Abstract
A novel dendritic cell (DC)–restricted molecule, Clec9A, was identified by gene expression profiling of mouse DC subtypes. Based on sequence similarity, a human ortholog was identified. Clec9A encodes a type II membrane protein with a single extracellular C-type lectin domain. Both the mouse Clec9A and human CLEC9A were cloned and expressed, and monoclonal antibodies (mAbs) against each were generated. Surface staining revealed that Clec9A was selective for mouse DCs and was restricted to the CD8+ conventional DC and plasmacytoid DC subtypes. A subset of human blood DCs also expressed CLEC9A. A single injection of mice with a mAb against Clec9A, which targets antigens (Ags) to the DCs, produced a striking enhancement of antibody responses in the absence of added adjuvants or danger signals, even in mice lacking Toll-like receptor signaling pathways. Such targeting also enhanced CD4 and CD8 T-cell responses. Thus, Clec9A serves as a new marker to distinguish subtypes of both mouse and human DCs. Furthermore, targeting Ags to DCs with antibodies to Clec9A is a promising strategy to enhance the efficiency of vaccines, even in the absence of adjuvants.
Introduction
The dendritic cell (DC) network is essential for the initiation and regulation of immune responses. Although all DCs are able to process antigens (Ags) and activate naive T cells, there are multiple subtypes of DCs differing in location and specialized functions.1,2 Some basic types of DCs are recognized in both mice and humans. Conventional DCs (cDCs) can be distinguished from the type I interferon-producing plasmacytoid DCs (pDCs).3-5 The cDCs include “inflammatory” DCs, not present in steady-state but which develop from monocytes on inflammation.6 The steady-state cDCs include “migratory” cDCs, which sample Ags in peripheral tissues but migrate into lymph nodes (LN) via the lymph, for interaction with T cells.1,7,8 Also found in lymphoid organs are the nonmigratory “lymphoid tissue–resident” cDCs, which arrive in a precursor form from the bloodstream and carry out all their functions within the one organ.2,6,9 However, these lymphoid tissue resident cDCs have been mainly studied in the mouse, where they have been divided into those expressing CD8α (CD8+ cDCs) and those that do not (CD8− cDCs). These differ in important functions, the CD8+ cDCs in particular being specialized for “cross-presentation” of exogenous Ags on MHC class I,10-12 as well as being the major producers of IL-12 on activation.13,14
Despite the important functional differences known between mouse lymphoid tissue resident cDC subtypes, there has been little translation of this knowledge to the human DC system. Limited access to human lymphoid organs and the fact that CD8 is not expressed on human DCs are the main reasons. There is evidence that human thymic DCs can be segregated into subsets equivalent to those in mouse thymus,15,16 and some markers that might segregate human DC subsets in other lymphoid organs have been proposed.17 DC subset-specific marker molecules conserved between species are needed to determine whether human DC biology is similar to the well-studied mouse model.
Molecules on the surface of DCs are important in the recognition, communication, and activation functions of DCs. Surface molecules that differ between DC subtypes are of particular interest, because they may underpin the functional specializations of these subtypes. Furthermore, surface molecules differing between DC subtypes may serve for selective delivery of Ags or therapeutic agents to the DCs to manipulate immune responses. Antibodies against DC surface molecules have been used to deliver Ags to DCs.18-30 Enhanced immune responses to the targeted Ags have been obtained, although in most studies only when the Ag complexed to the targeting antibody is administered with a DC-activating agent or adjuvant. To facilitate translation of these promising findings to enhancement of vaccine effectiveness in humans, it would be an advantage to use a mouse model where the surface molecule targeted is specific to DCs, is restricted to particular DC subtypes, and is conserved between species.
Here we describe a novel surface C-type lectin-like molecule, mouse Clec9A (mClec9A), which is largely restricted to DCs and is selectively expressed by mouse CD8+ cDCs and pDCs. Importantly, human CLEC9A (hCLEC9A) is also expressed on a human DC subtype. Targeted delivery of Ag to DCs using monoclonal antibodies (mAbs) against mClec9A induced a striking enhancement of humoral immunity, and enhanced CD4 and CD8 T-cell proliferative responses, all in the absence of additional adjuvants. Thus Clec9A serves as a new DC subtype marker crossing species barriers and as a promising new target for immune modulation and enhancement of vaccine effectiveness.
Methods
Identification and cloning of Clec9A
Clec9A was identified as a clone differentially expressed by splenic CD8+cDCs using gene expression profiling.31 Gene identification was performed by sequence comparison to the cDNA and protein databases of the National Center for Biotechnology Information.32 Genomic localization was performed by alignment to the mouse assembly (February 2006) and human assembly (March 2006) using University of California Santa Cruz Genome Browser.33 Full-length mClec9A and hCLEC9A were isolated by polymerase chain reaction (PCR) amplification from splenic DC cDNA using the following primers. mClec9A: 5′-gccatttcttgtaccaacctactcct-3′; 5′-cggtgtggtatggatcgtcactt-3′. hCLEC9A: 5′-agcctcctgtgtggactgcttt-3′; 5′-ttcatggcccacattttggttt-3′. mClec9A and hCLEC9A were expressed on the surface of chinese hamster ovary cells as C-terminal (extracellular) FLAG-tagged proteins and on the surface of mouse EL4 cells as a fusion protein where green fluorescent protein (GFP) was fused to the N-terminal cytoplasmic domain of Clec9A.
Quantitative RT-PCR
RNA was isolated, DNase treated, and reverse transcribed.31 The expression of mClec9A and Gapdh was determined using real-time reverse transcription (RT) PCR31 and the following specific primers. mClec9A: 5′-tgtgactgctcccacaactgga-3′; 5′-tttgcaccaatcacagcacaga-3′, Gapdh: 5′-catttgcagtggcaaagtggag-3′; 5′-gtctcgctcctggaagatggtg-3′. The expression level for each gene was determined using a standard curve and was expressed as a ratio relative to Gapdh.
Generation of mAb against Clec9A
Wistar rats were immunized 4 times with 50μg keyhole limpet hemocyanin-conjugated peptide: mClec9A peptide (H-DGSSPLSDLLPAERQRSAGQIC-OH), hCLEC9A peptide (H-RWLWQDGSSPSPGLLPAERSQSANQVC-OH), then given a final boost 4 days before fusion with Sp2/0 myeloma cells. Hybridomas secreting specific mAb (anti-mClec9A 24/04-10B4; isotype IgG2a and anti-hCLEC9A 20/05-3A4; isotype IgG2a) were identified by flow cytometric analysis of supernatants using FLAG-tagged and GFP-tagged Clec9A transfectant cells.
Mice
C57BL/6J Wehi, C57/BL6Ly5.1, and the ovalbumin (OVA)-specific CD8 (OT-I) and CD4 (OT-II) TCR-transgenic C57BL/6 background mice were bred under specific-pathogen–free conditions at The Walter and Eliza Hall Institute (WEHI). TRIF−/−34 and MyD88−/−35 backcrossed onto C57BL/6, were interbred to derive TRIF−/−MyD88−/− double-knockout mice. The FcRγ chain−/− mice, backcrossed onto C57BL/6,36 were obtained from the Burnet Institute (Melbourne, Australia). Female mice were used at 6 to 12 weeks of age; alternatively, gender- and age-matched cohorts were generated. Animals were handled according to the guidelines of the National Health and Medical Research Council of Australia. Experimental procedures were approved by the Animal Ethics Committee, WEHI.
Isolation and flow cytometric analysis of DCs
DC isolations from lymphoid organs were performed as previously described.37 Briefly, tissues were mechanically chopped, digested with collagenase and DNAse and treated with ethylenediamine tetraacetic acid (EDTA). Low-density cells were enriched by density centrifugation (1.077 g/cm3 Nycodenz; Axis-Shield, Oslo, Norway). Non-DC-lineage cells were coated with mAb (KT3-1.1, anti-CD3; T24/31.7, anti-Thy1; TER119, anti-erythrocytes; ID3, anti-CD19; and 1A8, anti-Ly6G) then removed using anti–rat Ig magnetic beads (Biomag beads, Qiagen, Victoria, Australia). Blood DCs were enriched by removing red blood cells (RBCs; 0.168M NH4Cl; 5 minutes at 4°C) and depletion of irrelevant cells as above, except the mAb cocktail also contained the mAb F4/80. DC-enriched populations were blocked using rat Ig and anti-FcR mAb (2.4G2), then stained with fluorochrome-conjugated mAb against CD11c (N418), CD205 (NLDC-145), CD4 (GK1.5), CD8 (YTS169.4), CD24 (M1/69), 120G8 or CD45RA (14.8), Sirpα (p84) and mClec9A (24/04-10B4-biotin). cDCs were selected as CD11chiCD45RA− or CD11chi120G8−; splenic cDC were further subdivided into CD4+cDC (CD11chiCD45RA−CD4+ CD8−), double-negative (DN) cDC (CD11chiCD45RA−CD4−CD8−) and CD8+cDC (CD11chiCD45RA−CD8+ CD4−); thymic DCs were subdivided into CD8− cDC (SirpαhiCD8lo) and CD8+ cDC (SirpαloCD8hi); and LN cDC were subdivided into CD8− cDC (CD11chiCD205−CD8−), dermal DC (CD11c+CD205intCD8−), Langerhans cells (CD11c+CD205hiCD8−) and CD8+ cDC (CD11c+CD205hiCD8+), as described previously.31 pDCs were separated as CD11cintCD45RA+ or CD11cint120G8+. Biotin staining was detected using streptavidin (SA)–phycoerythrin (PE). The expression of mClec9A on the various DC populations was analyzed and compared with isotype control staining (IgG2a; BD Pharmingen, San Diego, CA). Flow cytometric analysis was performed on an LSR II (Becton Dickinson, Franklin Lakes, NJ), excluding autofluorescent and propidium iodide (PI)–positive dead cells.
Isolation and flow cytometric analysis of human blood DCs and hemopoietic cells
Peripheral blood mononuclear cells (PBMCs) were isolated from human blood using Ficoll-Pacque-Plus (GE Healthcare, Rydalmere, Australia) density separation. Blood donors gave with informed consent and collection was approved by Human Research Ethics Committee, Melbourne Health in accordance with the Declaration of Helsinki. The PBMCs were blocked using rat Ig and anti-FcR mAb (2.4G2) then stained with mAb against HLA-DR (L243; Becton Dickinson), and a cocktail of PE-conjugated mAb against lineage markers, namely CD3 (BW264156; T cells), CD14 (Tuk4; monocytes), CD19 (6D5; B cells) and CD56 (AF12-7H3; NK cells). Blood DCs were gated as HLA-DRhi, lineage− cells and further segregated based on their expression of BDCA-1 (ADJ-8E3), BDCA-3 (AD5-14H2), BDCA-4 (AD5-17F6) and CD16 (VEP13). PBMCs were also used as a source of other hemopoietic cells that were isolated using mAbs against CD3 (BW264156; T cells), CD19 (6D5; B cells), CD56 (AF12-7H3), and NKp46 (9E2; CD56+NKp46+; NK cells) and CD14 (Tuk4; monocytes). Staining and flow cytometric analysis for the expression of hCLEC9A (20/05-3A4) was performed, excluding PI-positive dead cells. Unless otherwise specified, all anti–human mAb were purchased from Miltenyi Biotec (North Ryde, Australia).
Isolation and analysis of Clec9A on mouse hemopoietic cells
Spleen cell suspensions were prepared as for DC isolation.37 Cells were stained with mAb against CD3 (KT3-1.1), CD19 (ID3), NK1.1 (PK136), CD49b (Hmα2; eBioscience, San Diego, CA) then B cells (CD19+CD3−), T cells (CD19−CD3+) and NK cells (CD49b+NK1.1+CD3−) were selected. Splenic macrophages were first enriched by a 1.082 g/cm3 density centrifugation (Nycodenz) and immunomagnetic bead depletion of CD3+ T cells and CD19+ B cells; the enriched cells were stained with mAb against CD11b (M1/70) and F4/80, then macrophages were gated as CD11bhiF4/80+. Bone marrow macrophages and monocytes were first enriched as for spleen, then stained with CD11b (M1/70) and Ly6C (5075-3.6); monocytes were then gated as side-scatterloLy6ChiCD11bhi and macrophages as Ly6CintCD11bhi. All cells were blocked using rat Ig and anti-FcR mAb (2.4G2) before immunofluorescence staining with the various mAb cocktails including anti-Clec9A mAb (10B4-biotin). Biotin staining was detected using SA-PE. Samples were analyzed for their expression of Clec9A on an LSR II (Becton Dickinson), excluding PI-positive dead cells.
Immunization using anti-Clec9A mAb
Mice were normally injected intravenously with 10 μg of rat anti-Clec9A mAb (10B4) or isotype control mAb1 (IgG2a; eBioscience) or isotype control mAb2 (IgG2a, anti–β-gal; GL117). Serum samples were obtained at 2 to 8 weeks and the level of anti–rat Ig reactivity determined by enzyme-linked immunosorbent assay (ELISA). The anti-Clec9A mAb and the GL117 control mAb were also chemically conjugated to OVA.25 Capacity to recognize target Ag was verified by staining transfected cell lines with conjugates and detecting binding using biotinylated anti-OVA–specific sera (Calbiochem, Darmstadt, Germany) followed by SA-PE. Mice were immunized with 2.5 to 10 μg of anti–Clec9A-OVA (10B4 mAb conjugated to OVA) or control OVA (isotype control GL117 mAb conjugated to OVA).
ELISA for the detection of serum Ab
ELISA plates (Costar, Cambridge, United Kingdom) were coated overnight at 4°C with 2 μg/mL of rat GL117 mAb. Unbound mAb was washed away (phosphate-buffered saline [PBS], 0.05% Tween-20). Serially diluted serum samples were plated (PBS/5% milk powder) and incubated at 4°C overnight. Bound mouse anti–rat Ig antibodies were detected using donkey anti–mouse IgG HRP (Chemicon International, Temecula, CA) and visualized using 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sufonic acid)diammonium salt. Titers were considered positive when the optical density was more than 0.1. The isotype of the anti–rat response was assayed as above, but detected using anti–mouse IgG1-, IgG2b-, IgG2c- and IgG3-HRP conjugates (1/4000; Southern Biotech, Birmingham, AL). Anti-OVA responses were assayed by coating plates with 10 μg/mL OVA, and anti-OVA Ig antibodies were detected as above.
Purification of transgenic T cells and in vivo proliferation assays
Transgenic T cells were purified and labeled with carboxy fluorescein diacetate succinmidyl ester (CFSE).38 CFSE-labeled cells (106) were injected intravenously into C57BL6Ly5.1 mice. Three days later, spleens were removed, cell suspensions prepared and purged of RBCs, then stained with mAb against CD4 (GK1.5-APC) or CD8 (YTS169-APC) and Ly5.2 (S.450-15.2-PE). Proliferating OT-II (CD4+Ly5.2+) or OT-I (CD8+Ly5.2+) cells were visualized by the loss of CFSE fluorescence and enumerated by addition of a fixed number of calibration beads (BD Pharmingen). Dead cells were excluded using PI. Analysis was carried out on a FACSCalibur instrument (Becton Dickinson).
Results
Identification, cloning, and characterization of Clec9A
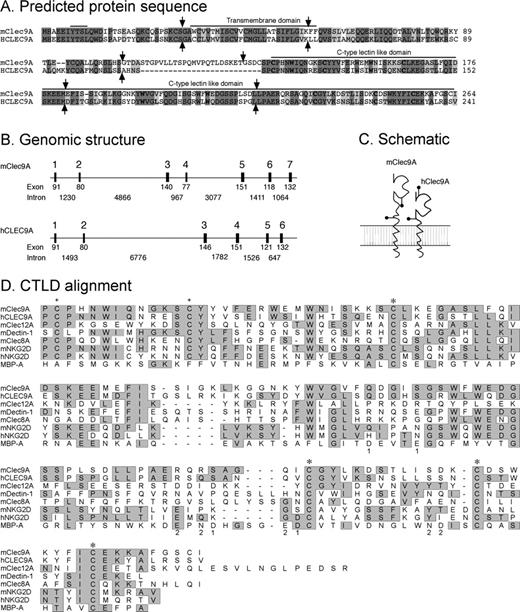
The 3 subsets of cDCs of mouse spleen differ in immunologic functions39 and in gene expression profiles.31,40,41 Here, we focused on the novel gene Clec9A, identified in our microarray analysis as preferentially expressed by CD8+ cDCs. We cloned and sequenced the full-length cDNA encoding mClec9A and hCLEC9A (Figure 1A).
The genomic structure and predicted protein structure encoded by mouse and human Clec9A genes. (A) Protein sequence alignment of the predicted protein sequence of mouse and human Clec9A. Sequence identity is highlighted in dark gray, similarity is shown in light gray. Invariant residues are shown in bold. Exon boundaries are denoted by  . (B) Schematic representation of the gene structures of mClec9A and hCLEC9A, determined by alignment of the cDNA to the genomic sequence databases of the C57BL/6J mouse and human databases, respectively. Exons encoding the coding region of Clec9A genes are denoted by black boxes and the size (bp) of the exons and introns are shown below. (C) A schematic representation of the mouse and human Clec9A proteins. Pins denote potential N-glycosylation sites. (D) Alignment of the CTLD of mouse and human Clec9A to proteins that share sequence homology. Rat mannose binding protein A (MBP-A) is included for comparison as a classical C-type lectin domain that has functional carbohydrate recognition domains. Gray boxes indicate conserved residues, + indicates additional pair of cysteine residues involved in protein homodimeration, and * marks the conserved cysteine residues that form disulfide bonds. The residues that ligate Ca2+ in the MBP-A are designated 1 and 2.
. (B) Schematic representation of the gene structures of mClec9A and hCLEC9A, determined by alignment of the cDNA to the genomic sequence databases of the C57BL/6J mouse and human databases, respectively. Exons encoding the coding region of Clec9A genes are denoted by black boxes and the size (bp) of the exons and introns are shown below. (C) A schematic representation of the mouse and human Clec9A proteins. Pins denote potential N-glycosylation sites. (D) Alignment of the CTLD of mouse and human Clec9A to proteins that share sequence homology. Rat mannose binding protein A (MBP-A) is included for comparison as a classical C-type lectin domain that has functional carbohydrate recognition domains. Gray boxes indicate conserved residues, + indicates additional pair of cysteine residues involved in protein homodimeration, and * marks the conserved cysteine residues that form disulfide bonds. The residues that ligate Ca2+ in the MBP-A are designated 1 and 2.
The genomic structure and predicted protein structure encoded by mouse and human Clec9A genes. (A) Protein sequence alignment of the predicted protein sequence of mouse and human Clec9A. Sequence identity is highlighted in dark gray, similarity is shown in light gray. Invariant residues are shown in bold. Exon boundaries are denoted by  . (B) Schematic representation of the gene structures of mClec9A and hCLEC9A, determined by alignment of the cDNA to the genomic sequence databases of the C57BL/6J mouse and human databases, respectively. Exons encoding the coding region of Clec9A genes are denoted by black boxes and the size (bp) of the exons and introns are shown below. (C) A schematic representation of the mouse and human Clec9A proteins. Pins denote potential N-glycosylation sites. (D) Alignment of the CTLD of mouse and human Clec9A to proteins that share sequence homology. Rat mannose binding protein A (MBP-A) is included for comparison as a classical C-type lectin domain that has functional carbohydrate recognition domains. Gray boxes indicate conserved residues, + indicates additional pair of cysteine residues involved in protein homodimeration, and * marks the conserved cysteine residues that form disulfide bonds. The residues that ligate Ca2+ in the MBP-A are designated 1 and 2.
. (B) Schematic representation of the gene structures of mClec9A and hCLEC9A, determined by alignment of the cDNA to the genomic sequence databases of the C57BL/6J mouse and human databases, respectively. Exons encoding the coding region of Clec9A genes are denoted by black boxes and the size (bp) of the exons and introns are shown below. (C) A schematic representation of the mouse and human Clec9A proteins. Pins denote potential N-glycosylation sites. (D) Alignment of the CTLD of mouse and human Clec9A to proteins that share sequence homology. Rat mannose binding protein A (MBP-A) is included for comparison as a classical C-type lectin domain that has functional carbohydrate recognition domains. Gray boxes indicate conserved residues, + indicates additional pair of cysteine residues involved in protein homodimeration, and * marks the conserved cysteine residues that form disulfide bonds. The residues that ligate Ca2+ in the MBP-A are designated 1 and 2.
Mouse C-type lectin domain family 9, member A (Clec9A) gene is on chromosome 6 (Riken 9830005G06; GenBank accession AK036399.1), and the human CLEC9A is on chromosome 12. The full-length coding sequence of mClec9A, encoded by 7 exons spanning 13.4 kb of genomic DNA (Figure 1B), contains a single open reading frame (ORF) encoding a protein of 264 amino acids (aa; Figure 1A). The hCLEC9A coding sequence, encoded by 6 exons spanning 12.9 kb of genomic DNA (Figure 1A), similarly contains a single ORF encoding a protein of 241 aa (Figure 1B).
The mouse and human Clec9A genes each encode a putative transmembrane protein with a single C-type lectin–like domain (CTLD) in its extracellular region, a cytoplasmic tail, and a transmembrane region containing the YXXL residues, which is a potential signaling motif42 (Figure 1A). hCLEC9A has shorter hinge region than mouse. Alignment of the mClec9A and hCLEC9A is presented in Figure 1A (53% identical, 69% similar), and a schematic representation is shown in Figure 1C. Using NCBI Blast protein analysis, it was determined that mClec9A shares most sequence similarity with mouse Dectin-1 (Clec7A), Clec12B, and NKG2D, whereas hClec9A is most similar to LOX-1 (Clec8A), Clec12B, and DCAL-2 (Clec12A; Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). The CTLD of Clec9A, like the classical C-type lectin the rat mannose binding protein A (MBP-A), has 4 conserved cysteine residues that form 2 disulfide bonds (Figure 1D). In addition, Clec9A possesses 2 additional cysteine residues in the neck region, allowing protein homodimerization.43 Critically, the residues involved in Ca2+ binding in classical C-type lectins are not present in mouse and human Clec9A (Figure 1D).
Gene expression of mClec9A
Microarray analysis predicted mClec9A to be expressed at higher levels in CD8+ cDCs relative to CD8− cDCs. Hence, we investigated the expression of mClec9A in mouse splenic cDC subsets by quantitative RT-PCR. CD8+ cDC expressed 22-fold more mClec9A mRNA than CD4+ and DN cDCs (Figure 2A).
Gene expression profiles of mouse Clec9A. Real-time PCR was used to determine the mRNA expression of the Clec9A gene relative to Gapdh using primers designed from the 3′ UTR region of mClec9A. (A) Expression in lymphoid organ steady state DCs including splenic cDC subsets (DN, CD4+, CD8+), thymic cDC subsets (CD8−, CD8+) LN cDC subsets (CD8−, CD8+), dermal and Langerhans cells (LC), and in thymic and splenic pDCs. (B) Expression in hemopoietic cells, including thymocytes (thym), LN B and T cells, spleen (spl) B and T cells, NK cells, immature macrophages (im mac; Mac-1+F4/80lo), mature macrophages (mat mac; Mac-1+F4/80hi), splenic pDCs and cDCs. The experiment was performed twice.
Gene expression profiles of mouse Clec9A. Real-time PCR was used to determine the mRNA expression of the Clec9A gene relative to Gapdh using primers designed from the 3′ UTR region of mClec9A. (A) Expression in lymphoid organ steady state DCs including splenic cDC subsets (DN, CD4+, CD8+), thymic cDC subsets (CD8−, CD8+) LN cDC subsets (CD8−, CD8+), dermal and Langerhans cells (LC), and in thymic and splenic pDCs. (B) Expression in hemopoietic cells, including thymocytes (thym), LN B and T cells, spleen (spl) B and T cells, NK cells, immature macrophages (im mac; Mac-1+F4/80lo), mature macrophages (mat mac; Mac-1+F4/80hi), splenic pDCs and cDCs. The experiment was performed twice.
We then examined the gene expression of mClec9A across a panel of hemopoietic cell types. Both cDCs and pDCs expressed high levels of mClec9A mRNA, but low level expression was also detected in NK cells (Figure 2B). It was preferentially expressed in CD8+ cDCs relative to CD8− cDCs, not only in spleen but also in thymus and LNs (Figure 2A).
Surface protein expression of mClec9A
To investigate the protein expression of mClec9A and hCLEC9A, we generated mAbs that recognized protein on the surface of Clec9A-transfected cells by flow cytometry. Staining of a panel of freshly isolated mouse hemopoietic cells with the mAb 10B4 indicated that mClec9A was expressed on a subset of cDCs and on most pDCs (Figure 3A). Strikingly, mClec9A protein was not detected on most other hemopoietic cells investigated, including T cells, most B cells, monocytes and macrophages. Nor was it detected on the NK cells that expressed some mRNA (Figure 3A). However, a small proportion (3%) of B cells, displayed clear positive staining for mClec9A. Only around 3% of all ungated bone marrow cells showed any staining with 10B4, and most of this was weak. Thus, in the hemopoietic system, mClec9A surface expression appears mainly restricted to DCs (Figure 3A). In addition, staining of frozen sections with the mAb 10B4 revealed no staining beyond that attributed to DCs (data not shown).
Surface expression of mClec9A protein on DCs and other hemopoietic cells. (A) The DCs were purified and surface labeled by 4-color immunofluorescent staining. DCs were stained with mAb against CD11c (N418-PeCy7), CD45RA (14.8-APC), CD8 (53-6.7-APC-Cy7) and mClec9A (10B4-biotin). Splenic DCs were also stained with CD4 (GK1.5-FITC), thymic DCs with Sirpα (p84-FITC), and subcutaneous LN DCs with CD205 (NLDC-145-FITC). Splenic cDCs were divided into CD4+cDCs (CD11hiCD45RA−CD4+CD8−), DN cDCs (CD11hiCD45RA−CD4−CD8−) and CD8+cDCs (CD11hiCD45RA−CD8+CD4−); thymic cDCs were divided into CD8−cDCs (SirpαhiCD8lo) and CD8+cDCs (SirpαloCD8+); and LN cDCs into CD8−cDC (CD11c+CD205−CD8−), dermal cDCs (CD11c+CD205intCD8−), Langerhans' cells (CD11c+CD205hiCD8−) and CD8+cDCs (CD11c+CD205hiCD8+), as described previously.31 pDCs were identified as CD11cintCD45RA+. Splenocytes were stained with mAb against CD3 (KT3-1.1-FITC), CD19 (1D3-PeCy7), NK1.1 (PK136-PeCy7), CD49b (Hmα2-APC) and B cells (CD19+CD3−), T cells (CD19−CD3+) and NK cells (NK1.1+CD49b+CD3−) were identified. Splenic macrophages were enriched as indicated in “Isolation and analysis of Clec9A on mouse hemopoietic cells” and stained with CD11b (M1/70-Cy5) and F4/80-FITC and defined as CD11bhiF4/80+. Bone marrow cells and splenocytes were stained with mAb against CD11b (M1/70-Cy5) and Ly6C (5075-3.6-FITC) and monocytes were defined as side-scatterloLy6ChiCD11bhi. Bone marrow macrophages were Ly6CintCD11bhi. Cell populations were counterstained with SA-PE and analyzed for mClec9A expression. The solid line represents mClec9A staining on gated cells, the dotted line represents staining of the gated cells with an isotype-matched control. (B) Enriched preparations of splenic DCs were stained with mAb against mClec9A (10B4-biotin), CD11c (N418-Quantum dot 655), CD8α (YTS-169-PercpCy5.5) and CD24 (M1/69-Alexa 633) and 120G8-FITC, then counterstained with SA-PE. pDCs (CD11cint120G8+) and cDCs (CD11chi120G8−) were analyzed for expression of mClec9A. mClec9A expression correlated with CD8α and CD24 expression on cDCs. Most splenic pDCs expressed mClec9A. (C) An enriched preparation of blood DCs was stained in parallel with the splenic DCs (B) using the same mAbs and analyzed using identical gating strategies. Blood DCs do not express CD8α, but do express CD24. Similar to splenic DCs, blood DCs expressing CD24 also coexpressed Clec9A. pDCs from the blood, like their splenic counterpart, expressed mClec9A.
Surface expression of mClec9A protein on DCs and other hemopoietic cells. (A) The DCs were purified and surface labeled by 4-color immunofluorescent staining. DCs were stained with mAb against CD11c (N418-PeCy7), CD45RA (14.8-APC), CD8 (53-6.7-APC-Cy7) and mClec9A (10B4-biotin). Splenic DCs were also stained with CD4 (GK1.5-FITC), thymic DCs with Sirpα (p84-FITC), and subcutaneous LN DCs with CD205 (NLDC-145-FITC). Splenic cDCs were divided into CD4+cDCs (CD11hiCD45RA−CD4+CD8−), DN cDCs (CD11hiCD45RA−CD4−CD8−) and CD8+cDCs (CD11hiCD45RA−CD8+CD4−); thymic cDCs were divided into CD8−cDCs (SirpαhiCD8lo) and CD8+cDCs (SirpαloCD8+); and LN cDCs into CD8−cDC (CD11c+CD205−CD8−), dermal cDCs (CD11c+CD205intCD8−), Langerhans' cells (CD11c+CD205hiCD8−) and CD8+cDCs (CD11c+CD205hiCD8+), as described previously.31 pDCs were identified as CD11cintCD45RA+. Splenocytes were stained with mAb against CD3 (KT3-1.1-FITC), CD19 (1D3-PeCy7), NK1.1 (PK136-PeCy7), CD49b (Hmα2-APC) and B cells (CD19+CD3−), T cells (CD19−CD3+) and NK cells (NK1.1+CD49b+CD3−) were identified. Splenic macrophages were enriched as indicated in “Isolation and analysis of Clec9A on mouse hemopoietic cells” and stained with CD11b (M1/70-Cy5) and F4/80-FITC and defined as CD11bhiF4/80+. Bone marrow cells and splenocytes were stained with mAb against CD11b (M1/70-Cy5) and Ly6C (5075-3.6-FITC) and monocytes were defined as side-scatterloLy6ChiCD11bhi. Bone marrow macrophages were Ly6CintCD11bhi. Cell populations were counterstained with SA-PE and analyzed for mClec9A expression. The solid line represents mClec9A staining on gated cells, the dotted line represents staining of the gated cells with an isotype-matched control. (B) Enriched preparations of splenic DCs were stained with mAb against mClec9A (10B4-biotin), CD11c (N418-Quantum dot 655), CD8α (YTS-169-PercpCy5.5) and CD24 (M1/69-Alexa 633) and 120G8-FITC, then counterstained with SA-PE. pDCs (CD11cint120G8+) and cDCs (CD11chi120G8−) were analyzed for expression of mClec9A. mClec9A expression correlated with CD8α and CD24 expression on cDCs. Most splenic pDCs expressed mClec9A. (C) An enriched preparation of blood DCs was stained in parallel with the splenic DCs (B) using the same mAbs and analyzed using identical gating strategies. Blood DCs do not express CD8α, but do express CD24. Similar to splenic DCs, blood DCs expressing CD24 also coexpressed Clec9A. pDCs from the blood, like their splenic counterpart, expressed mClec9A.
Surface levels of mClec9A were then compared on splenic, LN, and thymic cDCs. mClec9A was expressed by the CD8+ cDCs of spleen, thymus, and LN (Figure 3A). Most splenic, thymic, and LN CD8−cDCs and the migratory cDCs (dermal DCs and Langerhans cells) were negative for mClec9A expression (Figure 3A,B). However, a small proportion of CD8− cDCs showed above background staining; this could be attributed to a small proportion of DCs of the CD8+ cDC lineage not yet expressing CD8α, known to be present within this CD8− cDC gating.6,9,44,45 No mClec9A staining was detected on a preparation of inflammatory CD11intCD11bhi DCs from inflamed mouse spleens6 (data not shown). These DC surface expression profiles were consistent with the gene expression observed by quantitative RT-PCR (Figure 2).
Surface expression of mClec9A on mouse blood DC
Mouse blood contains very few fully developed DCs, and none that express CD8.46 However, mouse blood contains a small proportion of DCs expressing CD24, an alternative and early marker of the CD8 DC lineage6,44 ; presumably these CD24+ blood DCs are en route to becoming CD8+ cDCs. We isolated mouse blood DCs and stained them for CD11c, CD24 and mClec9A expression. A small proportion of blood DCs expressed mClec9A, and these were the subset expressing CD24 (Figure 3C).
Surface expression of hCLEC9A
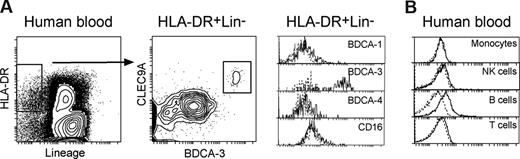
To determine whether hCLEC9A was also expressed on human DCs, we stained freshly isolated human blood PBMCs with the anti-hCLEC9A mAb 3A4. A small subset of HLADR+ DCs were positive for hCLEC9A (Figure 4A). The mAb 3A4 also stained a subset of DCs in macaque blood, indicating cross-reactivity with other primate CLEC9A molecules (Figure S2). However, analysis of the ungated PBMCs showed that most other human blood cells did not show positive staining, although low level staining was obtained on human blood B cells (Figure 4B). To determine whether the CLEC9A-expressing DCs resembled those seen in mouse blood, the blood DCs were also stained with BDCA-1, BDCA-3 and BDCA-4. Staining with mAb 3A4 was restricted to the minor BDCA-3+ DC subset (proposed equivalents of mouse CD8+ cDC17 ), and absent from BDCA-4+ subset. This suggests hCLEC9A is present on a cDC type similar to the mouse CD24+, CD8+ DC lineage,17 but in contrast to the mouse, not on pDCs (Figure 4A).
Expression of hCLEC9A on human blood DCs. (A) Human peripheral blood mononuclear cells (PBMCs) were isolated and surface immunofluorescence labeled with mAb against HLADR (L243-PercP), a cocktail of PE-conjugated mAb against Lineage markers including CD3 (BW264156; T cells), CD14 (Tuk4; monocytes), CD19 (6D5; B cells) and CD56 (AF12-7H3; NK cells), hCLEC9A (3A4-FITC), and either CD16 (VEP-APC-Cy7) or BDCA-1 (ADJ-8E7-APC), or BDCA-3 (AD5-14H12-APC), or BDCA-4 (AD5-17F6-APC; pDC). Blood DCs were gated as HLADR+Lineage− and further analyzed for their expression of hCLEC9A. (B) Other heamapoeitic cells were identified in preparations of PBMCs by immunofluorescence labeling with mAb against CD19 (6D5-PE; B cells), CD3 (BW264156-PE; T cells), CD56 (AF12-7H3-PE) and NKp46 (9E2-APC; CD56+NKp46+; NK cells), CD14 (Tuk4-PE; monocytes), together with 3A4-FITC, then analyzed for their expression of hCLEC9A. The dotted line represents the background staining of the gated cells with an isotype-matched control.
Expression of hCLEC9A on human blood DCs. (A) Human peripheral blood mononuclear cells (PBMCs) were isolated and surface immunofluorescence labeled with mAb against HLADR (L243-PercP), a cocktail of PE-conjugated mAb against Lineage markers including CD3 (BW264156; T cells), CD14 (Tuk4; monocytes), CD19 (6D5; B cells) and CD56 (AF12-7H3; NK cells), hCLEC9A (3A4-FITC), and either CD16 (VEP-APC-Cy7) or BDCA-1 (ADJ-8E7-APC), or BDCA-3 (AD5-14H12-APC), or BDCA-4 (AD5-17F6-APC; pDC). Blood DCs were gated as HLADR+Lineage− and further analyzed for their expression of hCLEC9A. (B) Other heamapoeitic cells were identified in preparations of PBMCs by immunofluorescence labeling with mAb against CD19 (6D5-PE; B cells), CD3 (BW264156-PE; T cells), CD56 (AF12-7H3-PE) and NKp46 (9E2-APC; CD56+NKp46+; NK cells), CD14 (Tuk4-PE; monocytes), together with 3A4-FITC, then analyzed for their expression of hCLEC9A. The dotted line represents the background staining of the gated cells with an isotype-matched control.
Enhancing antibody responses by targeting Ags to DC via mClec9A
Immune responses can be modulated by using mAb to target Ags to particular DC surface molecules.18-30 Because Clec9A is largely DC restricted, and common to human and mouse DCs, it was a promising candidate for delivery of Ags to DCs, with the mouse as a model for possible clinical applications. To determine whether Ab to Clec9A could be used to elicit humoral responses, mice were given a single intravenous injection of small amounts of anti-mClec9A, or with nontargeting isotype control mAbs. Because the rat IgG2a mAb is foreign to mice, the targeting mAb itself acts as an Ag and the response can be measured as the anti–rat IgG titer.
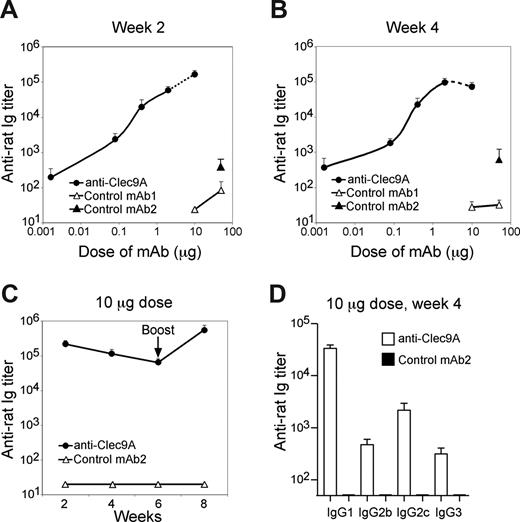
The injection of the anti-mClec9A mAb 10B4 alone, without any additional DC activation agents, produced a strong and prolonged anti–rat Ig response (Figure 5). Around 2 μg of 10B4 produced an optimal response, but as little as 16 ng gave a detectable titer (Figure 5A,B). The response to 10 μg of targeting mAb was around 5000-fold higher than 10 μg of a nontargeting isotype control mAb. To obtain a significant anti–rat Ig titer, at least a 3000-fold higher level of nontargeting rat Ig was required, compared with the targeted mAb (Figure 5A,B). Furthermore, once the anti–rat reactivity was established using the targeting anti-Clec9A mAb, nontargeting isotype control rat Ig gave a significant boost, suggesting a memory response had been generated (Figure 5C). The anti–rat Ig response induced by Clec9A targeting was dominated by the IgG1 isotype, but involved other isotypes including a significant IgG2c component (Figure 5D).
Targeting DC using anti-mClec9A mAb induces potent humoral responses. (A,B) Mice were injected intravenously with either 2 μg (n = 5), 0.4 μg (n = 5), 0.08 μg (n = 5), or 0.016 μg (n = 4) anti-mClec9A mAb (10B4) or with 50 μg (n = 5) and 10 μg (n = 5) of a nontargeting isotype control mAb-1 (eBioscience), or with 50 μg (n = 2) of an in-house isotype control mAb-2 (GL117). Serum anti-rat reactivity was measured by ELISA on week 2 (A) and week 4 (B). Mean titers plus or minus SEM are depicted. The titration experiment was performed twice. The 10-μg dose response represents the cumulative data of 5 experiments (week 2, n = 20; week 4, n = 19). (C) Mice (n = 5) were injected intravenously with 10 μg of either anti-mClec9A mAb or nontargeting isotype control mAb-1 (eBioscience). Serum samples were collected on weeks 2, 4, and 6, after which mice were injected with 10 μg of nontargeting isotype control mAb-2 (GL117). Serum anti–rat Ig reactivity was measured by ELISA on weeks 2, 4, 6, and 8 and is presented as mean titers plus or minus SEM (D) Mice were injected intravenously with 10 μg of either anti-mClec9A mAb (n = 7) or nontargeting isotype control mAb-2 (GL117; n = 4). The isotype of the serum anti–rat Ig reactivity was measured by ELISA on week 4. Bar graphs depict mean titers plus or minus SEM. The experiments were performed twice.
Targeting DC using anti-mClec9A mAb induces potent humoral responses. (A,B) Mice were injected intravenously with either 2 μg (n = 5), 0.4 μg (n = 5), 0.08 μg (n = 5), or 0.016 μg (n = 4) anti-mClec9A mAb (10B4) or with 50 μg (n = 5) and 10 μg (n = 5) of a nontargeting isotype control mAb-1 (eBioscience), or with 50 μg (n = 2) of an in-house isotype control mAb-2 (GL117). Serum anti-rat reactivity was measured by ELISA on week 2 (A) and week 4 (B). Mean titers plus or minus SEM are depicted. The titration experiment was performed twice. The 10-μg dose response represents the cumulative data of 5 experiments (week 2, n = 20; week 4, n = 19). (C) Mice (n = 5) were injected intravenously with 10 μg of either anti-mClec9A mAb or nontargeting isotype control mAb-1 (eBioscience). Serum samples were collected on weeks 2, 4, and 6, after which mice were injected with 10 μg of nontargeting isotype control mAb-2 (GL117). Serum anti–rat Ig reactivity was measured by ELISA on weeks 2, 4, 6, and 8 and is presented as mean titers plus or minus SEM (D) Mice were injected intravenously with 10 μg of either anti-mClec9A mAb (n = 7) or nontargeting isotype control mAb-2 (GL117; n = 4). The isotype of the serum anti–rat Ig reactivity was measured by ELISA on week 4. Bar graphs depict mean titers plus or minus SEM. The experiments were performed twice.
Requirements for enhancing antibody responses by targeting mClec9A
An important feature of the enhanced antibody responses obtained by targeting Ag to mClec9A on DCs was that no additional DC activation agents or adjuvants were used (Figures 5, 6A-C). The 10B4 mAb used was prepared under “endotoxin-free” conditions and the concentrated mAb contained no detectable endotoxin (less than 1 EU/mL). To confirm that the enhanced antibody response was not due to traces of endotoxin or other microbial products, the experiments were repeated using MyD88−/−TRIF−/− mice, which are unable to respond to Toll-like receptor (TLR) ligands.34,35 The induction of equivalent, potent antibody responses to rat Ig by injection of mAb to mClec9A was also seen in these mice (Figure 6A), indicating the response was independent of “danger” signals mediated by TLR ligands. When lipopolysaccharide was injected along with the targeting anti-Clec9A mAb, the antibody response was sometimes further enhanced (Figure 6D) but more often not (Figure 6E). Neither poly:IC not CpG when coinjected increased the anti–rat Ig response to the anti-Clec9A mAb (data not shown).
The nature of the humoral immunity induced by targeting DC using anti-mClec9A mAb. C57BL/6 or (A) C57BL/6 TRIF−/−MyD88−/− or (B) C57/BL6 FcRγ−/− or (C) C57/BL6 nu/nu mice were injected intravenously with 10 μg of the anti-mClec9A mAb (10B4) or the nontargeted isotype control mAb-2 (GL117). (D) C57BL/6 mice were injected intravenously with 10 μg of the anti-mClec9A mAb or isotype control mAb-2 (GL117), either with or without lipopolysaccharide (LPS; 10 ng). (E) Ten micrograms of OVA-conjugated anti-mClec9A mAb or OVA-conjugated isotype control mAb-2 (GL117) or (F) escalating doses of free OVA were injected intravenously into C57BL/6 mice. Serum anti–rat Ig Ab titers were measured by ELISA at week 4. Each circle represents an individual mouse, the geometric mean of the group is depicted by a line. Experiments were performed 2 to 4 times with similar results, with the exception of panel E, which was performed once.
The nature of the humoral immunity induced by targeting DC using anti-mClec9A mAb. C57BL/6 or (A) C57BL/6 TRIF−/−MyD88−/− or (B) C57/BL6 FcRγ−/− or (C) C57/BL6 nu/nu mice were injected intravenously with 10 μg of the anti-mClec9A mAb (10B4) or the nontargeted isotype control mAb-2 (GL117). (D) C57BL/6 mice were injected intravenously with 10 μg of the anti-mClec9A mAb or isotype control mAb-2 (GL117), either with or without lipopolysaccharide (LPS; 10 ng). (E) Ten micrograms of OVA-conjugated anti-mClec9A mAb or OVA-conjugated isotype control mAb-2 (GL117) or (F) escalating doses of free OVA were injected intravenously into C57BL/6 mice. Serum anti–rat Ig Ab titers were measured by ELISA at week 4. Each circle represents an individual mouse, the geometric mean of the group is depicted by a line. Experiments were performed 2 to 4 times with similar results, with the exception of panel E, which was performed once.
A possible reason for the enhanced responses might have been the binding of the anti-mClec9A mAb to FcR, as well as to mClec9A itself. This possibility was eliminated by injection of the 10B4 mAb into FcR γ chain deficient mice that cannot signal through activating FcγRI or FcγRIII36 (Figure 6B). These gave anti–rat Ig responses identical to control mice.
Because the enhancement of the humoral responses was obtained by a targeting strategy designed to deliver Ag to DCs, it was assumed the enhancement was mainly due to the activation of Ag-specific, CD4+ helper T cells. However, since some B cells expressed a little Clec9A, direct targeting to B cells could not be excluded. The role of T cells was tested by injecting the anti-Clec9A mAb 10B4 into nude mice, which lack all T cells. The enhanced antibody response to rat Ig was eliminated (Figure 6C), showing it was dependent on helper T cells.
The anti-Clec9A mAb could also serve to deliver a chemically linked OVA Ag and enhance the anti-OVA antibody response; an injection of a 100-fold higher level of free OVA was required to produce a positive anti-OVA titer and this was still several magnitudes lower than the titer induced by targeting OVA to DC via anti-Clec9A mAb (Figure 6E,F).
Enhanced T-cell responses on targeting mClec9A
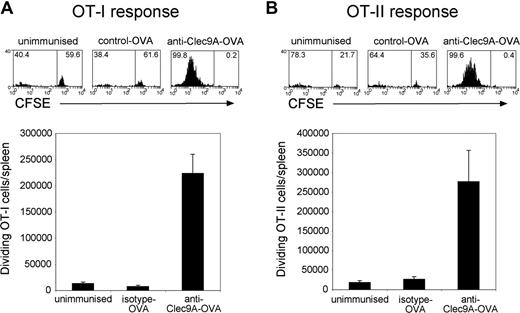
To directly determine whether T-cell responses to a specific Ag were enhanced by targeting Ag to Clec9A, a small number of CFSE-labeled, OVA-specific, CD8 (OT-I) or CD4 (OT-II) transgenic T cells were adoptively transferred into C57BL/6 mice, which were then immunized with OVA-conjugated anti-mClec9A (10B4) or with OVA-conjugated isotype control mAb (GL117). Three days later the proliferative response of the transferred Ag-specific T cells was enumerated as the reduction in CFSE fluorescence. Immunizing mice with the nontargeting control OVA-conjugated mAb failed to induce OT-I or OT-II proliferation, indicating insufficient Ag was presented to activate these T cells. By contrast, immunising mice with anti–mClec9A-OVA mAb conjugate induced extensive proliferation of both OT-I and OT-II T cells (Figure 7). Ag targeted to mClec9A resulted in enhanced activation of both CD4 and CD8 T cells.
Targeting Ag to DC using anti-mClec9A-OVA elicits both CD4 and CD8 T cell proliferative responses. OVA-specific transgenic CD8 (OT-I) or CD4 (OT-II) T cells (106) were adoptively transferred into naive C57/BL6 Ly5.1 mice. One day later mice were injected intravenously with 2.5 μg of anti–mClec9A-OVA (n = 3) or nontargeted isotype control-OVA mAb-2 (GL117; n = 3), or left unimmunized (n = 2). Three days after mAb injection, mice were killed and spleens harvested. Cells were stained with mAb against Ly5.2 (S.450-15.2-PE) and CD4 (GK1.5-APC) or CD8 (YTS169-APC) and proliferating CFSE-labeled transgenic T cells, (A) OT-I (Ly5.2+CD8+) or (B) OT-II (Ly5.2+CD4+), enumerated by flow cytometry. The proliferative response of OVA-specific T cells was seen as a loss of CFSE fluorescence by flow cytometry. The total number of OT-I cells and OT-II cells proliferating per spleen was enumerated as described in “Purification of transgenic T cells and in vivo proliferation assays.” One experiment with the 2.5-μg dose is presented as the mean response plus or minus SEM. The experiment was performed twice with 2.5 μg and once with 5 μg of OVA-conjugated mAb, with similar results.
Targeting Ag to DC using anti-mClec9A-OVA elicits both CD4 and CD8 T cell proliferative responses. OVA-specific transgenic CD8 (OT-I) or CD4 (OT-II) T cells (106) were adoptively transferred into naive C57/BL6 Ly5.1 mice. One day later mice were injected intravenously with 2.5 μg of anti–mClec9A-OVA (n = 3) or nontargeted isotype control-OVA mAb-2 (GL117; n = 3), or left unimmunized (n = 2). Three days after mAb injection, mice were killed and spleens harvested. Cells were stained with mAb against Ly5.2 (S.450-15.2-PE) and CD4 (GK1.5-APC) or CD8 (YTS169-APC) and proliferating CFSE-labeled transgenic T cells, (A) OT-I (Ly5.2+CD8+) or (B) OT-II (Ly5.2+CD4+), enumerated by flow cytometry. The proliferative response of OVA-specific T cells was seen as a loss of CFSE fluorescence by flow cytometry. The total number of OT-I cells and OT-II cells proliferating per spleen was enumerated as described in “Purification of transgenic T cells and in vivo proliferation assays.” One experiment with the 2.5-μg dose is presented as the mean response plus or minus SEM. The experiment was performed twice with 2.5 μg and once with 5 μg of OVA-conjugated mAb, with similar results.
Does binding to mClec9A activate DCs?
Because no additional DC-activating agents or adjuvants were required to obtain enhanced antibody responses or T-cell responses on targeting Ags to DCs with anti-mClec9A, it was possible that DC activating signals could be provided by Clec9A itself. To check if the DCs were activated, they were isolated from the spleens and LN of mice immunized with the targeting anti-Clec9A mAb 10B4, and from nonimmunized control mice. The DCs were stained with DC subset segregating markers, together with antibodies for the DC maturation markers MHC class II, CD80, CD86, and CD40. There was no evidence of any increase in any of these markers of DC activation in any DC subset, including the CD8+ cDCs, which are the primary targets for the mAb (data not shown). Targeting Ags to Clec9A appeared to enhance immune responses without the normal signs of DC activation.
Discussion
Clec9A is one of a family of C-type lectin-like molecules, encoded by genes on mouse chromosome 6 or human chromosome 12. Many of these are expressed on the surface of DCs. Clec9A expression appears more tightly restricted to DC in both mice and humans than the other C-type lectins, and more than most DC marker molecules. Although mouse NK cells expressed some mRNA for Clec9A, no surface protein was detected. Monocytes, macrophages, and T cells showed no Clec9A expression. However, a proportion of B cells in humans and a very small proportion in mice, showed positive staining.
In the mouse, Clec9A expression is DC subtype–restricted, limited to the CD8+CD24+Sirpα−cDCs, to the small subset of CD24+ blood cDCs probably related to the CD8+ cDC lineage, and to pDCs. The CD8−CD24−Sirpα+ lymphoid tissue resident cDCs, the migratory cDCs in lymphoid organs, and monocyte-derived inflammatory DCs were predominantly negative for mClec9A expression.
In humans, CLEC9A expression is likewise restricted to a DC subtype. It does not appear to be expressed on pDCs. hCLEC9A expression on a small subset of blood DCs is coincident with that of BDCA-3. It may mark the human equivalent of the mouse CD8+ cDC subtype, as has been suggested for BDCA-3 and Necl-2.17 Thus, Clec9A may help in the translation of results from the mouse DC system to human immunobiology. However, direct isolation and functional tests will be required to determine whether the human CLEC9A+ DCs have the same range of functions as the mouse CD8+Clec9A+ cDCs.
Clec9A shows great promise as a target for immune modulation, and its restricted expression is an advantage for this application. Targeting Ags to mClec9A on DCs via a single injection of anti-mClec9A mAb gave a marked enhancement of the normally poor mouse response to rat Ig, and good enhancement of responses to ovalbumin when conjugated to anti-mClec9A mAb. With targeting to mClec9A the dose of Ag required was at least 1000-fold less than with injection of free Ag. Although dominated by IgG1, the response involved a range of Ig isotypes. These are all promising features if the aim is to improve antibody responses to a vaccine.
The enhanced antibody response was entirely dependent on T cells, presumably on helper T-cell generation. Importantly, enhanced specific CD4 and CD8 T-cell proliferative responses were also obtained to ovalbumin conjugated to anti-mClec9A mAb. The antibody to mClec9A should target Ags to the CD8+cDC subtypes. The CD8+cDC have a special capacity to cross-present exogenous Ags on MHC class I and so activate CD8 T cells.10-12 CD8+ cDCs are also the major producers of bioactive IL-12 when activated,13,14 and so are able to induce inflammatory Th1 responses.47,48 However, the CD8+ cDCs are also effective activators of CD4 T cells, and this presumably is the role they play in enhancing antibody responses. Further studies are needed to determine whether targeting Ags to CD8+ cDCs via Clec9A using our mAb can be exploited to produce effective cytotoxic T-cell vaccines against tumor Ags. However, during the initial review of this manuscript, Sancho et al49 also reported the DC-restricted distribution of Clec9A (called by them DNGR-1), and they demonstrated clearly that targeting antigens to Clec9A, at least in the presence of DC-activating agents, can induce cytotoxic T cells that can eradicate tumors.
A special feature of the high antibody immune responses generated by targeting Clec9A is that they were obtained by a single injection of low doses of anti-Clec9A mAb in the absence of any additional adjuvants or DC activation agents. This finding appears counter to the generalization that presentation of Ags by steady-state immature DCs induces tolerance, whereas presentation by activated DCs induces immunity. Targeting of Ags to steady-state CD8+ mouse cDCs with mAb against CD205 had strongly supported this generalization, since tolerance was induced unless an additional activator of DCs was administered along with anti-CD205 mAb.28-30 We had obtained similar results in terms of antibody production by targeting CD205 using rat anti-CD205 mAb administration.19 Because the same CD8+cDC subtype was the primary target with either anti-Clec9A or anti-CD205, the difference must lie in the nature of the surface molecule targeted. Targeting Ags to other DC surface molecules, such CIRE and FIRE,19 has also given enhanced immune responses without the need for additional DC stimulants. However, these target the CD8− mouse cDC, and they do not have direct human orthologues.50,51
What is the reason for the enhancement of immune responses in the absence of added DC activation agents? Contamination of the anti-mClec9A mAb with microbial products seems unlikely, since the enhanced response was obtained even with TRIF−/− MyD88−/− mice, which lack the TLR signaling pathways. Some form of signaling, not using these pathways and given by ligation of Clec9A itself, is a possibility. However, there was no sign of DC activation, not even of CD8+cDC, after administration of anti-mClec9A, at least in terms of MHC Class II, CD80 or CD86 up-regulation (data not shown). A very subtle DC activation signal may be involved. Just before submission of this manuscript, Huysamen et al52 also reported Clec9A as a new DC surface molecule common to humans and mice, and expressed on human BDCA-3 DCs. They demonstrated using artificial constructs that Clec9A, via its intracellular activation-like motif, does have the potential to signal, although direct signaling, via Clec9A itself, was not reported. An alternative explanation follows from the fact that the steady-state “immature” CD8+cDCs already express a moderate level of MHC Class II, CD40, CD80 and CD86. The very efficient Clec9A-mediated entry to DC processing may result in a high level of MHC-peptide presentation, enough to induce immune responses with only the base level of costimulator molecules present on DCs in steady-state.
This study introduced Clec9A as a new DC subtype–specific marker that is conserved between species. Together with the findings of Sancho et al49 and Huysamen et al,52 our study should help translate the wealth of information on mouse DC biology to human clinical application. These studies indicate that Clec9A has particular promise as a target for enhancing the effectiveness of vaccines, improving both antibody and T cell responses to Ags. The optimization of conditions for vaccine enhancement by targeting the Ag to Clec9A can now be pursued using the mouse as an initial model, with some confidence that the results will have clinical application.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank V. Milovac, C. Tarlinton, and J. Garbe for assistance with flow cytometry and Prof Akira Shizuo for kindly providing the MyD88- and TRIF-deficient mice.
This study was supported by grants 454465, 406660, and 516727 from the National Health and Medical Research Council of Australia (Canberra, Australia), and in part by research funding from CSL Limited (Parkville, Australia).
Authorship
Contribution: I.C. and M.H.L. designed, performed, and analyzed research and wrote the paper; A.I.P., K.J., and A.R. performed research and analysis; F.A., S.K., J.S.T., J.C.Y.L., D.V., and S.L.H.v.D. performed research; L.W., E.M., A.M.L., W.R.H., and M.D.W. designed research and interpreted data; H.B., G.M.D., p.m., N.v.d.V., and I.K.C. contributed vital reagents; and K.S. designed research and cowrote the paper.
Conflict-of-interest disclosure: E.M. and H.B. are employed by CSL Limited; however, CSL does not own the intellectual property for this study. The remaining authors declare no competing financial interests.
Correspondence: Mireille H. Lahoud or Irina Caminschi, The Walter and Eliza Hall Institute of Medical Research, 1G, Royal Parade, Parkville, Victoria 3050, Australia; e-mail: lahoud@wehi.edu.au or caminschi@wehi.edu.au.
References
Author notes
*I.C. and A.I.P. contributed equally to this work.