Abstract
The histone deacetylase inhibitor SAHA enhances cell death stimulated by the proteasome inhibitor bortezomib (BZ) by disrupting BZ-induced aggresome formation. Here we report that Myc regulates the sensitivity of multiple myeloma (MM) cells to BZ + SAHA–induced cell death. In MM cells, Myc expression directly correlated with intracellular ER content, protein synthesis rates, the percentage of aggresome-positive cells, and the sensitivity to BZ + SAHA–induced cell death. Accordingly, Myc knockdown markedly reduced the sensitivity of MM cells to BZ + SAHA–mediated apoptosis. Furthermore, activation of Myc was sufficient to provoke aggresome formation and thus sensitivity to BZ + SAHA, and these responses required de novo protein synthesis. BZ + SAHA–mediated stimulation of apoptosis includes the induction of the proapoptotic BH3-only protein Noxa as well as endoplasmic reticular stress, a disruption of calcium homeostasis, and activation of capase-4. Finally, knockdown studies demonstrated that both caspase-4 and Noxa play significant roles in Myc-driven sensitivity to BZ + SAHA–induced apoptosis. Collectively, our results establish a mechanistic link between Myc activity, regulation of protein synthesis, increases in HDAC6 expression and aggresome formation, induction of Noxa, and sensitivity to BZ + SAHA–induced apoptosis. These data suggest that MM patients with elevated Myc activity may be particularly sensitive to the BZ + SAHA combination.
Introduction
Proteasome inhibitors represent a novel class of anticancer agents that have potent anticancer activity in both preclinical and clinical settings.1 Bortezomib (Velcade), the first proteasome inhibitor for cancer therapy, is the most clinically advanced of these agents and has received US Food and Drug Administration (FDA) approval for the treatment of multiple myeloma (MM) and mantle cell lymphoma (MCL). The great success of bortezomib in these cancers has prompted its evaluation in many other tumor models where it also displays promising in vivo antitumor activity.2-6 Furthermore, bortezomib enhances the anticancer activity of many other conventional and novel therapeutic modalities, thus demonstrating its potential broad-based applications in cancer therapy.7-11
Investigation of the mechanisms of action of bortezomib has revealed that it has pleiotropic cellular effects that contribute to its anticancer activity including growth inhibition, the induction of apoptosis in tumor and endothelial cells, the inhibition of tumor angiogenesis, and the disruption of survival signaling cascades. We have previously shown that bortezomib-mediated proteasomal inhibition results in the accumulation of large quantities of ubiquitin-conjugated proteins organized into perinuclear structures termed “aggresomes.”10 These ubiquitin aggresomes appear to form as a part of a cytoprotective response designed to handle the overwhelming protein accrual that occurs when the proteasome is inhibited in malignant but not in normal cells. The formation of aggresomes is an active process and requires histone deacetylase 6 (HDAC6) to form a bridge between microtubules and ubiquitin-conjugated proteins, enabling them to coalesce into a tightly bundled perinuclear structure.12
Our earlier studies in pancreatic cancer models showed that disruption of aggresomes by genetic (siRNA) or pharmacologic (SAHA) inhibition of HDAC6 activity augments the anticancer activity of bortezomib.10 The synergistic anticancer effects of simultaneous inhibition of HDACs and proteasomal activity have also been observed in other cancer models,13-16 and this therapeutic strategy is now moving forward into clinical trials for advanced solid malignancies.
The sensitivity to bortezomib and HDAC inhibitors such as SAHA relies upon active protein synthesis.10,17 To identify patients who may most benefit from their clinical use, we further investigated what factors might regulate sensitivity to these agents. Oncogenic activation of Myc is a hallmark of nearly all rapidly dividing malignancies and Myc activation markedly up-regulates the translational machinery and thus results in an increase in protein synthesis.18,19 Given these properties, we tested the hypothesis that the Myc family of transcription factors regulates sensitivity to the combination of bortezomib and SAHA.
Methods
Cells and cell culture
NCI-H929 and U266 MM cells and human foreskin fibroblasts (HFFs) were obtained from ATCC (Manassas, VA). MM cell lines and transformed human Epstein-Barr virus B cells (EBV-Bs) were maintained in RPMI-1640 media supplemented with 10% fetal bovine serum in a humidified incubator 37°C with 5% CO2. HFFs were infected with MSCV-IRES-puro retrovirus that expresses the puromycin resistance gene by virtue of an internal ribosome entry site (IRES), or with the MSCV-Myc-ERTAM-puro retrovirus, and transduced cells were cultured in phenol red–free DMEM with puromycin as previously described.20,21 HFFs were then treated with 1 μM 4-hydroxytamoxifen (4-HT) for 24 hours to activate Myc-ERTAM.
Antibodies and reagents
Antibodies were obtained from the following commercial sources: antiactin (Sigma-Aldrich, St Louis, MO); anti–active caspase-3, caspase-9, caspase-8, HDAC6, p-eif2α, eif2α, and eIF4E (Cell Signaling, Beverly, MA); anti–caspase-4 (MBL International, Woburn, MA); anti-Noxa (Calbiochem, Gibbstown, NJ); anti-Grp78/BiP (BD Biosciences, San Jose, CA); and anti–c-Myc, N-Myc, L-Myc, and ubiquitin (Santa Cruz Biotechnology, Santa Cruz, CA). Suberoylanilide hydroxamic acid (SAHA) was synthesized as previously described.10 Cycloheximide and 4-HT were purchased from Sigma-Aldrich. Bortezomib was obtained from the hospital pharmacy.
Quantification of drug-induced apoptosis
Apoptosis was measured by flow cytometry by propidium iodide (PI)/fluorescence-activated cell sorting (FACS) analysis of sub-G0/G1 DNA content and quantification of percentages of cells positive for annexin V–FITC (BD Biosciences) as previously described.22
Colony assays
Cells were treated with 2 nM bortezomib, 2 μM SAHA, or both for 24 hours. Drug-treated cells were washed twice in PBS. NCI-H929 and U266 cells were seeded in Methocult methylcellulose medium (StemCell Technologies, Vancouver, BC), and cultured for 10 days in a humidified incubator at 37°C with 5% CO2. Colonies were stained with 0.5% 2,3,5-triphenyltetrazolium chloride (TTC) and scored manually.23
Transmission electron microscopy
Cells were fixed as previously described to examine endoplasmic reticular content.17 Ultrathin sections were examined in a JEM 1010 transmission electron microscope (JEOL, Peabody, MA) at an accelerating voltage of 80 kV. Images were obtained using the AMT Imaging System (Advanced Microscopy Techniques, Danvers, MA) as previously described.17,22
siRNA transfection
Cells were transfected with 100 nM human c-Myc, HDAC6, caspase-4, or Noxa SMARTpool siRNA or with siCONTROL siRNA directed at luciferase (nontarget [NT]) from Dharmacon (Lafayette, CO). NCI-H929 cells were transfected using the Nucleofector II according to the manufacturer's instructions (Amaxa, Gaithersburg, MD). HFFs were transfected using OligofectAMINE (Invitrogen, Carlsbad, CA) for 40 hours according to the manufacturer's protocol. Knockdown of gene expression was confirmed by immunoblotting. Transfected cells were treated with the indicated concentrations of bortezomib and SAHA for 24 hours. Drug-induced apoptosis was quantified by PI/FACS.
Metabolic labeling
Global translation rates were measured as described.17 Briefly, an equal number of cells were incubated in [35S]cysteine- and [35S]methionine-free medium containing 10% dialyzed FBS. The cells were then labeled with [35S]cysteine and [35S]methionine (Translabel; ICN, Irvine, CA) for 30 minutes. Cells were subsequently harvested, lysed, and subjected to trichloroacetic acid precipitation. Scintillation counting was performed to measure [35S]cysteine and [35S]methionine incorporation into proteins.
Measurement of intracellular Ca2+ levels
HFFs were treated with bortezomib and SAHA for 12 hours. Cells were collected, washed in PBS, and incubated with 1 μM calcium green-1 (Molecular Probes, Eugene, OR) for 30 minutes. Cells were then harvested and washed in PBS. Fluorescence was quantified using a FACSCalibur with CellQuest Pro Software (BD Biosciences).
Immunoblot analyses
Confocal microscopy
NCI-H929, U266, and EBV-B cells were centrifuged onto glass slides after treatment with BZ, SAHA, or both and fixed in methanol, permeabilized, and stained as previously described.22 HFFs were plated onto chamber slides and treated with BZ, SAHA, or the combination. Cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton-X-100. Cells were stained with an antiubiquitin antibody overnight. Cells were then incubated with goat antimouse Alexa Flour 488 antibody (Molecular Probes) for 1 hour to visualize ubiquitin localization. ToPro-3 (Molecular Probes) was used to counterstain the nucleus. Coverslips were mounted using Prolong antifade reagent (Molecular Probes). Images were captured using a Zeiss LSM 510 Meta confocal microscope (Oberkochem, Germany) with a 40×/1.3 objective. Slides were covered with Zeiss Immersol 518F oil during image acquisition. Zeiss LSM 510 Version 3.2 SP2 software was used for image acquisition. The percentage of cells containing ubiquitin-conjugated aggresomes was quantified as described previously.10
Real-time PCR analysis
HFFs infected with MSCV-IRES-puromycin or MSCV-Myc-ERTAM-IRES-puromycin viruses were treated for 24 hours with 4-HT. RNA was isolated from HFFs using TRIzol reagent as per the manufacturer's directions (Life Technologies, Grand Island, NY). HDAC6 and GAPDH primers were obtained from Superarray (Frederick, MD) and Integrated DNA Technologies (Coralville, IA), respectively. The cDNA was prepared with the iScript cDNA synthesis kit and the real-time polymerase chain reaction (PCR) analysis was performed with the SYBR Green Supermix kit (BioRad, Hercules, CA) using a BioRad iCycler.
Statistical analyses
Statistical significance of differences observed between samples was determined using the Tukey-Kramer comparison test or the Student t test. Differences were considered significant in all experiments at P less than .05.
Results
Protein synthesis rates, ER content, and sensitivity to bortezomib and SAHA are linked to Myc expression in multiple myeloma cells
We and others have previously demonstrated that the anticancer activity of bortezomib (BZ) is augmented by the addition of HDAC inhibitors, yet the molecular mechanism(s) that regulates sensitivity to these agents has not been resolved.10,13-16,24 Given that BZ has earned FDA approval for the treatment of MM and MCL and has demonstrated promising activity in other preclinical models and clinical trials, we investigated the mechanism(s) that controls BZ sensitivity to identify patients who may most benefit from this therapeutic strategy. BZ-induced apoptosis is dependent upon active protein synthesis and the accumulation of proteins after proteasome inhibition.17 Since the members of the Myc family of transcription factors are major regulators of translation, we hypothesized that Myc might control sensitivity to BZ+ SAHA–induced apoptosis.
To address this question, we first measured the levels of the Myc proteins in 2 MM cell lines and EBV-immortalized B cells as a control. NCI-H929 and U266 cells expressed high levels of c-Myc and L-Myc, respectively (Figure 1A). In contrast, immortalized B cells expressed low levels of c-Myc (Figure 1A). N-Myc was not detected in any of the cell lines (data not shown). Consistent with high levels of c-Myc, NCI-H929 cells had a high intrinsic rate of translation (Figure 1B) and corresponding increased endoplasmic reticular (ER) content (Figure 1C) compared with the immortalized B cells. U266 cells exhibited lower levels of basal translation and ER content than NCI-H929 cells, but both were still much greater than what we observed in the immortalized B cells.
Myc expression correlates with BZ/SAHA-induced apoptosis. (A) Myc expression in MM and immortalized B cells. Levels of c-Myc, L-Myc, and β-actin were determined by immunoblotting. N-Myc was not expressed in these cells (data not shown). (B) Translation rates of MM and EBV-immortalized B cells. Equal numbers of cells were incubated with [35S]cysteine/[35S]methionine for 30 minutes. Protein was collected as described in “Metabolic labeling,” and [35S]cysteine/[35S]methionine incorporation was measured using a scintillation counter. n = 3 plus or minus SD. * indicates a significant difference compared with both MM cell lines, P < .05. (C) Determination of ER content. Cells were collected, fixed, and imaged using an electron microscope. (D-F) SAHA enhances bortezomib (BZ)–induced apoptosis. Cells were treated with 5 nM BZ, 2 μM SAHA, or this combination for 24 hours. Apoptosis was measured by (D) immunoblotting for active caspases-8, -9, and -3, (E) propidium iodide (PI) staining followed by FACS analysis, and (F) annexin V staining followed by FACS analysis as described in “Quantification of drug-induced apoptosis.” Mean (n = 3), SD. (G) SAHA augments bortezomib's ability to impair clonogenic survival. Cells were incubated with 2 nM BZ, 2 μM SAHA, or this combination for 24 hours. Clonogenic survival was determined as described in “Colony assays.” Mean (n = 3), SD. * indicates a significant difference from control and ** represents a significant difference from single-agent treatments, P < .05.
Myc expression correlates with BZ/SAHA-induced apoptosis. (A) Myc expression in MM and immortalized B cells. Levels of c-Myc, L-Myc, and β-actin were determined by immunoblotting. N-Myc was not expressed in these cells (data not shown). (B) Translation rates of MM and EBV-immortalized B cells. Equal numbers of cells were incubated with [35S]cysteine/[35S]methionine for 30 minutes. Protein was collected as described in “Metabolic labeling,” and [35S]cysteine/[35S]methionine incorporation was measured using a scintillation counter. n = 3 plus or minus SD. * indicates a significant difference compared with both MM cell lines, P < .05. (C) Determination of ER content. Cells were collected, fixed, and imaged using an electron microscope. (D-F) SAHA enhances bortezomib (BZ)–induced apoptosis. Cells were treated with 5 nM BZ, 2 μM SAHA, or this combination for 24 hours. Apoptosis was measured by (D) immunoblotting for active caspases-8, -9, and -3, (E) propidium iodide (PI) staining followed by FACS analysis, and (F) annexin V staining followed by FACS analysis as described in “Quantification of drug-induced apoptosis.” Mean (n = 3), SD. (G) SAHA augments bortezomib's ability to impair clonogenic survival. Cells were incubated with 2 nM BZ, 2 μM SAHA, or this combination for 24 hours. Clonogenic survival was determined as described in “Colony assays.” Mean (n = 3), SD. * indicates a significant difference from control and ** represents a significant difference from single-agent treatments, P < .05.
To assess whether Myc levels correlated with sensitivity to BZ + SAHA–induced cell death, the effects of this combination on hallmarks of apoptosis were determined. Immunoblotting analyses revealed that the combination of BZ + SAHA effectively promoted the activation of caspases-8, -9, and -3 in both U266 and NCI-H929 cells, indicating engagement of both the extrinsic and intrinsic apoptotic cascades (Figure 1D). Accordingly, analysis of DNA fragmentation by PI/FACS after 24-hour treatment with BZ, SAHA, or the combination revealed that both U266 and NCI-H929 cells had significantly higher percentages of apoptotic (Sub G0/G1) cells after exposure to BZ + SAHA versus single-agent treatments (Figure 1E). The strong effects of the combination of these agents on apoptosis induction were confirmed by annexin V/PI double staining (Figure 1F). Therefore, SAHA significantly augments the proapoptotic effects of BZ, and these effects are most pronounced in MM cells having high Myc expression. Finally, the combination of BZ and SAHA inhibited the clonogenic potential of MM cells much more effectively than single-agent treatments (Figure 1G), demonstrating the potential long-term benefit of this therapeutic strategy.
Bortezomib stimulates aggresome formation that can be disrupted by SAHA or reduced by decreasing protein synthesis with cycloheximide or c-Myc siRNA
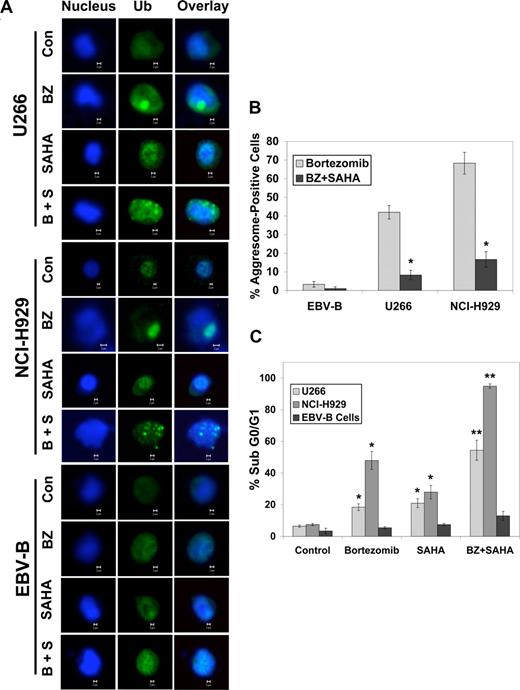
Malignant cells selectively form aggresomes of ubiquitin-conjugated proteins when faced with BZ-mediated proteasomal inhibition.10 Aggresomes appear to play a cytoprotective role since genetically or pharmacologically disrupting their formation augments BZ-induced cell death. We therefore assessed the role of aggresomes as a determinant of BZ- and BZ + SAHA–induced apoptosis in MM cells and whether their formation was controlled by Myc. Confocal microscopy revealed that both MM cell lines formed aggresomes after BZ exposure (Figure 2A,B) and that SAHA disrupted their formation and enhanced BZ-induced apoptosis (Figure 2C). In contrast, BZ treatment did not stimulate aggresome formation in immortalized B cells, and accordingly these cells were much less sensitive to BZ- and BZ + SAHA–induced apoptosis (Figure 2C).
SAHA disrupts bortezomib-induced aggresome formation and augments BZ-induced apoptosis. (A) Multiple myeloma cells form large aggresomes that are disrupted by SAHA. Cells were treated with 5 nM BZ, 2 μM SAHA, or both agents. Cells were then cytocentrifuged onto glass slides, fixed, and stained as described in “Confocal microscopy.” Images were obtained by confocal microscopy. Magnification = 40 ×. (B) Quantification of aggresome formation. Cells were either untreated or treated with SAHA, BZ, or the combination. Aggresomes were present only in cells treated with BZ or BZ + SAHA. Approximately 200 cells were counted and scored as aggresome positive or negative. Mean (n = 3), SD. * indicates a significant difference from values obtained for bortezomib treatment, P < .05. (C) EBV-immortalized B cells are unaffected by the bortezomib and SAHA combination. Cells were treated with 5 nM BZ, 2 μM SAHA, or both agents for 24 hours, and apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from control and ** represents a significant difference from single-agent treatments, P < .05.
SAHA disrupts bortezomib-induced aggresome formation and augments BZ-induced apoptosis. (A) Multiple myeloma cells form large aggresomes that are disrupted by SAHA. Cells were treated with 5 nM BZ, 2 μM SAHA, or both agents. Cells were then cytocentrifuged onto glass slides, fixed, and stained as described in “Confocal microscopy.” Images were obtained by confocal microscopy. Magnification = 40 ×. (B) Quantification of aggresome formation. Cells were either untreated or treated with SAHA, BZ, or the combination. Aggresomes were present only in cells treated with BZ or BZ + SAHA. Approximately 200 cells were counted and scored as aggresome positive or negative. Mean (n = 3), SD. * indicates a significant difference from values obtained for bortezomib treatment, P < .05. (C) EBV-immortalized B cells are unaffected by the bortezomib and SAHA combination. Cells were treated with 5 nM BZ, 2 μM SAHA, or both agents for 24 hours, and apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from control and ** represents a significant difference from single-agent treatments, P < .05.
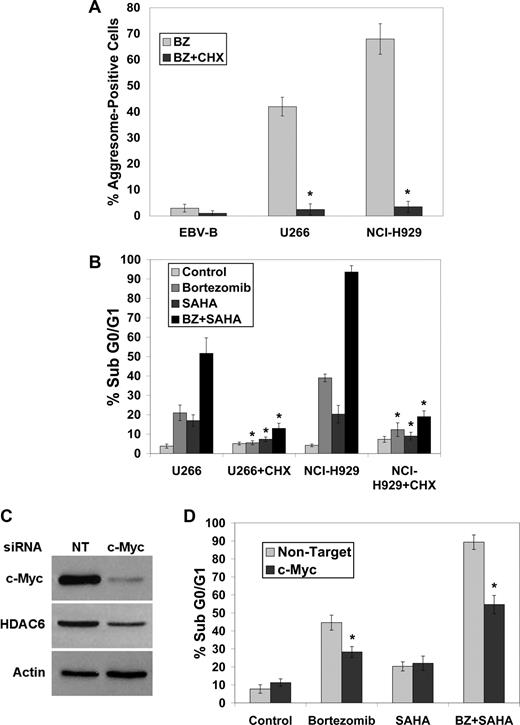
Since active protein synthesis appears to be required for aggresome formation and for sensitivity to BZ + SAHA–mediated cell death, the effects of the translation inhibitor cycloheximide on these responses were determined. Inhibition of translation with cycloheximide blocked aggresome formation (Figure 3A) and cell death (Figure 3B) stimulated by BZ or the BZ + SAHA combination in MM cells.
Blocking protein synthesis impairs bortezomib-induced aggresome formation and BZ + SAHA–mediated apoptosis. (A,B) Cycloheximide blocks bortezomib-induced aggresome formation and apoptosis. Cells were treated with bortezomib in the presence or absence of 1 μg/mL cycloheximide for 24 hours. Aggresomes were quantified by confocal microscopy and apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from cells treated without cycloheximide, P < .05. (C,D) Knockdown of c-Myc using siRNA reduces BZ + SAHA–induced apoptosis. Myc expression was decreased in NCI-H929 cells as described in “siRNA transfection,” and immunoblotting was performed to confirm silencing of c-Myc and demonstrate associated decrease in HDAC6 expression. After exposure to c-Myc or nontarget siRNA, cells were treated with 5 nM BZ, 2 μM SAHA, or both agents, and apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from transfection with nontargeted siRNA, P < .05.
Blocking protein synthesis impairs bortezomib-induced aggresome formation and BZ + SAHA–mediated apoptosis. (A,B) Cycloheximide blocks bortezomib-induced aggresome formation and apoptosis. Cells were treated with bortezomib in the presence or absence of 1 μg/mL cycloheximide for 24 hours. Aggresomes were quantified by confocal microscopy and apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from cells treated without cycloheximide, P < .05. (C,D) Knockdown of c-Myc using siRNA reduces BZ + SAHA–induced apoptosis. Myc expression was decreased in NCI-H929 cells as described in “siRNA transfection,” and immunoblotting was performed to confirm silencing of c-Myc and demonstrate associated decrease in HDAC6 expression. After exposure to c-Myc or nontarget siRNA, cells were treated with 5 nM BZ, 2 μM SAHA, or both agents, and apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from transfection with nontargeted siRNA, P < .05.
To address whether the increased sensitivity of MM cells to the BZ + SAHA combination was indeed due to their higher levels of Myc, the effects of knockdown of c-Myc with siRNA were evaluated in the NCI-H929 cells. Transfection of these MM cells with the c-Myc–selective siRNA led to a marked reduction (∼ 20-fold) in c-Myc protein (Figure 3C). Notably, knockdown of c-Myc reduced both protein synthesis (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and the sensitivity (Figure 3D) of NCI-H929 cells to both BZ- and BZ + SAHA–induced apoptosis. Although knockdown of c-Myc decreased protein synthesis, it did not completely block translation as observed with cycloheximide treatment (Figure S1). Therefore, this may account for the greater protection from BZ + SAHA–induced apoptosis observed with the cycloheximide treatment (Figure 3B). Collectively, these results suggest that cancer cells with high translation rates, such as those with Myc overexpression, may be especially sensitive to BZ + SAHA–induced cell death. In addition, the lower translation rates of normal cells may explain why BZ and BZ + SAHA preferentially kill tumor cells.
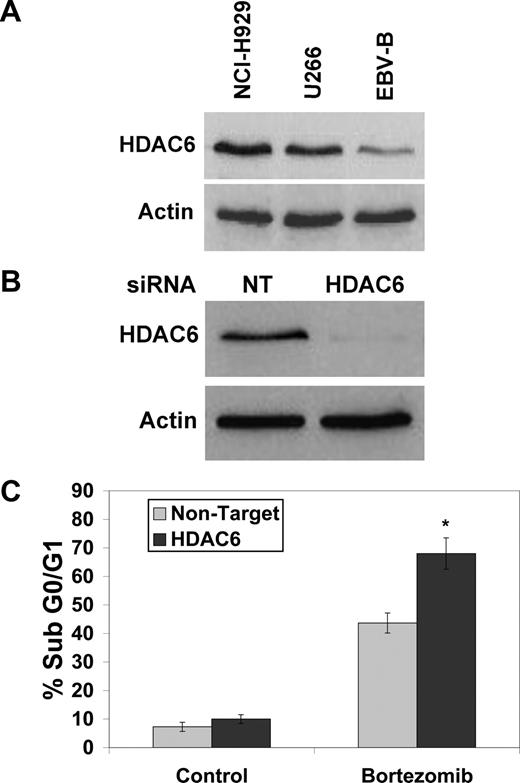
HDAC6 expression correlates with Myc levels in MM cells
HDAC6 is required for the formation of cytoprotective aggresomes,10,12,24 and HDAC6 has been reported to be a direct transcriptional target of Myc.25 Consistent with this notion, NCI-H929 cells expressed the highest levels of HDAC6, and EBV-immortalized B cells had the lowest levels of HDAC6 (Figure 4A). In addition, knockdown of c-Myc in NCI-H929 cells also led to a reduction in HDAC6 expression (Figure 3C). Similar to SAHA treatment, targeted knockdown of HDAC6 with siRNA (Figure 4B) was alone sufficient to sensitize NCI-H929 cells to BZ-induced apoptosis (Figure 4C). Therefore, high levels of Myc and HDAC6 in MM cells render these cells susceptible to the BZ + SAHA combination.
Knockdown of HDAC6 enhances bortezomib-induced apoptosis. (A) HDAC6 expression correlates with Myc expression in MM cell lines. HDAC6 expression was determined in MM cell lines and EBV-immortalized B cells by immunoblotting. (B,C) Knockdown of HDAC6 by siRNA sensitizes NCI-H929 cells to BZ-induced apoptosis. Cells were transfected with siRNA and knockdown was confirmed by immunoblotting. After exposure to siRNA, cells were treated with 5 nM BZ for 24 hours and apoptosis was measured by PI-FACS. Mean (n = 3), SD. * indicates a significant difference from nontarget siRNA–transfected cells treated with bortezomib, P < .05.
Knockdown of HDAC6 enhances bortezomib-induced apoptosis. (A) HDAC6 expression correlates with Myc expression in MM cell lines. HDAC6 expression was determined in MM cell lines and EBV-immortalized B cells by immunoblotting. (B,C) Knockdown of HDAC6 by siRNA sensitizes NCI-H929 cells to BZ-induced apoptosis. Cells were transfected with siRNA and knockdown was confirmed by immunoblotting. After exposure to siRNA, cells were treated with 5 nM BZ for 24 hours and apoptosis was measured by PI-FACS. Mean (n = 3), SD. * indicates a significant difference from nontarget siRNA–transfected cells treated with bortezomib, P < .05.
Myc activity increases translation rates, aggresome formation, and sensitivity to BZ/SAHA-induced apoptosis in normal human diploid fibroblast cells
To address whether Myc activation was sufficient to provoke increases in global protein synthesis and aggresome formation, and to confer sensitivity to BZ + SAHA–induced apoptosis, we transduced early-passage diploid human foreskin fibroblasts (HFFs) with the MSCV-Myc-ERTAM-IRES-puromycin (puro) retrovirus, which drives the expression of the puromycin resistance gene from an internal ribosome entry site (IRES) as well as a chimeric form of human c-Myc fused to the estrogen-binding domain of the estrogen receptor, modified so that it can be selectively activated by 4-hydroxytamoxifen (4-HT). This fusion is normally sequestered in heat shock complexes in the cytoplasm, but after the addition of 4-HT it rapidly relocalizes to the nucleus and activates Myc target genes.20,21 HFFs infected with the MSCV-Myc-ERTAM-puro retrovirus, or with the control MSCV-IRES-puro retrovirus, were cultured in puromycin-containing medium and then treated with 4-HT for 24 hours to activate Myc-ERTAM. As expected, Myc activation induced the expression of HDAC6 and eIF4E, a key translation initiation factor and a validated direct target of Myc26 (Figure 5A), and Myc activation also dramatically increased global protein synthesis (Figure 5B). Furthermore, real-time PCR analysis demonstrated that HDAC6 mRNA levels increase when Myc activity is induced by 4-HT treatment (Figure S2). We also found that the expression levels of HDAC6 were consistently higher as quantified by microarray analysis in B cells (an average of 33% greater) collected from the bone marrow of Eμ-myc transgenic mice compared with B cells harvested from wild-type mice (data not shown). Consistent with a marked increase in translation, Myc-ERTAM–expressing HFFs formed aggresomes when treated with BZ (Figure 5C). Furthermore, SAHA disrupted aggresome formation in Myc-ERTAM–expressing HFFs (Figure 5D) and strongly induced apoptosis in these cells when given in combination with BZ (Figure 5E). Finally, inhibition of translation with cycloheximide blocked aggresome formation in Myc-ERTAM–expressing HFFs (data not shown) and abrogated BZ + SAHA–induced apoptosis (Figure 5E). Therefore, Myc-induced translation is sufficient to sensitize primary fibroblasts to BZ-induced aggresome formation, and disruption of BZ-induced aggresomes by SAHA provokes death of Myc-expressing cells.
Myc activation increases global translation, stimulates BZ-mediated aggresome formation, and sensitizes HFFs to BZ + SAHA–induced apoptosis. (A) Activation of Myc increases the expression of the target genes eIF4E and HDAC6. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were treated with 1 μM 4-HT for 24 hours. Lysates were prepared and immunoblotting was performed as described in “Immunoblot analyses.” (B) Myc activation increases global translation rates of HFFs. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were treated with 1 μM 4-HT for 24 hours and equal cell numbers were then incubated with [35S]cysteine/[35S]methionine for 30 minutes. Protein was collected as described in “Metabolic labeling” and [35S]cysteine/[35S]methionine incorporation was measured using a scintillation counter. Mean (n = 3), SD. * indicates a significant difference from MSCV-IRES-puro–transfected cells, P < .05. (C) Myc activation stimulates aggresome formation in HFFs treated with BZ. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were plated on chamber slides and pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ, 2 μM SAHA, or both agents. After treatment, cells were fixed and stained and images were obtained by confocal microscopy. Magnification = 40×. (D) Quantification of aggresome formation. Approximately 200 cells were counted and scored as aggresome positive or negative. Mean (n = 3), SD. * indicates a significant difference from bortezomib-treated cells, P < .05. (E) Cycloheximide blocks BZ- and BZ + SAHA–induced apoptosis in Myc-expressing HFFs. The indicated cells were pretreated with 1 μM 4-HT and were then treated with 100 nM BZ, 2 μM SAHA, or both in the presence or absence of 50 μM cycloheximide for 24 hours. Apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from cells treated without cycloheximide, P < .05.
Myc activation increases global translation, stimulates BZ-mediated aggresome formation, and sensitizes HFFs to BZ + SAHA–induced apoptosis. (A) Activation of Myc increases the expression of the target genes eIF4E and HDAC6. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were treated with 1 μM 4-HT for 24 hours. Lysates were prepared and immunoblotting was performed as described in “Immunoblot analyses.” (B) Myc activation increases global translation rates of HFFs. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were treated with 1 μM 4-HT for 24 hours and equal cell numbers were then incubated with [35S]cysteine/[35S]methionine for 30 minutes. Protein was collected as described in “Metabolic labeling” and [35S]cysteine/[35S]methionine incorporation was measured using a scintillation counter. Mean (n = 3), SD. * indicates a significant difference from MSCV-IRES-puro–transfected cells, P < .05. (C) Myc activation stimulates aggresome formation in HFFs treated with BZ. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were plated on chamber slides and pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ, 2 μM SAHA, or both agents. After treatment, cells were fixed and stained and images were obtained by confocal microscopy. Magnification = 40×. (D) Quantification of aggresome formation. Approximately 200 cells were counted and scored as aggresome positive or negative. Mean (n = 3), SD. * indicates a significant difference from bortezomib-treated cells, P < .05. (E) Cycloheximide blocks BZ- and BZ + SAHA–induced apoptosis in Myc-expressing HFFs. The indicated cells were pretreated with 1 μM 4-HT and were then treated with 100 nM BZ, 2 μM SAHA, or both in the presence or absence of 50 μM cycloheximide for 24 hours. Apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from cells treated without cycloheximide, P < .05.
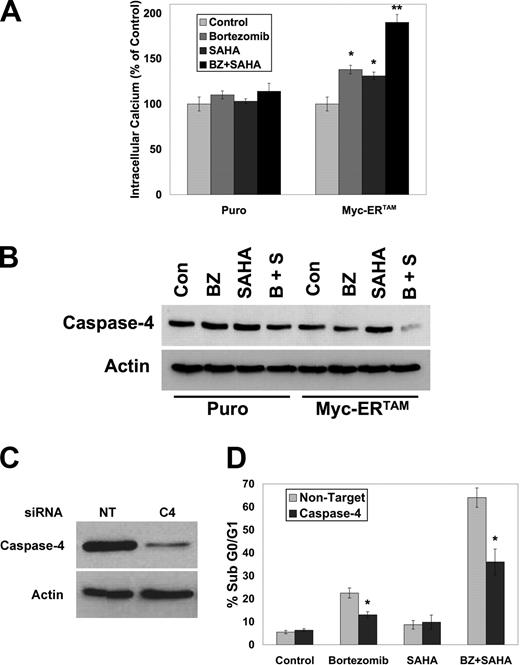
c-Myc sensitizes HFFs to BZ-mediated ER stress and apoptosis
A major mechanism by which BZ stimulates apoptosis is via the induction of ER stress.11,17,27-29 Since increased Myc activity enhances BZ + SAHA–induced apoptosis, the effects of Myc on ER stress were assessed in HFFs. We and others have previously demonstrated that bortezomib induces a unique form of ER stress characterized by an increase in Grp78/BiP, but an absence of eif2α phosphorylation.11,17,30 Activation of Myc caused an increase in Grp78/BiP after BZ treatment, whereas the BZ-stimulated decrease in eif2α phosphorylation was found to be independent of Myc expression (Figure S3). The lack of eif2α phosphorylation allows protein synthesis to continue during proteasomal inhibition. Since cells with high Myc activity intrinsically have higher translation rates, we hypothesized that BZ would induce aggresomes in these cells and this process could be disrupted with SAHA, leading to ER stress–mediated apoptosis. Consistent with this idea, BZ + SAHA treatment stimulated an increase in intracellular calcium levels (Figure 6A) and activation of caspase-4 (Figure 6B), 2 hallmarks of ER stress–mediated cell death. Further, knockdown of caspase-4 using siRNA (Figure 6C) significantly reduced both BZ- and BZ + SAHA–induced apoptosis in Myc-ERTAM–expressing cells (Figure 6D). Therefore, the BZ + SAHA combination induces ER stress that leads to apoptosis, and this response is augmented in Myc-expressing cells. Consistent with this notion, Myc-ERTAM–expressing HFFs also displayed enhanced apoptosis after treatment with the drugs tunicamycin and brefeldin A, which induce ER stress (Figure S4).
Myc sensitizes HFFs to bortezomib-mediated, ER stress–induced apoptosis. (A) Myc activation increases intracellular calcium levels induced by BZ + SAHA. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ and 2 μM SAHA for 12 hours. Cells were collected, stained, and analyzed as described in “Measurement of intracellular Ca2+ levels.” Mean (n = 3), SD. * indicates a significant difference from control and ** represents a significant difference from single-agent treatments, P < .05. (B) Activation of Myc promotes caspase-4 activation after treatment with BZ + SAHA. The indicated cells were pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ and 2 μM SAHA for 24 hours. Cells were collected and caspase-4 levels were detected by immunoblotting. (C,D) Caspase-4 contributes to BZ + SAHA–induced apoptosis. Caspase-4 levels were reduced using siRNA and knockdown was confirmed by immunoblotting. Cells were pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ and 2 μM SAHA for 24 hours. Apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from nontarget siRNA–treated cells, P < .05.
Myc sensitizes HFFs to bortezomib-mediated, ER stress–induced apoptosis. (A) Myc activation increases intracellular calcium levels induced by BZ + SAHA. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ and 2 μM SAHA for 12 hours. Cells were collected, stained, and analyzed as described in “Measurement of intracellular Ca2+ levels.” Mean (n = 3), SD. * indicates a significant difference from control and ** represents a significant difference from single-agent treatments, P < .05. (B) Activation of Myc promotes caspase-4 activation after treatment with BZ + SAHA. The indicated cells were pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ and 2 μM SAHA for 24 hours. Cells were collected and caspase-4 levels were detected by immunoblotting. (C,D) Caspase-4 contributes to BZ + SAHA–induced apoptosis. Caspase-4 levels were reduced using siRNA and knockdown was confirmed by immunoblotting. Cells were pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ and 2 μM SAHA for 24 hours. Apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from nontarget siRNA–treated cells, P < .05.
c-Myc regulates Noxa induction during BZ-induced apoptosis
Several recent studies have demonstrated that the BH3-only proapoptotic protein Noxa is induced after BZ treatment and that Noxa contributes to BZ-mediated apoptosis.4,5,31-33 We therefore evaluated potential mechanistic links between Myc activity, Noxa, and ER stress–induced apoptosis. Notably, Noxa was induced by BZ only in Myc-ERTAM–expressing HFFs, and Noxa expression was further increased by the combination of BZ + SAHA (Figure 7A). To assess whether Noxa was required for BZ + SAHA–induced apoptosis, the effects of Noxa knockdown by siRNA (Figure 7B) were evaluated. Reductions in Noxa impaired both BZ- and BZ + SAHA–induced apoptosis in Myc-ERTAM–expressing HFFs (Figure 7C). Furthermore, Noxa also appears to contribute to caspase-4 activation and ER stress in these cells, because Noxa knockdown led to a marked decrease in caspase-4 cleavage (Figure 7B).
Noxa is induced in Myc-expressing cells after bortezomib or BZ + SAHA treatment. (A) Myc activation promotes BZ- and BZ + SAHA–induced Noxa expression. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ, 2 μM SAHA, or both for 24 hours. Cells were collected and immunoblotting was performed as described in “Immunoblot analyses.” (B) Knockdown of Noxa blocks caspase-4 activation by BZ + SAHA. Cells were exposed to Noxa or nontarget (NT) siRNA and then pretreated with 1 μM 4-HT for 24 hours followed by treatment with 100 nM BZ, 2 μM SAHA, or both for 24 hours. Inhibition of Noxa induction and caspase-4 cleavage were determined by immunoblotting. (C) Knockdown of Noxa reduces BZ- and BZ + SAHA–induced apoptosis. Knockdown of Noxa and treatment of cells were performed as described in panel B. Apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from nontarget siRNA–treated cells, P < .05. (D) Noxa expression is induced by bortezomib and BZ + SAHA in NCI-H929 MM cells but not in EBV-immortalized B cells. Cells were treated with 5 nM BZ, 2 μM SAHA, or both agents for 24 hours. Noxa expression was determined by immunoblotting. (E,F) Noxa expression was knocked down in NCI-H929 cells using siRNA and the Nucleofector system. Cells were then treated with 5 nM BZ, 2 μM SAHA, or both agents for 24 hours. Inhibition of Noxa expression was confirmed by immunoblotting and apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from nontarget siRNA–treated cells, P < .05.
Noxa is induced in Myc-expressing cells after bortezomib or BZ + SAHA treatment. (A) Myc activation promotes BZ- and BZ + SAHA–induced Noxa expression. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ, 2 μM SAHA, or both for 24 hours. Cells were collected and immunoblotting was performed as described in “Immunoblot analyses.” (B) Knockdown of Noxa blocks caspase-4 activation by BZ + SAHA. Cells were exposed to Noxa or nontarget (NT) siRNA and then pretreated with 1 μM 4-HT for 24 hours followed by treatment with 100 nM BZ, 2 μM SAHA, or both for 24 hours. Inhibition of Noxa induction and caspase-4 cleavage were determined by immunoblotting. (C) Knockdown of Noxa reduces BZ- and BZ + SAHA–induced apoptosis. Knockdown of Noxa and treatment of cells were performed as described in panel B. Apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from nontarget siRNA–treated cells, P < .05. (D) Noxa expression is induced by bortezomib and BZ + SAHA in NCI-H929 MM cells but not in EBV-immortalized B cells. Cells were treated with 5 nM BZ, 2 μM SAHA, or both agents for 24 hours. Noxa expression was determined by immunoblotting. (E,F) Noxa expression was knocked down in NCI-H929 cells using siRNA and the Nucleofector system. Cells were then treated with 5 nM BZ, 2 μM SAHA, or both agents for 24 hours. Inhibition of Noxa expression was confirmed by immunoblotting and apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from nontarget siRNA–treated cells, P < .05.
To confirm that similar pathways were operational in multiple myeloma, we evaluated the regulation and role of Noxa in NCI-H929 cells. Noxa was also induced in the c-Myc–expressing NCI-H929 MM cells after their exposure to BZ and this response was augmented by BZ + SAHA treatment, but Noxa was not induced by either regimen in EBV-immortalized B cells (Figure 7D). Furthermore, knockdown of Noxa in NCI-H929 cells (Figure 7E) reduced BZ- and BZ + SAHA–induced apoptosis (Figure 7F). Collectively, these data indicate that Myc regulates BZ- and BZ + SAHA–induced triggered expression of Noxa, which is a critical event required for maximal apoptosis in response to these agents.
Discussion
The proteasome is an attractive anticancer target for therapeutic inhibition because it regulates the turnover of many proteins involved in cell-cycle progression, apoptosis, and angiogenesis.1 The proteasome inhibitor bortezomib has been FDA-approved for the treatment of multiple myeloma and mantle cell lymphoma and has displayed manageable toxicity at target doses. Since the proteasome plays roles in maintaining cellular protein homeostasis, bortezomib's ability to induce apoptosis selectively in neoplastic cells was unexpected. We previously demonstrated that bortezomib stimulates ER stress and that active protein synthesis in tumor cells is required for apoptosis induction.11,17 Therefore, the efficacy of the bortezomib/HDAC inhibitor combination could be directly related to the increased protein synthesis that is a hallmark of rapidly dividing tumor cells. Finally, inhibition of proteasomal degradation results in aggresome formation, which is cytoprotective, as disruption of this process using HDAC inhibitors, or as shown here by selective knockdown of HDAC6, augments bortezomib-induced apoptosis.10
Myc oncoproteins are overexpressed in nearly 70% of all rapidly diving human tumors, and a hallmark of Myc activation is coordinated increases in the protein translation machinery, which occurs through its ability to induce many genes involved in translation, including ribosomal and transfer RNAs.19,34,35 Myc enhances apoptosis in the presence of proteasome inhibitors,36 and we therefore hypothesized that the sensitivity of multiple myeloma cells to BZ and the BZ + SAHA combination would, at least in part, be a function of Myc levels in these tumor cells. Indeed, our studies established that Myc levels control sensitivity to BZ or the BZ + SAHA combination, where higher Myc is associated with markedly increased translation rates, abundant cellular ER content, and increased formation of aggresomes. Furthermore, our studies demonstrate that Myc activation is also sufficient to confer all of these responses, at least in the context of primary fibroblasts. We posit that instead of directly targeting Myc oncogene addiction, that high Myc activity may be an “Achilles heel” that can be exploited by treatment with the BZ + SAHA combination.
The ability of Myc to augment the formation of aggresomes appears largely due to its ability to induce HDAC6, which is an established, important regulator of aggresome formation in response to BZ treatment.10,12,24 Consistent with previous studies,25 our analyses suggest that HDAC6 is a direct Myc transcriptional target, and that in multiple myeloma cells HDAC6 expression requires sustained expression of Myc. Thus, because HDAC6 protects against bortezomib-induced apoptosis by stimulating aggresome formation, tumors with Myc involvement are probably best targeted using the BZ + SAHA combination.
Studies directed toward defining the mechanism by which Myc sensitizes cells to BZ + SAHA–induced apoptosis strongly suggest that increased translation directed by Myc leads to an ER stress response when functions of the proteasome are compromised, and that this includes the release of intracellular calcium and the activation of caspase-4, a known mediator of the ER stress–induced apoptosis,17,37 and of the proapoptotic protein Noxa. Furthermore, knockdown studies established that both caspase-4 and Noxa contribute to the apoptotic response. Precisely how these apoptotic effectors are selectively activated in Myc-expressing cells treated with BZ + SAHA is not entirely clear, but their selective induction does provide a reasonable accounting for the tumor-specific effects of this combination.31-33 Noxa induction by bortezomib is p53 independent,4,5 and in MM and primary fibroblasts it appears to be a proximal signal for the BZ + SAHA apoptotic response, as knockdown of Noxa reduces caspase-4 cleavage. Given that Noxa was recently identified to be a direct transcriptional target of Myc,38 and because Myc and signals that activate ER stress cooperate to induce apoptosis, we posit, as have others,39 that these signals converge to induce Noxa expression, which then triggers the intrinsic apoptotic pathway. Since Noxa can be directly induced by Myc, this may explain, in part, the anticancer activity of bortezomib observed in cancers with mutant p53. Consistent with this idea, HCT116 p53+/+ and HCT116 p53−/− cells are equally sensitive to apoptosis induced by the BZ/SAHA combination (data not shown).
Multiple myeloma cells have remarkably high levels of protein synthesis to produce large amounts of immunoglobulins. This may explain, in part, their increased sensitivity to bortezomib.40 However, the results of the current study demonstrate that elevated Myc activity is sufficient to increase translation to a level that yields normal fibroblasts sensitive to bortezomib-induced apoptosis. Although Myc sensitized cells to bortezomib-induced death, this effect was much more pronounced after treatment with the BZ + SAHA combination. This result may be due to the increased levels of HDAC6 induced by Myc, which leads to protection from bortezomib-mediated apoptosis via aggresome formation. We are planning to investigate the relationship between Myc expression and sensitivity to bortezomib and BZ + SAHA in primary cells obtained from MM patients. Chromosomal rearrangements leading to Myc dysregulation have been identified in 15% of newly diagnosed and approximately 40% of advanced MM patients in earlier investigations.41,42 Collectively, these findings provide additional rationale for planned clinical trials to evaluate the efficacy of the combination of bortezomib with vorinostat or other HDAC inhibitors that target HDAC6 in patients with MM as well as in other hematologic and solid malignancies where Myc is active. Finally, these findings indicate that Myc status should be explored as a biomarker predictive of clinical response to this drug combination.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the M. D. Anderson Specialized Programs of Research Excellence (SPORE) in Pancreatic Cancer grant (Houston, TX; D.J.M.), National Institutes of Health (Bethesda, MD) grant CA076379 (J.L.C.), Cancer Center Core grants CA054174 and CA021765, the American Lebanese Syrian Associated Charities (ALSAC) of St Jude Children's Research Hospital (Memphis, TN), and funds provided by the Institute for Drug Development, Cancer Therapy and Research Center at The University of Texas Health Science Center at San Antonio.
National Institutes of Health
Authorship
Contribution: S.T.N. and J.S.C. wrote the paper and were involved in all aspects of the study including experimental design, performing research, and data analysis; K.H.M. generated essential model systems; J.F.C. conducted real-time PCR analyses; P.H., J.A.H., J.L.C., and F.J.G. provided intellectual input regarding experimental design and data interpretation; and D.J.M. was involved in experimental design, data analysis/interpretation, and paper preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steffan T. Nawrocki, The Institute for Drug Development, Cancer Therapy and Research Center at The University of Texas Health Science Center at San Antonio, 14960 Omicron Drive, San Antonio, TX 78245; e-mail: nawrocki@uthscsa.edu.
References
Author notes
*S.T.N. and J.S.C. contributed equally to this work.

![Figure 1. Myc expression correlates with BZ/SAHA-induced apoptosis. (A) Myc expression in MM and immortalized B cells. Levels of c-Myc, L-Myc, and β-actin were determined by immunoblotting. N-Myc was not expressed in these cells (data not shown). (B) Translation rates of MM and EBV-immortalized B cells. Equal numbers of cells were incubated with [35S]cysteine/[35S]methionine for 30 minutes. Protein was collected as described in “Metabolic labeling,” and [35S]cysteine/[35S]methionine incorporation was measured using a scintillation counter. n = 3 plus or minus SD. * indicates a significant difference compared with both MM cell lines, P < .05. (C) Determination of ER content. Cells were collected, fixed, and imaged using an electron microscope. (D-F) SAHA enhances bortezomib (BZ)–induced apoptosis. Cells were treated with 5 nM BZ, 2 μM SAHA, or this combination for 24 hours. Apoptosis was measured by (D) immunoblotting for active caspases-8, -9, and -3, (E) propidium iodide (PI) staining followed by FACS analysis, and (F) annexin V staining followed by FACS analysis as described in “Quantification of drug-induced apoptosis.” Mean (n = 3), SD. (G) SAHA augments bortezomib's ability to impair clonogenic survival. Cells were incubated with 2 nM BZ, 2 μM SAHA, or this combination for 24 hours. Clonogenic survival was determined as described in “Colony assays.” Mean (n = 3), SD. * indicates a significant difference from control and ** represents a significant difference from single-agent treatments, P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/7/10.1182_blood-2007-12-130823/6/m_zh80200824890001.jpeg?Expires=1769105321&Signature=Kfv1QCrM3Jv8cf10Mx3bhqY3IrI4AczYy7-bo6VCx7v~3yDL1uKBFrPGYTR4u0uOdFabJGfJj8tAMQksWUC4CmHM~3T~ZSHyk1ruMmpw2rdzSwypbnqSnVuXX~PhkFX7QLKbj~lkZe4MV2RjSoZqZHIY-Mx9LeHqa~65paon8Y~9U~n5AJ9bJdEFuBGIrfL0d83pqhbLY9II6Q7gmmLA6iPq2x03fTFRJpDDkjgaf~LD0~QPIrLu2WSX1h3~0qZ033zu8N7H617xijwZPbysRFgPDEo7gsIy9TqQQQIFi5p77TUTIWRvD9Utu8Ks~MLyv1n-I7ld3uHfNccUAcJptA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Myc activation increases global translation, stimulates BZ-mediated aggresome formation, and sensitizes HFFs to BZ + SAHA–induced apoptosis. (A) Activation of Myc increases the expression of the target genes eIF4E and HDAC6. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were treated with 1 μM 4-HT for 24 hours. Lysates were prepared and immunoblotting was performed as described in “Immunoblot analyses.” (B) Myc activation increases global translation rates of HFFs. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were treated with 1 μM 4-HT for 24 hours and equal cell numbers were then incubated with [35S]cysteine/[35S]methionine for 30 minutes. Protein was collected as described in “Metabolic labeling” and [35S]cysteine/[35S]methionine incorporation was measured using a scintillation counter. Mean (n = 3), SD. * indicates a significant difference from MSCV-IRES-puro–transfected cells, P < .05. (C) Myc activation stimulates aggresome formation in HFFs treated with BZ. HFFs expressing the MSCV-IRES-puro or MSCV-Myc-ERTAM-IRES-puro retroviruses were plated on chamber slides and pretreated with 1 μM 4-HT for 24 hours and then treated with 100 nM BZ, 2 μM SAHA, or both agents. After treatment, cells were fixed and stained and images were obtained by confocal microscopy. Magnification = 40×. (D) Quantification of aggresome formation. Approximately 200 cells were counted and scored as aggresome positive or negative. Mean (n = 3), SD. * indicates a significant difference from bortezomib-treated cells, P < .05. (E) Cycloheximide blocks BZ- and BZ + SAHA–induced apoptosis in Myc-expressing HFFs. The indicated cells were pretreated with 1 μM 4-HT and were then treated with 100 nM BZ, 2 μM SAHA, or both in the presence or absence of 50 μM cycloheximide for 24 hours. Apoptosis was measured by PI-FACS analysis. Mean (n = 3), SD. * indicates a significant difference from cells treated without cycloheximide, P < .05.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/7/10.1182_blood-2007-12-130823/6/m_zh80200824890005.jpeg?Expires=1769105321&Signature=El8XIKSYxh7hwQpNF36oBzMtrMlml~hNX0UEuxX4xRpJ1bOluYslJU64RkBqdYULEf7QxmgDf3xlf5G~zCu4nfxaqTkvpGCaXZOvK-fQQk0rpJIb64hy1d9qv40vWb6ggAdDu1nL1VyoclN4bjYr8yJXHRO7fWwbhSSwvN6e4klEsITLvTl6ACGyHZ2vBneFa81AgS4VbvAID9ttitbFzeBkmdVd8TemotauJ2FY9RnjilavTOz6BWSDmLi-tX~58tSmjC90DR4~7eIfjyyYmAAzDe64HBIhbT1Vhy~y3zlZLmORztEg6DCsGWzdwMwVK0J7730pfTznlF6Kzg~GKA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal