Abstract
Although the in vitro expansion of the multiple myeloma (MM) clone has been unsuccessful, in a novel three-dimensional (3-D) culture model of reconstructed bone marrow (BM, n = 48) and mobilized blood autografts (n = 14) presented here, the entire MM clone proliferates and undergoes up to 17-fold expansion of malignant cells harboring the clonotypic IgH VDJ and characteristic chromosomal rearrangements. In this system, MM clone expands in a reconstructed microenvironment that is ideally suited for testing specificity of anti-MM therapeutics. In the 3-D model, melphalan and bortezomib had distinct targets, with melphalan targeting the hematopoietic, but not stromal com-partment. Bortezomib targeted only CD138+CD56+ MM plasma cells. The localization of nonproliferating cells to the reconstructed endosteum, in contact with N-cadherin–positive stroma, suggested the presence of MM-cancer stem cells. These drug-resistant CD20+ cells were enriched more than 10-fold by melphalan treatment, exhibited self-renewal, and generated clonotypic B and plasma cell progeny in colony forming unit assays. This is the first molecularly verified demonstration of proliferation in vitro by ex vivo MM cells. The 3-D culture provides a novel biologically relevant preclinical model for evaluating therapeutic vulnerabilities of all compartments of the MM clone, including presumptive drug-resistant MM stem cells.
Introduction
Hematopoiesis in bone marrow (BM) occurs in distinct microenvironmental niches. Discrete extracellular matrix (ECM) microenvironments within the BM help to separate endosteum, an interface between bone and BM, from the central marrow. Methods for studying hematopoiesis include clonal culture systems in semisolid media,1 short-term2 and long-term3,4 liquid cultures, and tissue culture systems where hematopoietic cells grow on feeder layers of BM stromal cells.5,6 However, cell culture systems involving growth on the surface of tissue culture plastic do not accurately represent tissue architecture7 or the complex interactions between cells and their micro-environment. Recently, a stromal spheroid coculture model8 and various scaffolds9,10 have been developed to recreate the three-dimensional (3-D) environment of the BM, but these models fail to recapitulate the physiologic conditions of the BM. To adequately study B-cell development,11 pathogenesis,12,13 and neoplasia,14 a culture system that places BM cells within their physiologic environment is required. For BM-localized malignancies, more effective culture systems must incorporate all compartments of the malignant clone, including cancer stem and progenitor cells, to identify their therapeutic vulnerabilities.
Multiple myeloma (MM), an incurable cancer with 3- to 5-year survival despite the development of potent new therapies,15 is characterized by monoclonal immunoglobulin (Ig), lytic bone lesions,16 and monoclonal plasma cells (PCs) in the BM. The immunoglobulin heavy chain gene rearrangement (IgH VDJ) provides a unique molecular signature, termed clonotypic.17,18 Because clonal expansion of primary MM cells outside their BM microenvironment has been unsuccessful,19 most preclinical studies have used MM cell lines derived from leukemic phase cells that have escaped BM dependence. Currently, only PCs are evaluated in preclinical studies to determine the impact of new drugs.20 However, nearly all MM patients relapse, suggesting that drug-resistant cancer stem cells escape conventional therapies.21-25 It seems likely that BM niches maintain MM cancer stem cells (MM-CSCs) in a quiescent, drug-resistant state. To date, preclinical models do not take into account adhesion-mediated drug resistance,26 and none allows testing of drug efficacy on MM-CSC populations. Mouse models of MM are inadequate for preclinical use because they cannot faithfully recapitulate human disease.27,28
Here we present a robust 3-D tissue culture model in which the human BM microenvironment is reconstructed in vitro. In 3-D, the MM clone expands within its native microenvironment providing a valuable preclinical model within which conventional (melphalan) and novel (bortezomib) therapeutics selectively kill their target cells. In 3-D cultures, nonproliferating cells from MM BM concentrate at a reconstructed endosteum-marrow junction (rEnd). Purified nonproliferating MM BM cells include MM-CSCs, as defined by their ability in a secondary culture to generate B/PC progeny harboring the unique MM clonotypic signature. Three-dimensional cultures of BM or mobilized blood autografts (MBAs) offer a preclinical model within which new therapies can be tested for their impact on all compartments of the MM clone, as well as providing access to the MM-CSC that underlie disease progression.
Methods
Materials
After approval from the Health Research Board (University of Alberta) and the Alberta Cancer Board, and after informed consent was obtained in accordance with the Declaration of Helsinki, BM samples (n = 48) were provided from patients undergoing BM biopsies at the Cross Cancer Institute. Aliquots of G-CSF MBAs were obtained at the time of harvest (n = 14). All of these samples were successfully cultured in 3-D. Plasma was obtained from blood of MM patients at routine clinic visits. Ficoll-Paque was purchased from Pfizer (New York, NY). Human fibronectin and rat tail collagen I were from Upstate Biotechnology (Charlottesville, VA). MethoCult-GF was from StemCell Technologies (Vancouver, BC). Matrigel, Cell Recovery Solution, fluorescently tagged annexin V, N-cadherin, CD20, CD33, CD34, CD45, CD38, and CD5 antibodies were from BD Biosciences (San Jose, CA). 5-6-Carboxyfluorescein diacetate succinimidyl ester (CFSE) and phalloidin were from Invitrogen Canada (Burlington, ON). Oil Red, Alizarin Red, SigmaFast BCIP/NBT tablets for alkaline phosphatase activity, and SigmaFast Red TR/Napthol AS-MX tablets for TRAP staining were from Sigma Diagnostics Canada (Mississauga, ON). DyNAmo HS SYBR Green qPCR kit was from New England Biolabs (Ipswich, MA). CD138 was from Beckman Coulter (Fullerton, CA), CD20 from Dako North America (Carpinteria, CA), and antifibronectin from Millipore (Billerica, MA). Fluorescent in situ hybridization (FISH) probes were from Vysis (Abbott Laboratories, Abbott Park, IL) or made in house. Melphalan and bortezomib were obtained from the Cross Cancer Institute.
Cell culture
Mononuclear cells were isolated from BM aspirates by Ficoll-Paque gradient centrifugation per the manufacturer's instructions. Surface coating (rEnd) was created by coating 48-well tissue culture plates (Corning, Corning, NY) with fibronectin/collagen I (1:1) in phosphate-buffered saline (PBS) at a final concentration of 5 μg/1 cm2 of each protein. Plates were incubated for 30 minutes or more at room temperature; after removal of excess fluid, the rEnd was overlaid with the rBM layer consisting of the BM mononuclear cells (BMCs) in an ECM mixture of Matrigel/fibronectin (2:1 vol/vol). For analysis of proliferation and identification of nonproliferating cells, cells were labeled with 0.25 μM CFSE for 15 minutes at 40°C. The labeling reaction was stopped with cold PBS for 10 minutes on ice, cells were centrifuged to remove excess dye, resuspended at 0.5 × 106 cells/1 cm2 in 10 μL PBS, mixed with 100 μL/1 cm2 of ECM mixture, and plated on top of the fibronectin/collagen I rEnd. The ECM proteins and their ratios were chosen based on the analysis of the ECM components from the human BM core biopsies of both normal donors and MM patients (J.K. and L.M.P., manuscript in preparation). The culture conditions were optimized to ensure maximum proliferation of the MM clone; controls established that the rEnd alone did not sustain ex vivo BM samples. The culture was allowed to solidify for 30 minutes in a 37°C, 5% CO2 incubator and overlaid with 1 mL growth medium (RPMI with l-glutamine, 20% MM patient plasma, 6.2 × 10−4 M of CaCl2, 10−6 M sodium succinate, 10−6 M hydrocortisone; Sigma Diagnostics Canada). Plasma from MM blood samples was an essential growth factor; neither plasma from healthy donors nor fetal bovine serum supported growth of MM cells. No differences were seen between autologous and nonautologous plasma samples. Cells were isolated from 3-D cultures with Cell Recovery Solution per the manufacturer's instructions. To obtain enough material for subsequent analyses, 6 to 10 × 106 cells from each patient were placed into 3-D culture. All patients were analyzed individually. In situ BM particles were grown as described for BMCs. BM particles in fresh BM aspirates are composed of speckles of the bone with attached BM. Particles were rinsed with PBS to remove red blood cells, and 1 to 4 particles were placed on the surface of fibronectin/collagen I-coated tissue culture plate and covered with 100 μL of Matrigel/fibronectin. The culture was allowed to solidify and was covered with growth medium as described.
Preclinical drug treatment studies
For preclinical studies, the rBM was grown for 14 days to allow for the expansion of the malignant and stromal compartments. A single treatment of either melphalan or bortezomib was administered in a fresh aliquot of growth medium incorporating MM plasma, overlaid on top of the 3-D culture. After 7 days of treatment, fresh growth medium was added and cells were allowed to recover for 7 days, to mimic clinical protocols involving a recovery period between chemotherapy treatment and measurement of tumor burden. Subsequently, cells were isolated from 3-D culture and treatment efficacy evaluated using real-time quantitative polymerase chain reaction (RQ-PCR) to detect genomic clonotypic IgH VDJ. For CFU activity, nonproliferating cells defined by their retention of high intensity of CFSE dye during the 3-D culture, termed label retaining cells (LRCs), were purified by sorting CFSEhigh cells within lymphocyte scatter gates using an EPICS ALTRA and transferred to a colony forming unit (CFU) assay.
CFU assay
The CFSEhigh or CFSEhighCD20+ cells were sorted by gating on forward and side scatter (lymphocyte gates) and on the CD20+ and CFSEhigh population based on the fluorescence intensity. CSFEhigh cells were defined as only those cells having an intensity of CSFE staining matching that at the time 3-D cultures were initiated. Sorted cells from individual patients were resuspended in 200 μL Iscove modified Dulbecco medium with 2% fetal bovine serum (Invitrogen Canada), mixed with 1.1 mL MethoCult GF medium (methylcellulose, 2% fetal bovine serum, bovine serum albumin, 2-mercaptoethanol, rhSCF, rhGM-CSF, rhIL-3, rhG-CSF, rh-erythropoietin in Iscove modified Dulbecco medium) in 35-mm plates. CFU cultures were incubated for 14 days, and the resulting colonies were counted. For serial replating experiments, a fragment of each colony was transferred to a fresh plate with MethoCult GF medium. This was done for each successive replating. The remainder of the colony was used for parallel phenotypic and genetic analysis. To quantify cells harboring the clonotypic IgH VDJ, individual colonies were plucked, transferred to PBS, centrifuged to remove MethoCult, and the resultant pellets resuspended in TRIzol (Invitrogen Canada), followed by purification of genomic DNA and RQ-PCR with patient-specific primers.
RQ-PCR
Total genomic DNA was isolated with TRIzol reagent per the manufacturer's instructions. RQ-PCR for patient-specific rearranged IgH VDJ clonotypic sequences was performed using the SYBR green method29 with product specificity confirmed by melting curve analysis30 per the manufacturer's instructions. Patient specific primers for CDR2 and CDR3 were designed as previously described.31,32 Because only one IgH VDJ allele has the clonotypic gene rearrangement, each malignant cell has only one genomic copy of this unique MM signature. To quantify the number of templates in unknown samples before and after 3-D culture, controls included cloned VDJ sequences for each patient for generating a standard curve for each specific clonotypic IgH VDJ and comparison with a standard curve for a housekeeping gene, β2 microglobulin (normalized to the 2 copies of B2M templates/cell). Percentage of clonal cells (% VDJ) was calculated based on the formula, %VDJ = [10(−0.2774c(t)) + 9.708]/[10(−0.2901c(t)) + 9.86] × 100, where the numerator of the equation corresponds to the number of clonal IgH VDJ molecules and the denominator corresponds to the number of β2-microglobulin molecules. Fluorescence was acquired at each cycle; the c(t) value corresponds to the number of cycles where the RQ-PCR curve crosses a threshold line set at the midpoint of the log fluorescence expansion.
Microscopy
For confocal microscopy, CFSE-labeled MM BM or MBAs were placed in 3-D cultures in coverslip-bottom (1-mm) 8-well chamber slides. When necessary, nuclei were stained by adding 1 μL 4,6-diamidino-2-phenylindole (DAPI; 1:4000) to the growth medium for 15 minutes at 37°C. Confocal z-slices were imaged on a Carl Zeiss confocal LSM 510 microscope (Toronto, ON) with a 10× objective. 3-D reconstruction used Zeiss 510 image analysis software. Quantification of LRC positioning was performed with Imaris 3.2.2 software (Bitplane, Zurich, Switzerland). The cellular identity was determined based on the coculture experiments where CFSE-labeled sorted B or plasma cells were added to unlabeled rBM. The cellular behavior was followed by confocal microscopy and was correlated with the DIC images to identify cell type in rBM. To identify cells in the stromal layer, the top matrix layer was carefully aspirated; cells remaining attached to the bottom of the culture plate were stained with TRAP, Oil Red, alkaline phosphatase, or Alizarin Red according to the manufacturer's instructions and imaged on Zeiss Axioskop digital microscope with a 20× objective. Brightfield microscopy was done using a Zeiss Axiovert 200M inverted microscope with 10× or 20× objectives and image analysis was completed using MetaMorph software (Molecular Devices, Downingtown, PA). For fibronectin, phalloidin, and N-cadherin staining, cells were fixed with 1% neutral buffered formalin for 10 minutes at room temperature, permeabilized with 0.5% Triton X-100, and nonspecific binding was blocked with 1% bovine serum albumin for 1 hour at room temperature. Cells were incubated with primary antibodies, followed by washing, incubation with secondary antibodies for 1 hour and confocal imaging.
Flow cytometry
For proliferation analysis, CFSE-labeled cells were isolated from 3-D cultures of individual patients, stained with CD20-phycoerythrin (PE), CD138-PC5, or CD56-PE/CD138-PC5, and 30 000 events were collected and analyzed on a FACSort (BD Biosciences). For apoptosis studies, cells were isolated from 3-D and stained with annexin V-PE in combination with CD3-fluorescein isothiocyanate (FITC), CD20-FITC, CD33-FITC, CD56-FITC, and CD138-PC5. To analyze the CFU composition, colonies from p4 and p5 were expanded to generate material sufficient for FACS analysis at p6. Data were analyzed with CellQuest Pro software (BD Biosciences).
Immunohistochemistry
Paraffin blocks were prepared by removing media from the culture and perfusing the matrix with 300 μL/well of 3% agar. Once solidified, the agar block was removed and fixed in 10% neutral buffered formalin overnight, before standard processing; 5-μ sections were stained for CD34, CD20, and CD138 expression using a standard avidin-biotin-peroxidase technique. Cytospins of sorted LRCs were stained for CD20 using immunohistochemistry. Imaging was done with a 40× objective on a Zeiss Axiovision microscope equipped with a color camera.
Fluorescent in situ hybridization
Cells before and after culture were immobilized onto a glass slide by cytospinning and stained with May-Grunwald for 3 minutes at room temperature, washed in H2O, stained with Giemsa in dH2O (1:20) for 10 minutes at room temperature, and rinsed with H2O. The position of all cells was recorded using the BioView Duet scanning system (Rehovot, Israel). The slide was then destained in methanol/acetic acid (3:1) for 60 minutes, and conventional interphase fluorescent in situ hybridization (FISH) analysis was performed as previously described.33 Three probe sets were used: a commercial Vysis LSI 13q34 probe for the detection of the deletion of 13q34 locus, Vysis LSI IGH/CCND1-XT dual color-dual fusion translocation probe for the detection of translocation (11,14)(q13;q32), and Vysis CEP1(D1Z5) as a control probe combined with a home-made probe targeting locus 1q21 for detecting amplified 1q21. Brightfield and fluorescence microscopy used the BiovVew Duet system to correlate the FISH staining (63× objective) pattern and morphology (40× objective) for each cell on the slide.
Statistical analysis
Data were presented as means plus or minus SEM. Statistical significance was measured by Student t test using Prism 4 software from GraphPad Software (San Diego, CA). Differences in the mean values were reported as P values with P less than .05 considered significant.
Results
Three-dimensional culture recapitulates the in vivo BM microenvironment
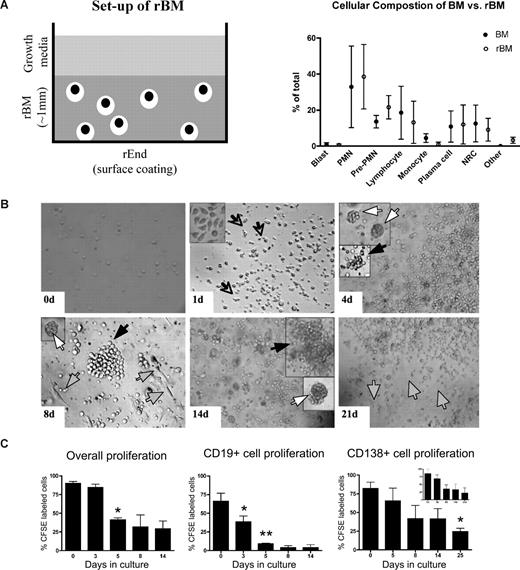
In 3-D cultures, endosteal (rEnd) and BM (rBM) matrices have been reconstructed to mimic the in vivo composition of BM34 (J.K. and L.M.P., manuscript in preparation). rEnd was reconstructed by surface coating of with collagen I/fibronectin mixture and overlaid with BMCs suspended in fibronectin/Matrigel mixture, which is representative of the in vivo central marrow composed of fibronectin, laminin, and collagen IV34 (Figure 1A). To confirm the similarity between the cellular composition of the preculture BM and the rBM postculture, the proportion of each cellular compartment was determined in BMCs and in cells from dissociated 3-D cultures. 3-D cultures maintain the cellular composition of in vivo BM (Figure 1B); no differences were seen between the percentages of hematopoietic cells in BM compared with rBM (P > .05). The only exceptions were a decreased proportion of monocytes in 3-D probably because of their differentiation to macrophages (P = .015) and an increase in osteoclasts (P = .001), defined by positive TRAP staining.
3-D culture model is designed to mimic the in vivo microenvironment of the BM. (A) Diagram of the 3-D BM culture setup with fibronectin/collagen I surface coating to reconstruct rEnd, overlaid with BMCs mixed with Matrigel/fibronectin ECM, covered with growth medium (left panel). Cellular composition of the 3-D culture assessed after 3 weeks (right panel). Cytospins were prepared from the preculture BM and from the same samples after culture (n = 6 individually analyzed patients) and stained with May-Grunwald-Giemsa. Nuclear and cytoplasmic staining and morphology were analyzed, and cells of various lineages were counted and reported as percentage of total cellularity. Blast indicates all blasts; PMN, polymorphonuclear (bands, neutrophils, eosinophils, basophils); pre-PMN, promyelocyte, myelocyte, metamyelocyte; lymphocyte, B and T cells; NRC, nucleated red cells (proerythroblast, basophilic erythroblast, polychromatophilic erythroblast, orthochromatic erythroblast); other, macrophages, stromal cells, reticular cells, osteoclastic cells, osteoblastic cells. (B) BMCs grown in rBM recapitulate multicellularity of the BM. BMCs were grown in 3-D for the indicated number of days, and the overall view of the culture was analyzed using brightfield microscopy (original magnification ×200; see “Microscopy”). Open arrows and inset represent migrating cells; large black arrows, B-cell colonies; large white arrows, PC colonies; striped arrows, stromal cells. Representative images from 48 individually analyzed patients. (C) Proliferation of the various cellular compartment of the rBM was measured by labeling BMCs with CFSE (n = 4 individually analyzed patients) and culturing in 3-D for the indicated number of days. Cells were isolated from 3-D, stained with fluorescent tagged antibodies as indicated, and proliferation capacity of the aggregate rBM (*P = .001), CD20+ B cells (*P = .024, **P = .006), and CD138+ PCs (*P = .004) was measured by multicolor flow cytometry (inset, proliferation profile of CD138+CD56+ cells). Percentage CFSE labeled cells represents cells with average amount of retained CFSE above the unlabeled control. Error bars in panels B and D represent SEM.
3-D culture model is designed to mimic the in vivo microenvironment of the BM. (A) Diagram of the 3-D BM culture setup with fibronectin/collagen I surface coating to reconstruct rEnd, overlaid with BMCs mixed with Matrigel/fibronectin ECM, covered with growth medium (left panel). Cellular composition of the 3-D culture assessed after 3 weeks (right panel). Cytospins were prepared from the preculture BM and from the same samples after culture (n = 6 individually analyzed patients) and stained with May-Grunwald-Giemsa. Nuclear and cytoplasmic staining and morphology were analyzed, and cells of various lineages were counted and reported as percentage of total cellularity. Blast indicates all blasts; PMN, polymorphonuclear (bands, neutrophils, eosinophils, basophils); pre-PMN, promyelocyte, myelocyte, metamyelocyte; lymphocyte, B and T cells; NRC, nucleated red cells (proerythroblast, basophilic erythroblast, polychromatophilic erythroblast, orthochromatic erythroblast); other, macrophages, stromal cells, reticular cells, osteoclastic cells, osteoblastic cells. (B) BMCs grown in rBM recapitulate multicellularity of the BM. BMCs were grown in 3-D for the indicated number of days, and the overall view of the culture was analyzed using brightfield microscopy (original magnification ×200; see “Microscopy”). Open arrows and inset represent migrating cells; large black arrows, B-cell colonies; large white arrows, PC colonies; striped arrows, stromal cells. Representative images from 48 individually analyzed patients. (C) Proliferation of the various cellular compartment of the rBM was measured by labeling BMCs with CFSE (n = 4 individually analyzed patients) and culturing in 3-D for the indicated number of days. Cells were isolated from 3-D, stained with fluorescent tagged antibodies as indicated, and proliferation capacity of the aggregate rBM (*P = .001), CD20+ B cells (*P = .024, **P = .006), and CD138+ PCs (*P = .004) was measured by multicolor flow cytometry (inset, proliferation profile of CD138+CD56+ cells). Percentage CFSE labeled cells represents cells with average amount of retained CFSE above the unlabeled control. Error bars in panels B and D represent SEM.
To better define rBM, the temporal organization of the 3-D culture was evaluated (Figure 1C). In 3-D cultures, day 0 (D0) BM cells were randomly distributed throughout the 3-D matrix. As early as D1 of culture, cells began to migrate within the ECM, marked by the appearance of leading edge protrusions. By D4, both diffuse and tight colonies were seen. By D14, the architecture of the rBM closely resembled that of the in vivo BM. Stromal cells appeared at the rEnd after a week in culture and grew to cover the surface of the culture vessel.
To determine whether rBM cellularity reflected cell proliferation, before culture in 3-D, BMCs were labeled with CFSE, a cytoplasmic fluorescent dye that is reduced in intensity by 50% with each cell division. Significant overall proliferation was observed from day 0 to day 5, with cells continuing to proliferate through day 14 (Figure 1D). Proliferation of the CD20+ B cells was rapid from day 0 through day 5. Consistent with their slow proliferation in vivo, CD138+ PCs, the majority of which were CD45dim, divided less rapidly than B cells, as defined by their higher intensity of CFSE, and their proliferation only reached significance by day 25 (Figure 1D). Proliferation of cells within rBM combined with the proportionate preservation of hematopoietic compartments (Figure 1B) implies that hematopoietic lineage relationships are preserved in 3-D cultures.
rBM maintains human BM architecture
Physical stratification of the BM includes distinct niches with dormant hematopoietic progenitor cells (HPCs) found at the bone surface in close association with the endosteum, and more differentiated hematopoietic cells such as B cells and PCs found in the central marrow, with the most differentiated cells localized farthest away from the endosteum.35-39 The interaction between HPCs, endosteal osteoblasts and osteoclasts, bone-forming and -resorbing cells, respectively, is essential for the maintenance of the HPC niche and mobilization of stem cells.35,36 In 3-D culture, after completion of redistribution and proliferation, rBM was stratified into approximately top, middle, and rEnd layers within the 1-mm 3-D cultures (Figure 2). Similar to the architecture of the in vivo BM, the endosteal niche of rBM was composed of fibroblastic stromal cells, adipocytes, and osteoclastic and osteoblastic cells capable of mineralizing calcium (Figure 2C). Exclusion of the rEnd coating from the culture greatly reduced the stromal compartment (not shown). Obtaining the entire stromal compartment in 3-D is a marked improvement over standard 2-D methods where only a single stomal cell type differentiates from BM, depending on the culture media.19,36 Mimicking their localization in intact BM, CD34+ HPCs were found at rEnd in contact with stromal cells, CD20+ B cells localized mainly to the middle layer, and CD138+ PCs mainly to the top layer of the rBM (Figure 2D). To verify that purified cells from BM aspirates maintain the growth properties of in vivo BM, we compared them with BM particles placed into 3-D culture within 2 hours of aspiration. BM particles remain intact to preserve the in situ integrity of the in vivo microenvironment. Grown in 3-D, cells expanding in situ from within BM particles had the same properties as the rBM cultures (Figure 2B), that is, proliferation, stratification, maintenance of LRCs, and expansion of the MM clone.
rBM maintains the architecture of in vivo BM. (A) BMCs were grown in rBM for 21 days and stained with DAPI (pseudocolored red) to mark nuclei. The stratification of the 3-D culture was assessed by confocal microscopy as a 3-D reconstruction from the confocal stack (original magnification 100×). Representative images from BM of 48 individually analyzed patient samples. (B) BM aspirated particle grown in rBM for 21 days and stained with DAPI (pseudocolored red) to mark nuclei. The stratification of the 3-D culture was assessed by confocal microscopy as a 3-D reconstruction from the confocal stack (original magnification 100×). Representative image from analysis of aspirated particles from 28 individually analyzed patients. (C) BM stromal cells redistribute to rEnd. The rBM layer was removed from 3-D culture, and the cells at rEnd were stained with markers to identify stromal components observed at rEnd (adipocytes, Oil Red; fibroblasts, fibronectin and phallodin; osteoblasts, alkaline phosphatase [ALP] and Alizarin Red [inset]; osteoclasts, tartarate-resistant acidic phosphatase [TRAP]). Brightfield microscopy (original magnification 200×). Representative images from 7 individually analyzed patients; see “Immunohistochemistry.” (D) BMCs were grown for 14 days and the percentage of CD34+ HPCs, B cells, or PCs in each layer (black represents fraction in bottom layer; gray, fraction in middle; white, fraction in top layer) of the 3-D culture was calculated based on the immunohistochemical staining for CD34, CD20, or CD138 (n = 4 individually analyzed patients). Error bars represent SEM. For complete image acquisition information, see “Microscopy.”
rBM maintains the architecture of in vivo BM. (A) BMCs were grown in rBM for 21 days and stained with DAPI (pseudocolored red) to mark nuclei. The stratification of the 3-D culture was assessed by confocal microscopy as a 3-D reconstruction from the confocal stack (original magnification 100×). Representative images from BM of 48 individually analyzed patient samples. (B) BM aspirated particle grown in rBM for 21 days and stained with DAPI (pseudocolored red) to mark nuclei. The stratification of the 3-D culture was assessed by confocal microscopy as a 3-D reconstruction from the confocal stack (original magnification 100×). Representative image from analysis of aspirated particles from 28 individually analyzed patients. (C) BM stromal cells redistribute to rEnd. The rBM layer was removed from 3-D culture, and the cells at rEnd were stained with markers to identify stromal components observed at rEnd (adipocytes, Oil Red; fibroblasts, fibronectin and phallodin; osteoblasts, alkaline phosphatase [ALP] and Alizarin Red [inset]; osteoclasts, tartarate-resistant acidic phosphatase [TRAP]). Brightfield microscopy (original magnification 200×). Representative images from 7 individually analyzed patients; see “Immunohistochemistry.” (D) BMCs were grown for 14 days and the percentage of CD34+ HPCs, B cells, or PCs in each layer (black represents fraction in bottom layer; gray, fraction in middle; white, fraction in top layer) of the 3-D culture was calculated based on the immunohistochemical staining for CD34, CD20, or CD138 (n = 4 individually analyzed patients). Error bars represent SEM. For complete image acquisition information, see “Microscopy.”
rBM from MM patients supports expansion of the MM clone
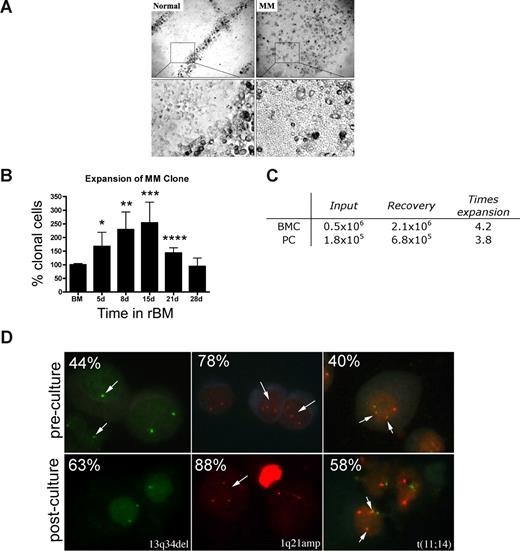
Osteoclasts occupied distinct positions in the rBM of healthy donors. In contrast, rBM from MM patients was disorganized, with osteoclast-like cells throughout (Figure 3A). The ordered osteoclast organization of rBM disappeared when plasmacytoma progressed to MM (not shown).
MM rBM has an abnormal tissue architecture and exhibits clonal expansion of MM cells with chromosomal abnormalities. (A) BMCs from normal donors (n = 5) and MM patients (n = 10) were grown in rBM for 21 days followed by assessment of the overall culture architecture by brightfield microscopy (top, original magnification 50×; bottom, original magnification 200×). Cells localized to “tracks” are TRAP+ osteoclasts (not shown). Representative images are shown with the plane of focus set to the bottom of the plate representing reconstructed endosteum. (B) BM cells (n = 16 patients) were grown in rBM for the indicated number of days, and the extent of the malignant outgrowth of the MM clone was measured by RQ-PCR using patient specific primers. Each malignant cell has only one copy of the IgH VDJ rearranged template; RQ-PCR determines the number of rearranged IgH VDJ templates compared with a β2-microglobulin standard curve and an IgH VDJ positive control curve. Percent clonal cells corresponds to the percentage clonal VDJ templates present in the sample normalized to the β2-microglobulin gene (*P = .006, **P = .001, ***P = .002, ****P = .003). Error bars represent SEM. (C) Absolute numbers of BMCs and PCs, which were originally placed into and recovered from rBM (mean values for 3 different patients). (D) FISH analysis of cells harvested from 3-D culture. Cells from 3-D cultures were screened for the chromosomal abnormalities (marked with arrows, and percentage plasma cells with each abnormality stated at the top left) detected in preculture BM PCs, using cytospins from the matching D21 rBM (n = 5 patients) stained by interphase FISH. Abnormal cells are indicated by arrows. (Left panel) PCs with a deletion of 13q34 locus (1 green signal) and a normal cell (2 green signals). (Middle panel) PCs with an amplification of 1q21 (3 red signals) and a normal cell (2 red signals). (Right panel) PCs with t(11;14) translocation with 2 derivative chromosomes (2 yellow signals), 1 normal chromosome 11 (1 red signal), and 1 normal chromosome 14 (1 green signal; original magnification 630×). For additional image acquisition information, see “Microscopy” and “Fluorescent in situ hybridization.”
MM rBM has an abnormal tissue architecture and exhibits clonal expansion of MM cells with chromosomal abnormalities. (A) BMCs from normal donors (n = 5) and MM patients (n = 10) were grown in rBM for 21 days followed by assessment of the overall culture architecture by brightfield microscopy (top, original magnification 50×; bottom, original magnification 200×). Cells localized to “tracks” are TRAP+ osteoclasts (not shown). Representative images are shown with the plane of focus set to the bottom of the plate representing reconstructed endosteum. (B) BM cells (n = 16 patients) were grown in rBM for the indicated number of days, and the extent of the malignant outgrowth of the MM clone was measured by RQ-PCR using patient specific primers. Each malignant cell has only one copy of the IgH VDJ rearranged template; RQ-PCR determines the number of rearranged IgH VDJ templates compared with a β2-microglobulin standard curve and an IgH VDJ positive control curve. Percent clonal cells corresponds to the percentage clonal VDJ templates present in the sample normalized to the β2-microglobulin gene (*P = .006, **P = .001, ***P = .002, ****P = .003). Error bars represent SEM. (C) Absolute numbers of BMCs and PCs, which were originally placed into and recovered from rBM (mean values for 3 different patients). (D) FISH analysis of cells harvested from 3-D culture. Cells from 3-D cultures were screened for the chromosomal abnormalities (marked with arrows, and percentage plasma cells with each abnormality stated at the top left) detected in preculture BM PCs, using cytospins from the matching D21 rBM (n = 5 patients) stained by interphase FISH. Abnormal cells are indicated by arrows. (Left panel) PCs with a deletion of 13q34 locus (1 green signal) and a normal cell (2 green signals). (Middle panel) PCs with an amplification of 1q21 (3 red signals) and a normal cell (2 red signals). (Right panel) PCs with t(11;14) translocation with 2 derivative chromosomes (2 yellow signals), 1 normal chromosome 11 (1 red signal), and 1 normal chromosome 14 (1 green signal; original magnification 630×). For additional image acquisition information, see “Microscopy” and “Fluorescent in situ hybridization.”
rBM supported the expansion of the MM clone as measured by the RQ-PCR analysis of cells harvested from 3-D, using patient-specific primers to amplify the clonotypic IgH VDJ,31 providing a unique clonal marker to identify all MM cells.17,32 For rBM from 16 patients, by day 15 there was a mean increase of 2.5-fold (1.3- to 9.9-fold) in the percentage clonotypic MM cells in rBM compared with autologous precultured BM (P = .009; Figure 3B). Sequencing confirmed that the IgH VDJ of MM cells arising in 3-D was identical to that of autologous ex vivo MM PCs. BM cells from aspirated particles exhibited similar levels of MM clonal expansion to those of purified BMC, with a 3 times increase in clonotypic cells in 3-D cultures (not shown). To determine whether the increase in the percentage of clonotypic cells is related to numerical expansion rather than the death of nonmalignant cells, we compared the absolute number of cells at the time of rBM setup (day 0) with that at the time of harvest (day 21). We determined that malignant PC undergo a mean 3.8 times expansion in 3-D culture, from 1.8 × 105 PCs at day 0 to 6.8 × 105 PCs at day 21 (Figure 3C).
MM is also characterized by chromosomal abnormalities.40 To confirm that PCs proliferating in 3-D cultures were part of the MM clone, the chromosomal abnormalities in PCs from rBM were compared with those present in autologous ex vivo PCs, using interphase FISH. PCs from 3-D had the same chromosomal abnormalities as ex vivo PCs (Figure 3D), with a 1.3 times increase in the absolute number of chromosomally abnormal PCs by day 15 of culture. In contrast, the total number of clonotypic cells in the 3-D culture, as measured the RQ-PCR, was increased 2.5 times (Figure 3B). This suggests that, in addition to PCs, other clonotypic MM compartments have also expanded in rBM. The proliferation of the MM clone in 3-D is clinically significant because MM cells do not proliferate in any of the currently existing systems, rendering the 2-D culture models unsuitable for preclinical drug testing using ex vivo BM cells.
Three-dimensional culture allows preclinical testing of therapeutic targeting to all compartments of the MM clone
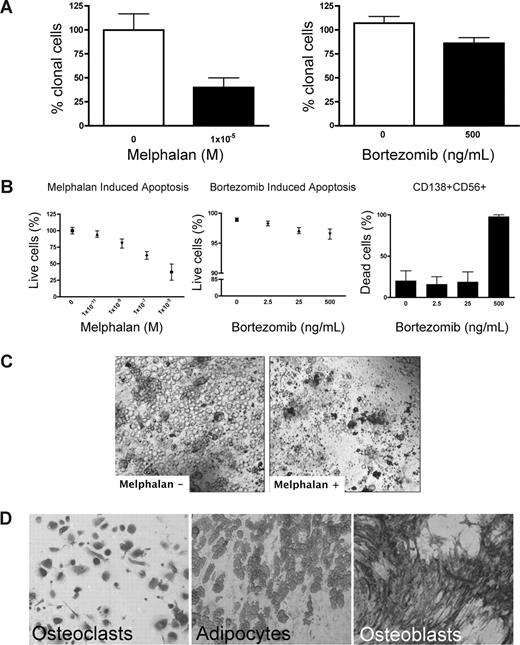
Preclinical testing in MM has been hindered by the lack of systems to sustain growth of the MM clone. Because rBM contains the full complement of BM cells found in vivo, chemotherapeutic (melphalan) or biologically based (bortezomib) agents were used to validate the 3-D model for preclinical use. Treatment of rBM with melphalan led to a 60% decrease in the number of clonotypic cells as measured by RQ-PCR (Figure 4A). Consistent with its myeloablative properties, melphalan induced apoptosis of all cell types within the hematopoietic hierarchy present in the rBM (Figure 4B,C) but spared the stromal compartment (Figure 4D). Although bortezomib induced a significant (21%) reduction in clonotypic cells (Figure 4A), overall apoptosis was not significant (Figure 4B) in rBM. Closer examination revealed that bortezomib-induced apoptosis was exclusive to a minor population of CD138+CD56+ PCs (Figure 4B). Neither CD20+, nor CD138+56−, nor CD56+138−, nor CD33+, nor CD3+ cells of the hematopoietic compartment nor the stromal compartment was affected by bortezomib (not shown). A clinical reduction in BM plasmacytosis marked by the disappearance of monoclonal Ig, coupled with inevitable relapse, suggests that bortezomib targets a PC subset responsible for disease symptoms but leaves other generative compartments intact. Unlike the conventional drug evaluation, which measures, the impact only on MM PCs, 3-D cultures allow monitoring of all BM compartments in the context of microenvironment-mediated drug resistance to establish clinically relevant drug dosing. The apparent agreement between clinical impact and the preclinical studies in rBM validates the 3-D model as relevant to events in vivo. However, given the inevitable relapse of MM and the likelihood that MM-CSCs escape current modes of therapy, a valid preclinical model must also monitor the impact of therapy on cancer stem cells.
Melphalan and bortezomib affect the hematopoietic, but not the stromal, compartment of rBM. (A-D) MM BMCs were grown in 3-D for 14 days and treated with melphalan or bortezomib (“Preclinical drug treatment studies”). (A) The reduction of the clonal cells was measured by RQ-PCR with patient specific primers (melphalan, P = .016; bortezomib, P = .033; n = 5 individually analyzed patients). (B) Apoptosis within rBM was assessed by flow cytometry measuring annexin V reactivity after treatment with melphalan (left panel) or bor-tezomib (middle panel). MM specific cell killing by bor-tezomib was monitored in the CD138+/CD56+ population by annexin V reactivity (right panel; n = 5 individually analyzed patients). Error bars represent SEM. (C) rBM was grown and treated with melphalan as in panel A. Cell kill was monitored by brightfield microscopy (original magnification 200×; see “Microscopy”). Representative images from 5 individually analyzed patients. (D) The rBM layer was removed, and the cells at rEnd were stained with TRAP (osteoclasts), Oil Red (adipocytes), and ALP (osteoblasts). Brightfield microscopy (original magnification 200×; see “Immunohistochemistry”). Representative images from 5 individually analyzed patients.
Melphalan and bortezomib affect the hematopoietic, but not the stromal, compartment of rBM. (A-D) MM BMCs were grown in 3-D for 14 days and treated with melphalan or bortezomib (“Preclinical drug treatment studies”). (A) The reduction of the clonal cells was measured by RQ-PCR with patient specific primers (melphalan, P = .016; bortezomib, P = .033; n = 5 individually analyzed patients). (B) Apoptosis within rBM was assessed by flow cytometry measuring annexin V reactivity after treatment with melphalan (left panel) or bor-tezomib (middle panel). MM specific cell killing by bor-tezomib was monitored in the CD138+/CD56+ population by annexin V reactivity (right panel; n = 5 individually analyzed patients). Error bars represent SEM. (C) rBM was grown and treated with melphalan as in panel A. Cell kill was monitored by brightfield microscopy (original magnification 200×; see “Microscopy”). Representative images from 5 individually analyzed patients. (D) The rBM layer was removed, and the cells at rEnd were stained with TRAP (osteoclasts), Oil Red (adipocytes), and ALP (osteoblasts). Brightfield microscopy (original magnification 200×; see “Immunohistochemistry”). Representative images from 5 individually analyzed patients.
Three-dimensional culture reveals presumptive MM-CSCs
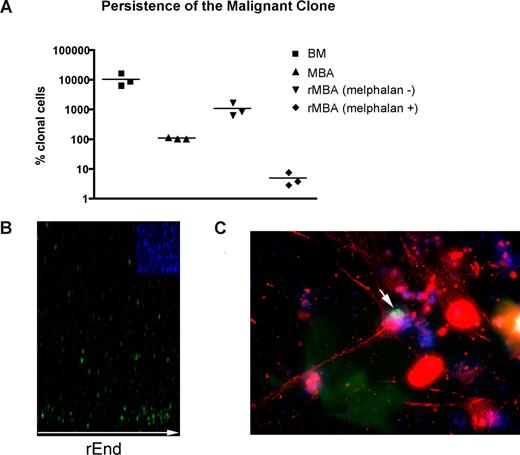
Although residual disease almost certainly contributes to relapse after high-dose chemotherapy and stem cell rescue, MBAs have been shown to harbor malignant cells capable of regenerating MM.23 As measured by RQ-PCR, MBA samples exhibit 100 times reduction in the tumor burden compared with BM samples collected before myeloablative therapy and autologous stem cell transplantation (P = .003; Figure 5A). MBAs from MM patients reconstituted a BM-like 3-D microenvironment (rMBA), generating 6.3- to 16.6-fold increases in the number of clonotypic MM cells, compared with autologous preculture MBAs (P = .04). Melphalan treatment of rMBA resulted in depletion of clonotypic MM cells (P = .03; Figure 5A). We hypothesized that putative MM-CSCs capable of initiating relapse are found in the MBA.
LRCs redistribute to rEnd. (A) Mononuclear cells from MBA (n = 3 patients) were treated with melphalan (“Preclinical drug treatment studies”) and grown in rBM for 21 days, and the extent of the malignant outgrowth was measured by RQ-PCR. MBA generated MM progeny and LRCs comparable with those from BM; the majority of MBAs also generated stromal layers. Preculture ex vivo BMCs and MBA cells were used as controls. Horizontal lines represent medians. (B,C) BMCs were labeled with CFSE (green) and cultured for 21 days. (B) rBM was stained with DAPI (blue inset) to detect total cells in the cultures, and the position of the brightly staining CFSE+ LRCs was analyzed by confocal microscopy (original magnification 100×) as indicated in “Microscopy.” Representative images from 8 individually analyzed patient samples. (C) The rBM layer was removed, and the cells at rEnd were fixed and stained with N-cadherin. CFSE+ LRCs (green) shown in contact with an N-cadherin+ stromal cell (red), DAPI stained nuclei (blue). Representative 10 images from 3 individually analyzed patients.
LRCs redistribute to rEnd. (A) Mononuclear cells from MBA (n = 3 patients) were treated with melphalan (“Preclinical drug treatment studies”) and grown in rBM for 21 days, and the extent of the malignant outgrowth was measured by RQ-PCR. MBA generated MM progeny and LRCs comparable with those from BM; the majority of MBAs also generated stromal layers. Preculture ex vivo BMCs and MBA cells were used as controls. Horizontal lines represent medians. (B,C) BMCs were labeled with CFSE (green) and cultured for 21 days. (B) rBM was stained with DAPI (blue inset) to detect total cells in the cultures, and the position of the brightly staining CFSE+ LRCs was analyzed by confocal microscopy (original magnification 100×) as indicated in “Microscopy.” Representative images from 8 individually analyzed patient samples. (C) The rBM layer was removed, and the cells at rEnd were fixed and stained with N-cadherin. CFSE+ LRCs (green) shown in contact with an N-cadherin+ stromal cell (red), DAPI stained nuclei (blue). Representative 10 images from 3 individually analyzed patients.
A defining property of cancer stem cells, their proliferative quiescence, was measured here by the retention of CFSE fluorescent label (LRCs), to distinguish them from proliferating cells, which progressively lose CSFE with each cell division.41 To identify putative MM-CSCs, we characterized CFSEhigh LRCs from 3-D cultures of rMBA. Quiescent cells cannot be distinguished or accessed in conventional cultures because the MM clone as a whole fails to proliferate in these cultures. In contrast, 3-D culture enables identification and sorting of nonproliferating CFSEhigh LRCs, including putative MM-CSCs, for further analysis. Selection of CSFEhigh cells excludes all proliferating compartments of the rBM; thus, the LRCs represent populations derived directly from the patient after a period of stasis in the 3-D microenvironment.
Overall, 60% of LRCs localized to the rEnd (Figure 5B) in close contact with osteocalcin (not shown) and N-cadherin–expressing osteoblastic cells (Figure 5C). This interaction has been shown to maintain dormancy of normal HPCs.42-45 To determine whether LRCs localized specifically to the rEnd, as opposed to a simple preference for a rigid substratum, the rEnd was eliminated from a set of 3-D cultures. Without rEnd, LRCs were dispersed evenly throughout the matrix (not shown) and the stromal layer was only minimal, confirming the requirement for rEnd in attracting LRC and promoting stromal development.
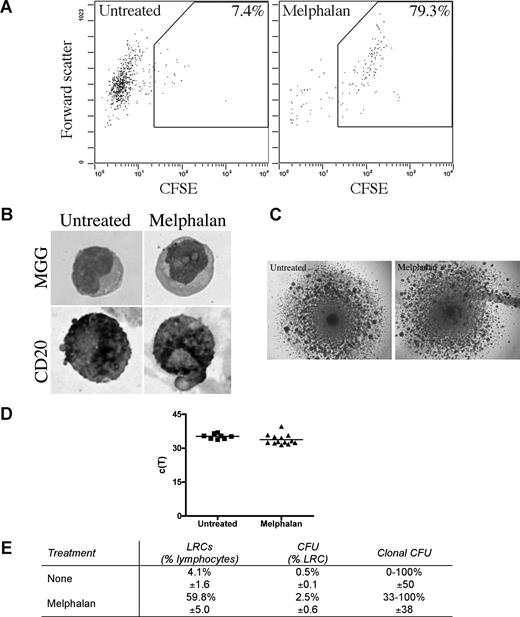
LRCs were isolated from 3-D by sorting CFSEhigh cells having lymphocyte scatter properties; lymphocytic LRCs were drug-resistant as defined by their persistence in 3-D cultures treated with melphalan or bortezomib, and were enriched 10 times in melphalan-treated cultures (Figure 6A). The largest subset of LRCs (mean, 46%) were B lymphocytes, as assessed by May-Grunwald-Giemsa staining and expression of CD20 (Figure 6B). The remainder of LRCs were terminally differentiated myelocytes (3%), bands (3%), polymorphonuclear cells (21%), monocytes (3%), red blood cells (14%), and stromal cells (10%), which were excluded from further analysis by the sort parameters; no morphologically identifiable PCs were detected among LRC.
LRCs include drug-resistant progenitors able to generate clonotypic progeny. (A-D) Mononuclear cells were isolated from the mobilized blood preparations, labeled with CFSE, grown in 3-D for 14 days, and treated with melphalan (“Preclinical drug treatment studies”). (A) Flow cytometric profile of the LRCs. The gates for sorting were set to isolate cells with lymphocyte scatter properties (scatter plot shown is gated on the lymphocytes) and CFSEhigh fluorescence, comparable with that in the stained population preculture, as expected for nonproliferating or slowly proliferating cells. Percentage of LRCs was calculated as the percentage of CFSEhigh cells in the lymphocyte fraction of the 3-D culture. Representative FACS plots from 5 individually analyzed patients. (B) May-Grunwald-Giemsa (MGG) staining of the sorted LRCs (top) and immunohistochemical staining of sorted LRCs for CD20 (bottom; original magnification ×400); see “Immunohistochemistry.” Representative images from 3 individually analyzed patients. (C) Representative colonies from the CFU assay (original magnification 40×; n = 10 individually analyzed patients) as indicated in “Microscopy.” (D) Progeny generated from sorted LRCs from both untreated and melphalan-treated cultures were analyzed by RQ-PCR to quantify the number of clonotypic cells. These c(T) values, compared with control values, indicate that approximately 100% of the progeny were clonotypic. No RQ-PCR amplification was detectable for nonclonotypic colonies. Similar c(T) values were observed for clonotypic colonies from both treatment groups (n = 8 colonies from untreated group and n = 11 colonies from melphalan group). Horizontal lines represent medians. (E) Quantification of the LRCs as a percentage of total cells in the culture, CFU as a percentage of LRCs, and clonal CFU as a percentage of total colonies obtained in the CFU assay (mean ± SD of 8 individually analyzed patients).
LRCs include drug-resistant progenitors able to generate clonotypic progeny. (A-D) Mononuclear cells were isolated from the mobilized blood preparations, labeled with CFSE, grown in 3-D for 14 days, and treated with melphalan (“Preclinical drug treatment studies”). (A) Flow cytometric profile of the LRCs. The gates for sorting were set to isolate cells with lymphocyte scatter properties (scatter plot shown is gated on the lymphocytes) and CFSEhigh fluorescence, comparable with that in the stained population preculture, as expected for nonproliferating or slowly proliferating cells. Percentage of LRCs was calculated as the percentage of CFSEhigh cells in the lymphocyte fraction of the 3-D culture. Representative FACS plots from 5 individually analyzed patients. (B) May-Grunwald-Giemsa (MGG) staining of the sorted LRCs (top) and immunohistochemical staining of sorted LRCs for CD20 (bottom; original magnification ×400); see “Immunohistochemistry.” Representative images from 3 individually analyzed patients. (C) Representative colonies from the CFU assay (original magnification 40×; n = 10 individually analyzed patients) as indicated in “Microscopy.” (D) Progeny generated from sorted LRCs from both untreated and melphalan-treated cultures were analyzed by RQ-PCR to quantify the number of clonotypic cells. These c(T) values, compared with control values, indicate that approximately 100% of the progeny were clonotypic. No RQ-PCR amplification was detectable for nonclonotypic colonies. Similar c(T) values were observed for clonotypic colonies from both treatment groups (n = 8 colonies from untreated group and n = 11 colonies from melphalan group). Horizontal lines represent medians. (E) Quantification of the LRCs as a percentage of total cells in the culture, CFU as a percentage of LRCs, and clonal CFU as a percentage of total colonies obtained in the CFU assay (mean ± SD of 8 individually analyzed patients).
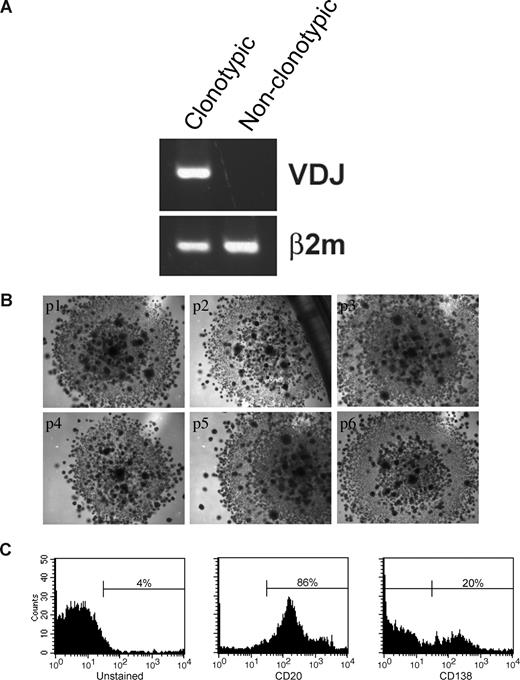
Sorted LRCs were transferred to conventional carboxymethocellulose cultures normally used for counting CFUs, to determine the number of CFU in the population, representing the number of the stem-like cells among the LRCs. CFU colonies from 11 MM patients (Figure 6C) were individually harvested and assessed for the MM clonotypic signature (Figure 6D), by RQ-PCR with patient-specific primers. Approximately 0.5% of LRCs gave rise to colonies, approximately half of which were composed of clonotypic progeny (Figure 6E). Furthermore, sorted CFSEhighCD20+ LRCs gave rise to colonies in a CFU assay, but CFSEhighCD20− LRCs did not. LRCs capable of generating CFU colonies comprised 0.003% of the total cell population in the 3-D culture, and 0.5% to 2.5% of lymphocytic LRCs. Based on quantitation by RQ-PCR, progeny within a given colony were either 100% clonotypic or had no clonotypic cells (Figures 6D, 7A), indicating that MM-CSCs among the LRCs are fully committed to the myeloma lineage.
LRCs can be serially passaged and can generate MM plasma cells. (A) Mononuclear cells were isolated from the mobilized blood preparations, labeled with CFSE, grown in 3-D for 14 days, after which CFSEhigh lymphocytic cells were sorted from the harvested cells, and cultured in a CFU assay (no morphologic differences between clonotypic and nonclonotypic colonies were observed). Clonotypic or nonclonotypic progeny were generated from LRCs in CFU assay and tested for clonotypic IgH VDJ signatures by PCR and by RQ-PCR. Representative single-stage PCR analysis of the clonotypic IgH VDJ and β2-microglobulin from representative CFU progeny is shown; these results were confirmed using RQ-PCR (Figure 6D and data not shown). (B) LRCs were sorted and placed into CFU culture, followed by serial replating for 6 generations. Each panel is a representative image from the consecutive passage of a colony (p1) from a single patient (n = 6 patients). (C) FACS analysis of the LRC progeny after the sixth serial passage of a clonotypic colony (culture p6 from panel B). The entirety of p4 and p5 was used for clonal expansion to obtain sufficient cells for the p6 FACS analysis. Histograms are representative of those obtained for 6 different patients. All of the secondary colonies from clonotypic CFU were clonotypic. In contrast, secondary colonies from the nonclonotypic CFU were all nonclonotypic. Self-renewal capability within colonies was further supported by the morphology of the CFU arising from LRCs, which at all passages included small secondary colonies scattered throughout the primary colony, as shown in the figure.
LRCs can be serially passaged and can generate MM plasma cells. (A) Mononuclear cells were isolated from the mobilized blood preparations, labeled with CFSE, grown in 3-D for 14 days, after which CFSEhigh lymphocytic cells were sorted from the harvested cells, and cultured in a CFU assay (no morphologic differences between clonotypic and nonclonotypic colonies were observed). Clonotypic or nonclonotypic progeny were generated from LRCs in CFU assay and tested for clonotypic IgH VDJ signatures by PCR and by RQ-PCR. Representative single-stage PCR analysis of the clonotypic IgH VDJ and β2-microglobulin from representative CFU progeny is shown; these results were confirmed using RQ-PCR (Figure 6D and data not shown). (B) LRCs were sorted and placed into CFU culture, followed by serial replating for 6 generations. Each panel is a representative image from the consecutive passage of a colony (p1) from a single patient (n = 6 patients). (C) FACS analysis of the LRC progeny after the sixth serial passage of a clonotypic colony (culture p6 from panel B). The entirety of p4 and p5 was used for clonal expansion to obtain sufficient cells for the p6 FACS analysis. Histograms are representative of those obtained for 6 different patients. All of the secondary colonies from clonotypic CFU were clonotypic. In contrast, secondary colonies from the nonclonotypic CFU were all nonclonotypic. Self-renewal capability within colonies was further supported by the morphology of the CFU arising from LRCs, which at all passages included small secondary colonies scattered throughout the primary colony, as shown in the figure.
The self-renewal properties of LRCs and the CFU morphology were maintained through 6 consecutive cycles of replating (Figure 7B). Colonies at p1 to p3 consisted of 100% clonal cells as measured by RQ-PCR (not shown). Self-renewal potential of LRCs was further confirmed by their ability to maintain CD20+ phenotype in serial replating experiments (Figure 7C). A population of CD138+ (20%) cells differentiated from the CD20+ LRCs at passage 6, indicating that LRCs retain the ability to generate MM plasma cells (Figure 7C). Taken together, these results demonstrate that 3-D reconstructions of BM or MBA maintain putative MM-CSCs in a dormant state, from which they can be harvested and released from dormancy under CFU culture conditions. Therapeutic reductions in the number of CFUs from 3-D LRC may provide a clinically feasible means to monitor the impact of new drugs on cancer stem cells from MM and other malignancies of the hematopoietic system.
Discussion
This work describes a unique preclinical model that supports in vitro expansion of the MM clone and allows access to putative MM-CSCs. Because cancer cells are highly drug-resistant in 3-D microenvironments,46 adhesion-mediated drug-resistance26,47 is a confounding factor for conventional MM preclinical models. 3-D reconstruction of BM delivers a powerful model wherein the effects of therapies can be tested on the entire MM clone. This is the first model in which MM B and plasma cells proliferate and undergo 2- to 17-fold clonal expansion, confirmed to reflect the ex vivo MM clone by quantitative analysis of clonotypic IgH VDJ and identification of chromosomal abnormalities. Although both melphalan and bortezomib reduced the burden of clonotypic cells in 3-D, substantial numbers survived, and neither drug appeared to affect the stromal layer formed in 3-D. We also show that MM includes a dormant, nonproliferative compartment of stem-like MM cells. Putative MM-CSCs, here defined as quiescent, drug-resistant CD20+ LRCs that generate clonotypic CD20+ MM B cells and CD138+ MM PCs, exhibit self-renewal, and reside at the reconstructed endosteum, a niche known to maintain dormancy of normal progenitors.43-45 In colony-forming assays, all progeny have the MM molecular signature, based on clontypic VDJ analysis. Multiple serial passages of MM-CSCs show that they maintain the ability to generate clonotypic progeny, as measured by RQ-PCR, and acquire the ability to generate CD138+ plasma cells. Although differentiation occurs in MM-CSC CFU cultures, self-renewal also appears to occur, as evidenced by continued growth in serial CFU cultures of individual clonotypic colonies.
Sorted lymphocytic LRCs are heterogeneous, including both normal and malignant progenitor cells as defined by the presence of both clonotypic and nonclonotypic colonies in secondary CFU cultures of LRCs. Both types give rise to predominantly lymphocyte progeny, suggesting that this system also enriches for normal lymphoid progenitors within MM BM or MBAs, a speculation to be evaluated in the future. For analysis of MM, harvesting of CD20+ LRCs enriches for and provides access to dormant MM-CSCs, and their progeny, when released from dormancy, possibly because of the presence of IL-3 in the CFU medium.48-50
The identity of the MM-CSCs remains controversial, with some studies suggesting that the disease arises from PCs and others indicating a B lymphocyte origin of MM. Data obtained from the SCID-hu mouse model suggest that MM arises from a PC (CD38+CD45−).51 However, in this model, MM cells are forced to colonize a microenvironment provided by fetal bone, which may not support growth of populations from older adults, as occurs in MM, a concern supported by our observation that plasma from young normal donors cannot sustain 3-D expansion of the MM clone (J.K. and L.M.P., manuscript in preparation), and may compromise xenografting of LRCs or progeny of LRCs into mice. Generative cells in another model52 express CD20 and lack CD138 but have not been shown to harbor the MM clonotypic signature53 characteristic of MM patients.17,54 Xenografting of CD34-enriched cells from MM MBAs leads to lytic bone lesions and clonotypic progeny in murine BM, and early-stage clonotypic MM B cells are self-renewing in immunodeficient mice,22-24 but it is difficult to resolve the fine details of cell-microenvironment interactions in a murine model. The combined use of 3-D culture to promote MM clonal expansion and CFU cultures to release quiescent CD20+ MM-CSCs from dormancy, thereby generating CD20+ MM-B cells and CD138+ MM-PCs, provides a novel model that enables direct analysis malignant cells from MM patients.
The work presented here highlights a previously underexplored but slowly emerging aspect of preclinical testing, evaluating malignant stem cells and their clonal expansion within the context of the aggregate microenvironment. The comprehensive reconstruction of normal BM elements in 3-D culture allows expanded monitoring of drug impacts on both malignant and healthy cells, to evaluate the efficacy and potential side effects of novel therapies. Most importantly, this is the first model within which the entirety of the MM clone, including MM-CSCs, can be accessed, subjected to molecular validation and evaluated for susceptibility to therapeutic targeting.
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was funded by grants from the Canadian Institutes of Health Research and the Alberta Cancer Board Research Initiatives Program. J.K. was funded by a fellowship award from the Alberta Heritage Foundation for Medical Research (AHFMR). L.D.M. and K.J.T. hold AHFMR studentship awards, and T.R. holds an AHFMR Clinical Investigator Award. L.M.P. is the Canada Research Chair in Biomedical Nanotechnology, and this work was funded in part by the Chairs program.
Authorship
Contribution: J.K. conceived the study, designed and executed experiments, performed data analysis and interpretation, and prepared the manuscript; K.J.T. assisted with RQ-PCR; L.D.M. performed FISH experiments; C.D.M. quantified cellular composition of the 3-D culture; A.R.B. collected bone marrow samples and edited the manuscript; T.R. collected bone marrow samples, consulted on statistical analysis, and edited the manuscript; and L.M.P. directed research and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Linda M. Pilarski, Cross Cancer Institute, Division of Oncology, 11560 University Avenue, Edmonton, AB T6G 1Z2, Canada; e-mail: lpilarsk@ualberta.ca.

![Figure 2. rBM maintains the architecture of in vivo BM. (A) BMCs were grown in rBM for 21 days and stained with DAPI (pseudocolored red) to mark nuclei. The stratification of the 3-D culture was assessed by confocal microscopy as a 3-D reconstruction from the confocal stack (original magnification 100×). Representative images from BM of 48 individually analyzed patient samples. (B) BM aspirated particle grown in rBM for 21 days and stained with DAPI (pseudocolored red) to mark nuclei. The stratification of the 3-D culture was assessed by confocal microscopy as a 3-D reconstruction from the confocal stack (original magnification 100×). Representative image from analysis of aspirated particles from 28 individually analyzed patients. (C) BM stromal cells redistribute to rEnd. The rBM layer was removed from 3-D culture, and the cells at rEnd were stained with markers to identify stromal components observed at rEnd (adipocytes, Oil Red; fibroblasts, fibronectin and phallodin; osteoblasts, alkaline phosphatase [ALP] and Alizarin Red [inset]; osteoclasts, tartarate-resistant acidic phosphatase [TRAP]). Brightfield microscopy (original magnification 200×). Representative images from 7 individually analyzed patients; see “Immunohistochemistry.” (D) BMCs were grown for 14 days and the percentage of CD34+ HPCs, B cells, or PCs in each layer (black represents fraction in bottom layer; gray, fraction in middle; white, fraction in top layer) of the 3-D culture was calculated based on the immunohistochemical staining for CD34, CD20, or CD138 (n = 4 individually analyzed patients). Error bars represent SEM. For complete image acquisition information, see “Microscopy.”](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/112/7/10.1182_blood-2008-02-142430/6/m_zh80160822700002.jpeg?Expires=1769159765&Signature=b4YIla3vedzcqtQVMj7MSrqOyaPSHfQZHk54AtdjQLYDcmr9NU8esdsrlfbnNreET4zEJ14BaZ8KTChweNKEbiXBqkneniB4dnyTspWxwAwN0cwwGzGP8Cc91vdlm4O1DDFXkwrsckGqOeQfH0yh0TRct4XMmauV~npiuGFpCIR1ceKc3~dbdukha2bM1AE0a9wGMFxdr9lx59hq4pLA0-w1FRltOmvKX93pMKiUKMaTJ0arVQo9QeDRwJ1lCg8feSJcc6Qsdqchdr5lcyVnKcrKe6FzyfP~gAY42Ug19XiROV-E6MzJLUNeRyOy0v7LBJ9e4mcrf~tfZ6E9cj4dMA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal