnic Institute and State University, Blacksburg; http://www.bioinfo.tsinghua.edu.cn/SubLoc/eu_predict.htm), P2SL (Institute of Bioinformatics, Tsinghua University, Beijing, China; http://www.i-cancer.org/p2sl/), and the subnuclear compartments prediction system (Bioinformatics Program, Department of Bioengineering, University of Illinois at Chicago; http://array.bioengr.uic.edu/subnuclear.htm).
Results
Pyrin is cleaved by caspase-1
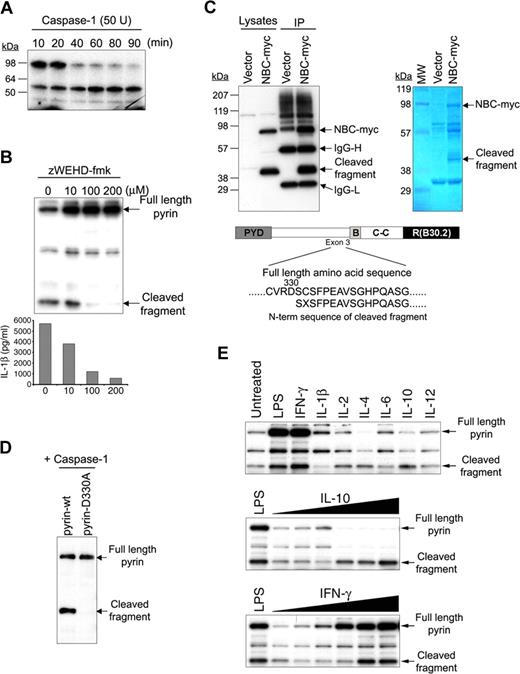
We have previously demonstrated a direct interaction between pyrin and caspase-1.18 Since caspase-1 is a proteolytic enzyme, it was possible that pyrin could also be a substrate of caspase-1. To test this hypothesis, 35S-labeled pyrin was synthesized by in vitro transcription and translation, and incubated with 50 units of caspase-1 for various time intervals (Figure 1A). Increasing caspase-1 incubation times produced additional smaller molecular weight fragments, and the amount of full-length pyrin was decreased. To examine the caspase-1–mediated cleavage of pyrin in vivo, Cos7 cells were cotransfected with constructs expressing pyrin, caspase-1, and pro–IL-1β and treated with various amounts of the specific caspase-1 inhibitor z-WEHD-fmk (Figure 1B). Western blots were probed with a polyclonal Ab developed against the N-terminal 374 aa's of human pyrin.18 Without the inhibitor, cells produced an approximately 50-kDa pyrin cleavage fragment in addition to full-length pyrin, and secreted high levels of IL-1β. However both the generation of cleaved pyrin and the secretion of IL-1β were reduced by the caspase-1 inhibitor in a dose-dependent manner.
Cleavage of pyrin by caspase-1. (A) 35S-labeled pyrin was incubated with 50 units recombinant caspase-1 at 37°C for various periods as indicated, subjected to SDS-PAGE, and visualized by autoradiography. (B) Pyrin was cotransfected into Cos-7 cells with caspase-1 and pro–IL-1β. Cells were treated with increasing amounts of z-WEHD-fmk as indicated. After 24 hours, cell lysates underwent SDS-PAGE and Western blotting with a polyclonal Ab developed against the N-terminal 374 aa of pyrin (top). Cell culture supernatants were also collected and assayed for IL-1β by ELISA (bottom). (C) PT67 cells were cotransfected with pNBC-myc (construct expressing B30.2 domain–deleted pyrin) and with caspase-1. After 24 hours, cell lysates were immunoprecipitated using antimyc Ab, and subjected to SDS-PAGE followed by Western blotting with antimyc Ab (top left panel) or Coomassie blue staining (top right panel). The result of N-terminal Edman sequencing of the eluted, Coomassie-stained band corresponding to the C-terminal cleaved fragment is shown under the schematic diagram of pyrin. (D) WT or D330A pyrin was cotransfected with caspase-1 into PT67 cells. Lysates were subjected to SDS-PAGE and Western blotting with the same antipyrin Ab as in panel B. (E) PBMCs from healthy controls were cultured in RPMI supplemented with 10% fetal bovine serum in 6-well culture plates, treated with LPS (1 μg/mL) from E coli 0127:B7 (Sigma-Aldrich), IFN-γ (100 ng/mL), IL-1β (3.75 ng/mL), IL-2 (1.6 ng/mL), IL-4 (12.5 ng/mL), IL-6 (8.5 ng/mL), IL-10 (100 ng/mL) or IL-12 (10 ng/mL; BD Biosciences, San Jose, CA). After 24 hours, cells were lysed and 12 μg total protein was subjected to SDS-PAGE followed by Western blot with the same antipyrin Ab as in panel B (top). PBMCs were also stimulated with LPS or various amounts (from 10 pg/mL to 100 ng/mL) of IL-10 (middle) or IFN-γ (bottom) for 24 hours. Cells were lysed and analyzed as in top panel.
Cleavage of pyrin by caspase-1. (A) 35S-labeled pyrin was incubated with 50 units recombinant caspase-1 at 37°C for various periods as indicated, subjected to SDS-PAGE, and visualized by autoradiography. (B) Pyrin was cotransfected into Cos-7 cells with caspase-1 and pro–IL-1β. Cells were treated with increasing amounts of z-WEHD-fmk as indicated. After 24 hours, cell lysates underwent SDS-PAGE and Western blotting with a polyclonal Ab developed against the N-terminal 374 aa of pyrin (top). Cell culture supernatants were also collected and assayed for IL-1β by ELISA (bottom). (C) PT67 cells were cotransfected with pNBC-myc (construct expressing B30.2 domain–deleted pyrin) and with caspase-1. After 24 hours, cell lysates were immunoprecipitated using antimyc Ab, and subjected to SDS-PAGE followed by Western blotting with antimyc Ab (top left panel) or Coomassie blue staining (top right panel). The result of N-terminal Edman sequencing of the eluted, Coomassie-stained band corresponding to the C-terminal cleaved fragment is shown under the schematic diagram of pyrin. (D) WT or D330A pyrin was cotransfected with caspase-1 into PT67 cells. Lysates were subjected to SDS-PAGE and Western blotting with the same antipyrin Ab as in panel B. (E) PBMCs from healthy controls were cultured in RPMI supplemented with 10% fetal bovine serum in 6-well culture plates, treated with LPS (1 μg/mL) from E coli 0127:B7 (Sigma-Aldrich), IFN-γ (100 ng/mL), IL-1β (3.75 ng/mL), IL-2 (1.6 ng/mL), IL-4 (12.5 ng/mL), IL-6 (8.5 ng/mL), IL-10 (100 ng/mL) or IL-12 (10 ng/mL; BD Biosciences, San Jose, CA). After 24 hours, cells were lysed and 12 μg total protein was subjected to SDS-PAGE followed by Western blot with the same antipyrin Ab as in panel B (top). PBMCs were also stimulated with LPS or various amounts (from 10 pg/mL to 100 ng/mL) of IL-10 (middle) or IFN-γ (bottom) for 24 hours. Cells were lysed and analyzed as in top panel.
Aspartate is an important residue for caspase-1 recognition, and the pyrin protein contains several potential caspase-1 recognition sites between residues 300 and 335. To determine the actual cleavage site, we performed myc-specific IPs on lysates of PT67 cells cotransfected with caspase-1 and a B30.2-truncated pyrin tagged with myc at its C-terminus (NBC-myc), chosen to separate the myc-tagged C-terminal cleaved fragment from IgG heavy chain, which has a molecular weight similar to the C-terminal cleaved fragment produced from full-length pyrin. Western analysis with antimyc Abs (Figure 1C) demonstrated the presence of both the intact B30.2-truncated pyrin (NBC-myc) and its expected C-terminal approximately 40 kDa cleavage fragment (which is smaller than the ∼ 60 kDa C-terminal cleaved fragment of full-length pyrin because the B30.2 domain is absent). The band corresponding to the cleaved fragment was excised from a Coomassie-stained membrane and subjected to Edman N-terminal sequencing, which demonstrated that the primary caspase-1 cleavage site of pyrin is between Asp330 and Ser331, encoded within exon 3. When Asp330 was mutated to alanine (pyrin-D330A) and cotransfected with caspase-1, no cleavage was detected (Figure 1D).
We also examined the cleavage of endogenous pyrin in unstimulated and LPS- or cytokine-treated PBMCs from several individuals. Endogenous pyrin can also be cleaved (Figure 1E), although there is considerable individual variation (Figure 2C). Moreover, we observed complete removal of full-length pyrin in the cells treated with increasing doses of IL-10, and increases in the amounts of both full-length and cleaved pyrin in cells treated with IFN-γ.
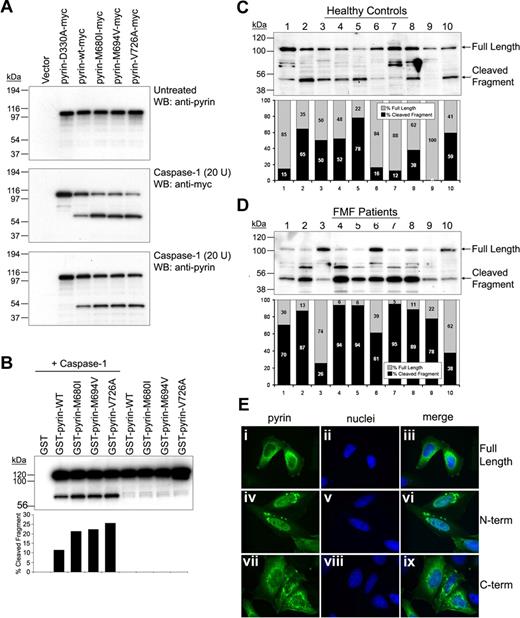
Cleavage of FMF-associated mutant pyrins by caspase-1 and cellular localization of cleaved fragments. (A) Myc-tagged D330A, WT, or FMF-associated mutant pyrin was transfected into PT67 cells. After 24 hours, cell lysates (10 μg total protein) underwent Western blotting with antipyrin Ab (top). The same amounts of lysates were incubated with 20 units of recombinant caspase-1 for 10 minutes at room temperature. The incubated cell lysates were subjected to Western blotting with antimyc, detecting the C-terminal cleavage fragment (middle), and antipyrin Abs, detecting the smaller N-terminal cleavage fragment (bottom). (B) GST-fused WT or mutant pyrin was cotransfected into PT67 cells with (lanes 1-5) or without (lanes 6-9) caspase-1. Cell lysates were analyzed by Western blots using antipyrin Ab (top panel). The cleaved and uncleaved pyrin fragments were quantified by densitometry and represented as percentage of total pyrin (bottom panel). (C,D) Lysates of PBMCs from 10 healthy individuals (C) and 10 FMF patients (D) were subjected to SDS-PAGE, and Western blotting with antipyrin Ab. Exposure times were the same for panels C and D. The full-length and cleaved pyrin were quantified by densitometry and represented as percentage of total pyrin. (E) HeLa cells were transfected with myc-tagged full-length pyrin (i-iii), N-terminal cleaved pyrin (iv-vi), or C-terminal cleaved pyrin (vii-ix). After 24 hours, cells were fixed with 4% paraformaldehyde in PBS, and stained with AlexaFluor 488–conjugated antimyc Ab (green), counterstained with DAPI (blue), and visualized on a Leica DMR microscope. Pyrin variants are shown in the first column (i, iv, and vii); nuclei are in the second column (ii, v, and viii); merged images are in the third column (iii, vi, and ix).
Cleavage of FMF-associated mutant pyrins by caspase-1 and cellular localization of cleaved fragments. (A) Myc-tagged D330A, WT, or FMF-associated mutant pyrin was transfected into PT67 cells. After 24 hours, cell lysates (10 μg total protein) underwent Western blotting with antipyrin Ab (top). The same amounts of lysates were incubated with 20 units of recombinant caspase-1 for 10 minutes at room temperature. The incubated cell lysates were subjected to Western blotting with antimyc, detecting the C-terminal cleavage fragment (middle), and antipyrin Abs, detecting the smaller N-terminal cleavage fragment (bottom). (B) GST-fused WT or mutant pyrin was cotransfected into PT67 cells with (lanes 1-5) or without (lanes 6-9) caspase-1. Cell lysates were analyzed by Western blots using antipyrin Ab (top panel). The cleaved and uncleaved pyrin fragments were quantified by densitometry and represented as percentage of total pyrin (bottom panel). (C,D) Lysates of PBMCs from 10 healthy individuals (C) and 10 FMF patients (D) were subjected to SDS-PAGE, and Western blotting with antipyrin Ab. Exposure times were the same for panels C and D. The full-length and cleaved pyrin were quantified by densitometry and represented as percentage of total pyrin. (E) HeLa cells were transfected with myc-tagged full-length pyrin (i-iii), N-terminal cleaved pyrin (iv-vi), or C-terminal cleaved pyrin (vii-ix). After 24 hours, cells were fixed with 4% paraformaldehyde in PBS, and stained with AlexaFluor 488–conjugated antimyc Ab (green), counterstained with DAPI (blue), and visualized on a Leica DMR microscope. Pyrin variants are shown in the first column (i, iv, and vii); nuclei are in the second column (ii, v, and viii); merged images are in the third column (iii, vi, and ix).
FMF-associated mutants are cleaved more than WT pyrin by caspase-1
Pyrin inhibits the catalytic activity of caspase-1 through its B30.2 domain,18 and, although still a matter of debate,19 the inhibitory activity may be diminished in pyrin variants with B30.2 domain mutations. Thus it was possible that the cleavage of pyrin by caspase-1 could be affected by mutations in the B30.2 domain (aa's 577-757), although it is remote from the cleavage site, Asp330. Figure 2A demonstrates increased C-terminal (middle panel) and N-terminal (bottom panel) cleavage products of FMF pyrin mutants subjected to in vitro catalysis by caspase-1. Figure 2B shows increased susceptibility of mutant pyrins to caspase-1 cleavage in a cotransfection system, using antipyrin Ab that detects the GST-conjugated N-terminal fragment.
We also compared the cleavage of endogenous pyrin in leukocytes from either healthy controls (Figure 2C) or FMF patients (Figure 2D). Lysates from freshly isolated PBMCs were electrophoresed and Western blots were probed with an antiserum specific for N-terminal pyrin and analyzed by densitometry. In 16 FMF patients with B30.2 mutations sampled while not in an attack (10 shown in Figure 2D), 71.47% plus or minus 7.33% of the pyrin signal was in the cleaved band, versus 36.77% plus or minus 9.74% in 15 healthy controls (10 shown in Figure 2C; P = .004). The degree of pyrin cleavage did not correlate with specific mutations. Variability in the amounts of cleaved pyrin in leukocyte lysates from both healthy controls and FMF patients may be the consequence of the in vivo cytokine milieu, as seen in Figure 1E, or may be due to as yet unidentified genetic factors.
N-terminal pyrin localizes to the nucleus and potentiates NF-κB activation
To examine the consequences of pyrin cleavage, we transfected constructs expressing myc-tagged N-terminal pyrin (aa's 1-330, N330), C-terminal pyrin (aa's 331-781, C330), or full-length pyrin into PT67 cells, and the expressed proteins were visualized by immunostaining with antimyc Ab. Consistent with previous observations, full-length transfected pyrin localized in the cytoplasm, as did the C330 (Figure 2E). In contrast, N330 was found in both the nucleus and the cytoplasm, appearing as a distinctive filamentous network in the nucleus. Similar results were obtained with PT67 cells expressing GFP-tagged N330 construct (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
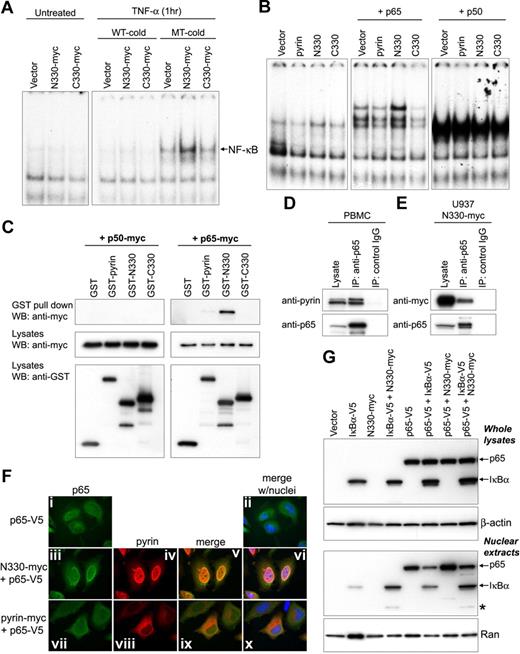
The nuclear localization of the N-terminal fragment suggested a possible role of pyrin as a nuclear factor. Moreover, it has been reported that pyrin modulates the effect of ASC on NF-κB activation when coexpressed in HEK293T cells.31,32,34 We therefore examined the effect of N330 or C330 cleaved pyrin on NF-κB DNA-binding activity from nuclear lysates of HeLa cells, as measured by EMSAs using a NF-κB consensus oligonucleotide. After 1 hour of treatment with TNF-α, the transfection of N330 resulted in more NF-κB activation than vector control or C330, whereas no NF-κB activation was detected without any stimulation (Figure 3A). To identify the NF-κB subunits involved in pyrin-mediated activation, pyrin constructs were cotransfected with p65 or p50. N330 increased the binding activity of p65, but not p50, relative to vector control, and there was no activation within the cells cotransfected with full-length pyrin or C330 (Figure 3B).
N-terminal cleaved pyrin activates NF-κB and interacts with p65. (A) NF-κB DNA-binding activity was measured by EMSA in nuclear lysates prepared from HeLa cells that had been transfected with myc-tagged empty vector, N330, or C330, and incubated with or without TNF-α (12.5 ng/mL) for 1 hour. As indicated, either unlabeled excess NF-κB wt (WT-cold) or mutant (MT-cold) oligonucleotides were added to the EMSA reactions as a competitor to assay binding specificity. (B) HeLa cells were cotransfected with the indicated pyrin constructs and either p65 or p50. From the nuclear lysates, NF-κB DNA-binding activity was measured by EMSA as in panel A. (C) Myc-tagged p50 (left panels) or p65 (right panels) was cotransfected into PT67 cells with GST vector, GST-fused pyrin, GST-N330, or GST-C330. Lysates were subjected to GST pull-down assay followed by Western blotting with antimyc Ab (top panels). Expression of transfected constructs is shown in the bottom 2 panels (cell lysates) probed with antimyc or anti-GST Abs. (D) Lysates from PBMCs of a healthy donor were immunoprecipitated with anti-p65 Ab or control IgG. Cell lysates and eluted proteins were analyzed by Western blotting with antipyrin (top) or anti-p65 Abs (bottom). Data are from a representative experiment from 3 separate subjects. (E) U937 cells were transduced with retroviral constructs expressing N330-myc or with empty vector. After G418 selection, cell lysates were immunoprecipitated with anti-p65 Ab or control IgG, and analyzed as in panel D. (F) HeLa cells were transfected with V5-tagged p65 (p65-V5) alone (i-ii), or cotransfected with p65-V5 and N330-myc (iii-vi), or with p65-V5 and myc-tagged full-length pyrin (vii-xi). After 24 hours, p65 and pyrin variants were stained with AlexaFluor 488–conjugated anti-V5 Ab (green) and AlexaFluor 568–conjugated antimyc Ab (red), respectively. Cells were counterstained with DAPI (blue) and visualized on a Leica DMR microscope. Expressed p65 is shown in the first column (i, iii, and vii); pyrin variants are shown in the second column (iv and viii); merged images for p65 and pyrin variants are in the third column (v and ix); merged images for p65, pyrin variants, and nuclei are in the fourth column (ii, vi, and x). (G) IκB-α–V5, p65-V5, and N330-myc were transfected individually, or cotransfected as indicated into HeLa cells. After 24 hours, total cell lysates (top 2 panels) and nuclear extracts (bottom 2 panels) were prepared and subjected to SDS-PAGE for Western blotting with anti-V5 Ab (top panel and third panel), anti–β-actin Ab (for loading control of whole-cell extracts), or anti-Ran Ab (for loading control of nuclear extracts). Asterisk denotes degraded 30-kDa IκB-α fragment that is explained in “N330 interacts with p65 and enhances the nuclear localization of p65.”
N-terminal cleaved pyrin activates NF-κB and interacts with p65. (A) NF-κB DNA-binding activity was measured by EMSA in nuclear lysates prepared from HeLa cells that had been transfected with myc-tagged empty vector, N330, or C330, and incubated with or without TNF-α (12.5 ng/mL) for 1 hour. As indicated, either unlabeled excess NF-κB wt (WT-cold) or mutant (MT-cold) oligonucleotides were added to the EMSA reactions as a competitor to assay binding specificity. (B) HeLa cells were cotransfected with the indicated pyrin constructs and either p65 or p50. From the nuclear lysates, NF-κB DNA-binding activity was measured by EMSA as in panel A. (C) Myc-tagged p50 (left panels) or p65 (right panels) was cotransfected into PT67 cells with GST vector, GST-fused pyrin, GST-N330, or GST-C330. Lysates were subjected to GST pull-down assay followed by Western blotting with antimyc Ab (top panels). Expression of transfected constructs is shown in the bottom 2 panels (cell lysates) probed with antimyc or anti-GST Abs. (D) Lysates from PBMCs of a healthy donor were immunoprecipitated with anti-p65 Ab or control IgG. Cell lysates and eluted proteins were analyzed by Western blotting with antipyrin (top) or anti-p65 Abs (bottom). Data are from a representative experiment from 3 separate subjects. (E) U937 cells were transduced with retroviral constructs expressing N330-myc or with empty vector. After G418 selection, cell lysates were immunoprecipitated with anti-p65 Ab or control IgG, and analyzed as in panel D. (F) HeLa cells were transfected with V5-tagged p65 (p65-V5) alone (i-ii), or cotransfected with p65-V5 and N330-myc (iii-vi), or with p65-V5 and myc-tagged full-length pyrin (vii-xi). After 24 hours, p65 and pyrin variants were stained with AlexaFluor 488–conjugated anti-V5 Ab (green) and AlexaFluor 568–conjugated antimyc Ab (red), respectively. Cells were counterstained with DAPI (blue) and visualized on a Leica DMR microscope. Expressed p65 is shown in the first column (i, iii, and vii); pyrin variants are shown in the second column (iv and viii); merged images for p65 and pyrin variants are in the third column (v and ix); merged images for p65, pyrin variants, and nuclei are in the fourth column (ii, vi, and x). (G) IκB-α–V5, p65-V5, and N330-myc were transfected individually, or cotransfected as indicated into HeLa cells. After 24 hours, total cell lysates (top 2 panels) and nuclear extracts (bottom 2 panels) were prepared and subjected to SDS-PAGE for Western blotting with anti-V5 Ab (top panel and third panel), anti–β-actin Ab (for loading control of whole-cell extracts), or anti-Ran Ab (for loading control of nuclear extracts). Asterisk denotes degraded 30-kDa IκB-α fragment that is explained in “N330 interacts with p65 and enhances the nuclear localization of p65.”
N330 interacts with p65 and enhances the nuclear localization of p65
We also examined the interactions of full-length or cleaved pyrin fragments with NF-κB subunits using in vivo GST pull-down analysis of lysates from PT67 cells cotransfected with GST-tagged pyrin constructs and myc-tagged p50 or p65. The p65 subunit interacted with pyrin, and specifically bound to N330 much more than full-length pyrin, whereas p50 did not interact with any of the pyrin constructs (Figure 3C). The interaction of endogenous pyrin and p65 was confirmed by co-IP from the lysates of healthy donor PBMCs (Figure 3D). The interaction of transduced N330 with endogenous p65 was also demonstrated in differentiated U937 cells that stably expressed N330 (Figure 3E).
The foregoing data showing the nuclear localization of N330 and the interaction of N330 with p65 suggested the possibility that N330 might facilitate the entrance of p65 into the nucleus. To test this hypothesis, myc-tagged N330 or full-length pyrin was cotransfected with V5-tagged p65 into HeLa cells. Immunostaining of the cells showed the colocalization of p65 and N330 within the nucleus, whereas p65 remained mostly in the cytoplasm with full-length pyrin (Figure 3F). The increased nuclear localization of p65 by N330 was confirmed by Western blot from the nuclear extracts of transfected cells (Figure 3G). Higher levels of p65 were detected in the nuclear extract from cells cotransfected with p65 and N330 (Figure 3G lane 7) than p65 alone (lane 5). Moreover, although the amount of p65 in the nucleus was decreased by the cotransfection of IκB-α (Figure 3G lane 6 vs lane 5), the relative amount of p65 in the nucleus was increased when N330 was present (lane 8 vs lane 6).
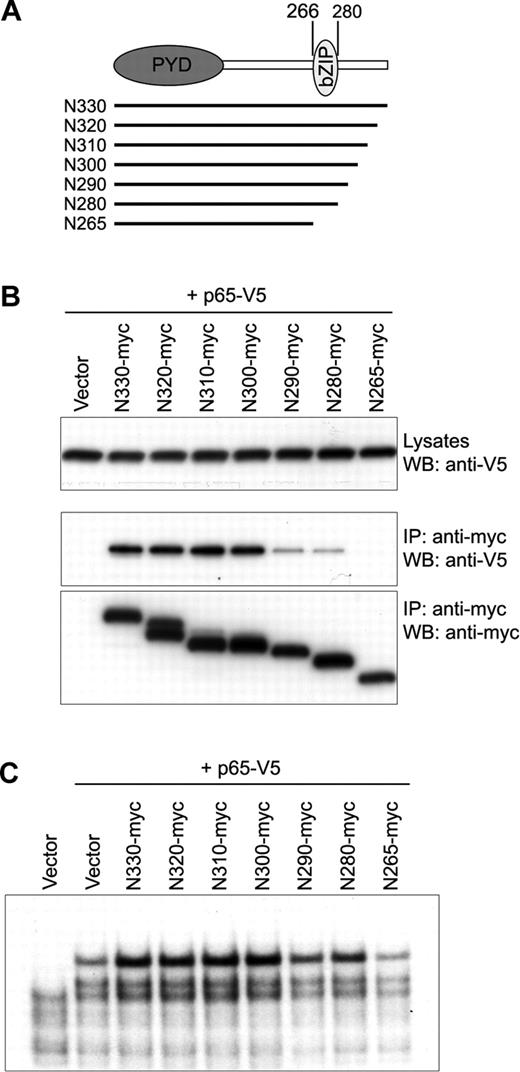
Within N330, 2 known domains have been identified—the PYD and bZIP basic (aa's 266-280) domains. Because of the known interactions between bZIP proteins and Rel family transcription factors,35 we examined the role of the bZIP basic domain of N330 in the interaction with p65. PT67 cells were cotransfected with V5-tagged p65 and various myc-tagged, 10–amino acid (aa) deleted N-terminal pyrin fragments from aa 330 to aa 266, where the bZIP basic domain starts (Figure 4A). Co-IP showed that the interaction started decreasing when C-terminal 40 aa had been deleted from the N-terminal fragment (N290), and completely disappeared in the cells cotransfected with N-terminal constructs in which the whole bZIP basic domain had been deleted (N265; Figure 4B). We also examined the NF-κB activation induced by the interactions of cotransfected p65 and N-terminal pyrin deletion fragments by EMSA of HeLa cell nuclear extracts. As shown in Figure 4C, the binding activity of p65 on DNA was reduced in the cells cotransfected with the N290 or N280, which showed reduced binding with p65, and decreased to the level of vector control in the cells transfected with N265.
Interaction of the bZIP basic domain of N330 with p65. (A) Sequential 10-aa deleted N-terminal fragments from aa's 330 to 266 are indicated by lines under the schematic of N330 in which the PYRIN domain (PYD) and bZIP basic domain (bZIP) reside. The first and last aa numbers of bZIP basic domain are indicated on the top. (B) PT67 cells were cotransfected with p65-V5 and the 10-aa deleted N-terminal fragments depicted in panel A. Lysates were immunoprecipitated with antimyc Ab followed by Western blotting with anti-V5 (middle) and antimyc (bottom) Abs. Expression of p65-V5 in whole lysates is shown by anti-V5 Ab (top). (C) HeLa cells were cotransfected with p65-V5 and the 10-aa deleted N-terminal fragments depicted in panel A. From the nuclear lysates, NF-κB DNA-binding activity was measured by EMSA as in Figure 3A.
Interaction of the bZIP basic domain of N330 with p65. (A) Sequential 10-aa deleted N-terminal fragments from aa's 330 to 266 are indicated by lines under the schematic of N330 in which the PYRIN domain (PYD) and bZIP basic domain (bZIP) reside. The first and last aa numbers of bZIP basic domain are indicated on the top. (B) PT67 cells were cotransfected with p65-V5 and the 10-aa deleted N-terminal fragments depicted in panel A. Lysates were immunoprecipitated with antimyc Ab followed by Western blotting with anti-V5 (middle) and antimyc (bottom) Abs. Expression of p65-V5 in whole lysates is shown by anti-V5 Ab (top). (C) HeLa cells were cotransfected with p65-V5 and the 10-aa deleted N-terminal fragments depicted in panel A. From the nuclear lysates, NF-κB DNA-binding activity was measured by EMSA as in Figure 3A.
N-terminal pyrin interacts with IκB-α and induces its proteolysis
The foregoing data suggested that N330 enhances nuclear localization of p65. However, NF-κB proteins are bound by IκBs that prevent their entry into the nucleus and thereby prevent binding to target DNA sequences. Indeed, as shown in Figure 3G, the amount of p65 in the nucleus was decreased by transfected IκB-α. To bind target DNA sequences (κB sites), p65 must first be dissociated from IκB, which is then degraded. Thus, we also examined the role of pyrin in the degradation of IκB-α. We first tested pyrin–IκB-α interactions by IP analysis of lysates from PT67 cells cotransfected with myc-tagged pyrin constructs and V5-tagged IκB-α. Similar to its interactions with p65, N330 interacted with IκB-α more than full-length pyrin (Figure 5A). The interaction of endogenous pyrin and IκB-α was also confirmed by co-IP assay from healthy donor PBMCs (Figure 5B). Moreover, immunostaining of the cells cotransfected with N330 and IκB-α showed enhanced nuclear localization of IκB-α as well as colocalization of N330 and IκB-α (Figure 5C).
Interaction of N330 with IκB-α and the N330-induced proteolysis of IκB-α. (A) V5-tagged IκB-α was cotransfected into PT67 cells with myc-tagged pyrin, N330, or C330 or empty vector. Lysates were immunoprecipitated with antimyc Ab and analyzed as in Figure 4B. (B) Lysates from PBMCs of a healthy donor were immunoprecipitated with anti–IκB-α Ab or control IgG. Cell lysates and eluted proteins were analyzed by Western blotting with antipyrin (top) or anti–IκB-α Abs (bottom). Data are from a representative experiment from 3 separate donors. (C) HeLa cells were transfected with V5-tagged IκB-α (IκB-α–V5) alone (i,ii), or cotransfected with IκB-α–V5 and N330-myc (iii-vi). After 24 hours, IκB-α and N330 were stained with AlexaFluor 488–conjugated anti-V5 Ab (green) and AlexaFluor 568–conjugated antimyc Ab (red), respectively. Cells were counterstained with DAPI (blue) and visualized on a Leica DMR microscope. Expressed IκB-α is shown in panels i, ii (merged with nuclei), and iii, and N330 is in panel iv. Merged images for IκB-α and N330 without and with nuclei are in panels v and vi, respectively. (D) HeLa cells were cotransfected with IκB-α–V5 and myc-tagged pyrin variants or empty vector. Cell lysates were analyzed by Western blotting with anti-V5 (top) or antimyc Abs (bottom). (E) HeLa cells were cotransfected with IκB-α–V5 and increasing amount of N330-myc, and also transfected with empty vector or N330-myc alone. Cell lysates were analyzed by Western blotting with anti–IκB-α (top) or antimyc Abs (bottom). (F) PT67 cells were cotransfected with IκB-α–V5 and the 10-aa deleted N-terminal fragments depicted in Figure 4A. Lysates were immunoprecipitated with antimyc Ab and analyzed as in Figure 4B. (G) HeLa cells were cotransfected with IκB-α–V5 and N330-myc, N265-myc, or empty vector, and also transfected with N330-myc alone. Cell lysates were analyzed by Western blotting with anti-V5 (top) or antimyc Abs (bottom).
Interaction of N330 with IκB-α and the N330-induced proteolysis of IκB-α. (A) V5-tagged IκB-α was cotransfected into PT67 cells with myc-tagged pyrin, N330, or C330 or empty vector. Lysates were immunoprecipitated with antimyc Ab and analyzed as in Figure 4B. (B) Lysates from PBMCs of a healthy donor were immunoprecipitated with anti–IκB-α Ab or control IgG. Cell lysates and eluted proteins were analyzed by Western blotting with antipyrin (top) or anti–IκB-α Abs (bottom). Data are from a representative experiment from 3 separate donors. (C) HeLa cells were transfected with V5-tagged IκB-α (IκB-α–V5) alone (i,ii), or cotransfected with IκB-α–V5 and N330-myc (iii-vi). After 24 hours, IκB-α and N330 were stained with AlexaFluor 488–conjugated anti-V5 Ab (green) and AlexaFluor 568–conjugated antimyc Ab (red), respectively. Cells were counterstained with DAPI (blue) and visualized on a Leica DMR microscope. Expressed IκB-α is shown in panels i, ii (merged with nuclei), and iii, and N330 is in panel iv. Merged images for IκB-α and N330 without and with nuclei are in panels v and vi, respectively. (D) HeLa cells were cotransfected with IκB-α–V5 and myc-tagged pyrin variants or empty vector. Cell lysates were analyzed by Western blotting with anti-V5 (top) or antimyc Abs (bottom). (E) HeLa cells were cotransfected with IκB-α–V5 and increasing amount of N330-myc, and also transfected with empty vector or N330-myc alone. Cell lysates were analyzed by Western blotting with anti–IκB-α (top) or antimyc Abs (bottom). (F) PT67 cells were cotransfected with IκB-α–V5 and the 10-aa deleted N-terminal fragments depicted in Figure 4A. Lysates were immunoprecipitated with antimyc Ab and analyzed as in Figure 4B. (G) HeLa cells were cotransfected with IκB-α–V5 and N330-myc, N265-myc, or empty vector, and also transfected with N330-myc alone. Cell lysates were analyzed by Western blotting with anti-V5 (top) or antimyc Abs (bottom).
The N330-induced translocation of IκB-α into the nucleus was also observed in the Western blot of the nuclear fraction from the cotransfected HeLa cells (Figure 3G). In addition, in the Western blot for IκB-α using anti-V5, lower molecular weight bands (∼30 kDa) were detected only in the lysates of the cells cotransfected with approximately 50 kDa N330 (Figure 3G lanes 4 and 8, band denoted with an asterisk). We next compared the levels of the 30-kDa fragments that were produced in the cells cotransfected with full-length pyrin, N330, or C330 (Figure 5D). The 30-kDa fragment was produced at low level in the absence of pyrin (Figure 5D lane 1). The levels of the 30-kDa fragment were increased with full-length pyrin and to an even greater degree with N330, but not with C330. To confirm the identity of the 30-kDa fragment, IκB-α was cotransfected into HeLa cells with increasing amounts of N330. The 30-kDa fragment was readily detectable with anti–IκB-α Abs (Figure 5E), and was proportionate to the amount of N330 pyrin transfected. These data suggest that N330 pyrin induces IκB-α proteolysis.
To determine the binding domain of N330 with IκB-α, we performed co-IP using the N-terminal deletion fragments shown in Figure 4A. In contrast with the results for p65, the interaction of IκB-α with N-terminal pyrin started to decrease with the first 10-aa deletion (N320), and was completely abolished after 40-aa deletion (N290, N280, and N265) (Figure 5F). These data indicate that the binding region of N330 for IκB-α is located in the C-terminal side of the bZIP basic domain. We then studied the proteolysis of IκB-α in cells cotransfected with IκB-α and N265, which does not interact with IκB-α. The level of 30-kDa fragment was significantly decreased in the cells cotransfected with N265, relative to the cells cotransfected with N330 (Figure 5G).
The degradation of IκB-α is inhibited by calpain inhibitors and colchicine
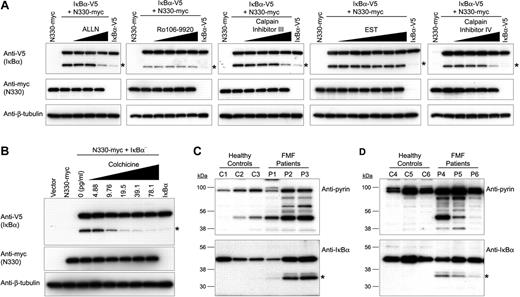
Although IκB-α degradation is attributed primarily to the ubiquitin-proteosome mechanism, there have been several alternate proteolytic pathways described. One well-defined mechanism is calpain-mediated IκB-α proteolysis, in which an intermediate of degraded IκB-α can be detected as a 30-kDa fragment.36-38 Thus, the induction of 30-kDa IκB-α fragment by N330 pyrin implicated the possible involvement of calpain. Indeed, inhibitors of calpain I and II (N-acetyl-leucyl-leucyl-norleucinal [ALLN], calpain inhibitor III, calpeptin, and PD15060), or of calpain II alone (calpain inhibitor IV), but not of calpain I alone ([2S,3S]-trans-epoxysuccinyl-l-leucylamido-3-methylbutane ethyl ester [EST] and calpastatin), decreased the production of 30-kDa IκB-α fragments in cells that were cotransfected with V5-tagged IκB-α and myc-tagged N330 (Figure 6A; Figure S2). On the other hand, we could not observe any inhibition of the production of the 30-kDa IκB-α fragment from cells treated with the ubiquitination inhibitor, Ro106-9920, which inhibits upstream of the ubiquitin-proteosome pathway.
Effects of calpain inhibitors, colchicine, and FMF mutations on N330-induced IκB-α degradation. (A) HeLa cells were cotransfected with IκB-α–V5 and N330-myc, and treated with various inhibitors, ALLN (0.3-1.25 μg/mL, proteosome, calpain I and II inhibitor), Ro106-9920 (0.39-3.125 μM, ubiquitination inhibitor), calpain inhibitor III (0.625-5 μg/mL, calpain I and II inhibitor), EST (3.125-100 mM, calpain I inhibitor), or calpain inhibitor IV (1.25-5 μg/mL, calpain II inhibitor). Cells were also transfected with N330-myc or IκB-α–V5 alone as controls. Cell lysates were analyzed by Western blotting with anti-V5 Ab for IκB-α (top), antimyc Ab for N330 (middle), or anti–β-tubulin Ab for loading controls (bottom). The approximately 30-kDa IκB-α fragment is denoted with an asterisk. (B) HeLa cells were cotransfected or transfected as in panel A. Cotransfected cells were treated with various amounts of colchicine (5-80 pg/mL). Cell lysates were analyzed as in panel A. (C) PBMCs from 3 FMF patients who had at least one mutation in the B30.2 domain, as well as from 3 healthy controls, were lysed immediately after purification. Cell lysates were subjected to Western blotting with antipyrin (top panel) or anti–IκB-α Abs (bottom panel). Asterisks denote the approximately 30-kDa IκB-α fragment. (D) PBMCs from 3 FMF patients who have 2 mutations on B30.2 domains of both alleles, as well as from 3 healthy controls, were treated with IFN-γ for 24 hours. Cells were lysed and analyzed as in panel C.
Effects of calpain inhibitors, colchicine, and FMF mutations on N330-induced IκB-α degradation. (A) HeLa cells were cotransfected with IκB-α–V5 and N330-myc, and treated with various inhibitors, ALLN (0.3-1.25 μg/mL, proteosome, calpain I and II inhibitor), Ro106-9920 (0.39-3.125 μM, ubiquitination inhibitor), calpain inhibitor III (0.625-5 μg/mL, calpain I and II inhibitor), EST (3.125-100 mM, calpain I inhibitor), or calpain inhibitor IV (1.25-5 μg/mL, calpain II inhibitor). Cells were also transfected with N330-myc or IκB-α–V5 alone as controls. Cell lysates were analyzed by Western blotting with anti-V5 Ab for IκB-α (top), antimyc Ab for N330 (middle), or anti–β-tubulin Ab for loading controls (bottom). The approximately 30-kDa IκB-α fragment is denoted with an asterisk. (B) HeLa cells were cotransfected or transfected as in panel A. Cotransfected cells were treated with various amounts of colchicine (5-80 pg/mL). Cell lysates were analyzed as in panel A. (C) PBMCs from 3 FMF patients who had at least one mutation in the B30.2 domain, as well as from 3 healthy controls, were lysed immediately after purification. Cell lysates were subjected to Western blotting with antipyrin (top panel) or anti–IκB-α Abs (bottom panel). Asterisks denote the approximately 30-kDa IκB-α fragment. (D) PBMCs from 3 FMF patients who have 2 mutations on B30.2 domains of both alleles, as well as from 3 healthy controls, were treated with IFN-γ for 24 hours. Cells were lysed and analyzed as in panel C.
We also examined the effect of colchicine on the N330-induced proteolysis of IκB-α, because colchicine is the standard treatment for FMF and its mechanism of action is still incompletely understood. When cotransfected cells were treated with increasing doses of colchicine (5-80 pg/mL), the levels of the 30-kDa IκB-α fragment decreased in a dose-dependent manner. Minimal concentrations of colchicine (10 and 20 pg/mL), which were effective for inhibition of the proteolysis of IκB-α, were much less than the serum levels of colchicine (750 ± 380 pg/mL; n = 8) in FMF patients who had received therapeutic doses of colchicine 12 hours earlier (N.T.C., unpublished observations, August 2007).
Finally, we examined the proteolytic cleavage of IκB-α in PBMCs from FMF patients. As shown in Figure 6C (lysates from cells immediately after isolation) and 6D (lysates from PBMCs treated with IFN-γ), the absolute and relative quantities of cleaved pyrin were substantially increased in PBMCs from FMF patients relative to healthy controls, and the 30-kDa IκB-α fragment was detected only in the PBMCs of FMF patients. Moreover, the amount of 30-kDa IκB-α proteolytic intermediate was directly proportional to the quantity of cleaved pyrin.
Discussion
Based on the dramatic inflammatory manifestations of FMF, pyrin appears to be a major regulator of inflammation and innate immunity. There is now a large body of data indicating an important role for pyrin in the regulation of caspase-1 and IL-1β activation that may be context dependent,17-21 mediated both through homotypic interactions of the N-terminal PYD with ASC and through the inhibitory effects of the C-terminal B30.2 domain on caspase-1 and other components of the inflammasome.18,19 Following up on our recent findings of a direct interaction of pyrin with caspase-1, here we show that pyrin is cleaved by this protease, leading to NF-κB activation. Analysis of the mechanism of this latter effect reveals at least 2 separable events, the potentiation of calpain-dependent IκB-α degradation and the augmentation of p65 nuclear translocation, both by N330 pyrin. The differential sensitivity of mutant and WT pyrin to cleavage suggests a pathway by which pyrin mutations lead to heightened potential for inflammation through NF-κB.
In addition to providing a new link between the inflammatory caspases and NF-κB activation, our data expand the spectrum of caspase-1 substrates. Heretofore, caspase-1 has been shown to mediate the proteolytic processing of precursors of the IL-1 family of cytokines, including IL-1β,39,40 IL-18,41 IL-1F7b,42 and IL-33.43 Caspase-1 has also been shown to process proteins not in the IL-1 family, including actin,44 the kinase PITSLRE,45 and parkin46 in vitro. Recent evidence indicates that caspase-1 also cleaves MyD88 adapter-like (Mal), a protein that, like pyrin, is important in innate immunity.47 Pyrin cleavage occurs at an aspartate residue typical of caspase substrates. This site is remote from the B30.2 domain that binds caspase-1.18,19 There are other well-documented examples of enzyme-substrate interactions remote from the cleavage site of the substrate, including the activities of the IdeS streptococcal endopeptidase,48 tetanus toxin light chain,49 and thrombin.50 Our data indicate that FMF-associated mutations in pyrin increase its sensitivity to caspase-1 cleavage. One possible explanation is the differential binding and inhibitory effect of B30.2 mutant pyrins for caspase-1,18 which has not been observed under all experimental conditions.19 Alternatively, mutant pyrins may assume a conformation that is more susceptible to caspase-1 cleavage.
Our current data introduce a new level of complexity in the subcellular localization of pyrin. When MEFV was cloned, pyrin was computationally predicted to be a nuclear factor, with 2 overlapping nuclear localization signals, a bZIP transcription factor basic domain, and a B-box zinc finger domain.1 Somewhat unexpectedly, in transfected cells, full-length pyrin as well as a C-terminal deletion construct exclusively localized in the cytoplasm, whereas a rare splice isoform lacking exon 2 was found in the nucleus.8,51 Endogenous pyrin localizes predominantly in the nucleus in synovial fibroblasts, dendritic cells, and polymorphonuclear cells, and in the cytoplasm in monocytes.4
Here we provide evidence for N-terminal (N330) and C-terminal (C330) pyrin cleavage fragments, and demonstrate the nuclear localization of N330, but not full-length pyrin or C330, in transfection systems. Although the Robbins-Dingwall nuclear localization signal predicted in the full-length sequence is in C330, computational analysis supports the nuclear localization of N330 (data not shown). N330 includes sites that, when phosphorylated, may bind members of the 14.3.3 family and cause cytoplasmic retention,52 but it is possible that deletion of more C-terminal residues abrogates this effect. It is still unclear whether the endogenous pyrin found in the nucleus of synovial fibroblasts, dendritic cells, and polymorphonuclear leukocytes is full-length or the N-terminal cleaved fragment, because the aforementioned immunostaining experiments4 were performed with an antibody to the N-terminal cleaved fragment. Nevertheless, the data presented in this paper are consistent with a predominance of cleaved pyrin in these cell types.
Given the nuclear localization of N330, and recent studies implicating pyrin and related proteins in the regulation of NF-κB activation, we examined the effect of N330 on NF-κB activity. In previous reports, the impact of full-length pyrin on NF-κB activation has ranged from an inhibitory effect,33,34 to a potentiating effect,32 to a context-dependent effect,31 to no effect at all.20 Our results may clarify this issue, since we found that only N330, but not C330 or full-length pyrin, enhances NF-κB activation, and thus raise the possibility that discrepancies in the literature result from differing experimental conditions that may or may not favor pyrin cleavage.
In the classical NF-κB activation pathway, NF-κB is complexed with IκB proteins and sequestered in the cytoplasm in nonstimulated cells, whereas inducers of NF-κB cause phosphorylation and degradation of IκB, allowing free NF-κB to translocate to the nucleus. In addition, IκB-α as well as NF-κB:IκB-α complexes continuously shuttle between the nucleus and cytoplasm.53-56 In the present study, we also observed that N330 induces the nuclear entrance of IκB-α as well as p65 (Figures 3G and 5C).
Enhanced p65 nuclear translocation may be potentiated by a second effect of N330, the induction of the calpain-mediated degradation of IκB-α. Although IκB-α degradation is attributed primarily to the ubiquitin-proteosome pathway, several calpain-mediated proteolytic mechanisms have been described.36-38,57 Calpains are a family of calcium-dependent, nonlysosomal cysteine proteases composed of 2 subunits, a large 80-kDa subunit and a smaller 30-kDa subunit. The large subunit contains the catalytic function and a calmodulin-like domain that can bind to the PEST domain of IκB-α for proteolytic cleavage.58 Calpain-mediated cleavage takes place at the N-terminus of IκB-α and produces an approximately 30-kDa intermediate fragment that cannot be detected as a product of the classical ubiquitin-proteosome pathway.37,38,59 The presence of this 30-kDa intermediate in N330-transfected cells, as well as the proportionate relationship of this intermediate to cleaved pyrin in PBMCs, corroborates a role for N330-induced calpain-mediated IκB-α degradation. However, it is unclear whether N330 directly interacts with calpain to trigger its activation, since co-IP did not demonstrate the direct interaction of N330 with calpain I or calpain II (data not shown), even as the direct interaction of N330 with IκB-α was easily demonstrable.
We also found that colchicine inhibits N330-induced calpain-mediated IκB-α degradation. These data suggest yet another mechanism, beyond its well-documented effects on integrin expression and leukocyte migration, by which colchicine may be effective in the treatment of FMF. This pathway is a potential target for novel therapies directed toward those FMF patients who manifest an incomplete response to colchicine or cannot tolerate its side effects.
Our findings also contribute to an emerging body of data on the interactions of pyrin with other proteins. The binding of the PYD motif with ASC,14 the B-box/coiled-coil domains with PSTPIP1,20,60 and the B30.2 domain with caspase-118,19 are all well documented. The present study suggests an important function for the bZIP transcription factor basic domain (residues 266-2801 ) and the C-terminal adjacent sequences. The bZIP domain is found in a large number of eukaryotic transcription factors as a bipartite structure consisting of a DNA-binding region enriched in basic amino acids (14 to 20 aa's) followed by a leucine zipper motif. This latter region usually mediates interactions with other transcription factors that provide the specificity of the binding on the target DNA.61 In this context, it is somewhat surprising that pyrin does not have a leucine-zipper domain. Instead, our data indicate that the bZIP basic domain and the C-terminal adjacent sequences directly mediate binding to p65 and IκB-α, respectively, but not to p50. It is possible that, in addition to facilitating the transit of p65 into the nucleus, pyrin may also bind specific DNA sequences and thereby modulate p65 targeting.
In summary, our data demonstrate that caspase-1 initiates a pyrin-dependent NF-κB activation pathway, thus establishing another link between the inflammatory caspases and autoinflammatory disease.62 The increased susceptibility of mutant pyrin to cleavage may confer heightened resistance to microbial pathogens, possibly accounting for the high FMF carrier frequencies in certain populations. In addition, the gain of function conferred by FMF mutations described herein may account for the inflammatory phenotype—whether biochemical63 or clinical64 —observed in many individuals carrying a single MEFV mutation, and thus account for the relatively large number of patients with clinical FMF and only a single demonstrable mutation.65 Finally, N330 pyrin is a potentially attractive target for anti-inflammatory drug discovery.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the Intramural Research Program of the National Institute of Arthritis and Musculoskeletal and Skin Diseases (Bethesda, MD).
National Institutes of Health
Authorship
Contribution: J.J.C. designed the study, performed experiments, and wrote the paper; G.W., K.R., H.J., and N.T.C. performed experiments; S.L.M. contributed to data analysis and paper preparation; D.L.G. designed the study; N.G.S. designed the study and performed experiments; and D.L.K. designed the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jae Jin Chae, GGB, NIAMS, NIH, Building 10, Room 9N-214, 10 Center Drive MSC 1820, Bethesda, MD 20892-1820; e-mail: chaej@exchange.nih.gov.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal