Abstract
Gene expression profiling (GEP) on frozen tissues has identified genes predicting outcome in patients with diffuse large B-cell lymphoma (DLBCL). Confirmation of results in current patients is limited by availability of frozen samples and addition of monoclonal antibodies to treatment regimens. We used a quantitative nuclease protection assay (qNPA) to analyze formalin-fixed, paraffin-embedded tissue blocks for 36 previously identified genes (N = 209, 93 chemotherapy; 116 rituximab + chemotherapy). By qNPA, 208 cases were successfully analyzed (99.5%). In addition, 15 of 36 and 11 of 36 genes, representing each functional group previously identified by GEP, were associated with survival (P < .05) in the 2 treatment groups, respectively. In addition, 30 of 36 hazard ratios of death trended in the same direction versus the original studies. Multivariate and variable cut-off point analysis identified low levels of HLA-DRB (< 20%) and high levels of MYC (> 80%) as independent indicators of survival, together distinguishing cases with the worst prognosis. Our results solve a clinical research problem by demonstrating that prognostic genes can be meaningfully quantified using qNPA technology on formalin-fixed, paraffin-embedded tissues; previous GEP findings in DLBCL are relevant with current treatments; and 2 genes, representing immune escape and proliferation, are the common features of the most aggressive DLBCL.
Introduction
Diffuse large B cell lymphoma (DLBCL) is a heterogeneous disease with variable patient survival. It accounts for nearly 35% of all cases of lymphoma. Gene expression profiling (GEP) studies of DLBCL have been performed by different research groups and have identified largely nonoverlapping gene sets associated with patient survival. The Leukemia and Lymphoma Molecular Profiling Project reported on 17 genes that could be used to determine an outcome predictor score using a competitive microarray platform, that when divided into quartiles was able to predict patient overall survival.1 A reanalysis of these data identified a “redox” signature score, which could also predict survival.2 Another group of investigators using a different microarray technology platform identified 13 different genes predictive of overall survival.3 A third group evaluated genes reported to be of prognostic interest in the literature to create a 6-gene model using quantitative reverse-transcribed polymerase chain reaction (RT-PCR).4 The genes identified by these different researchers represent largely nonoverlapping gene sets with the exception of 2 genes, BCL6 and fibronectin, as having prognostic significance. No actual side-by-side comparisons have yet been published. These conflicting data in the literature make it difficult to determine which genes are the most prognostically important to evaluate in new studies. Furthermore, progress is often limited by the numbers of cases for which frozen materials are available.
To overcome the limitation of using snap-frozen tissues, we used a multiplexed quantitative nuclease protection assay, the ArrayPlate, qNPA, useful for measuring mRNA levels in fixed paraffin-embedded samples. The assay was customized to measure all of the genes of interest from 4 previously described gene expression papers of DLBCL.5 This assay's performance demonstrated excellent reproducibility, applicability to archived paraffin blocks, and quantitative results that correlated well with GEP.
Most patients are now treated with monoclonal antibody therapy, most commonly rituximab, combined with chemotherapy, raising the question of whether the prognostic genes identified in the setting of chemotherapy treatment alone retain prognostic significance. The results of several randomized trials and a population-based registry experience have clearly indicated that rituximab plus cyclophosphamide, hydroxydaunorubicin, oncovorin (vincristine) and prednisione (R-CHOP) is the new treatment standard for DLBCL.6-8 There is some evidence to indicate that, indeed, the importance of some factors may be affected with new treatment, in particular BCL6 and BCL2. In one study, the authors found a reduction in treatment failure and death with the addition of rituximab to CHOP only for the BCL6− cases and that addition of rituximab did not benefit BCL6+ cases.9 Another group demonstrated that the addition of rituximab to chemotherapy for DLBCL patients overcame the negative prognostic value of the BCL2 protein.10 Thus, the question of how combined immunochemotherapy modifies the prognostic ranking of specific genes measured by expression levels remains pertinent.
In this paper, we demonstrate the robust use of the qNPA assay on formalin-fixed, paraffin-embedded tissue (FFPET) blocks, that the results can be related to patient overall survival, that prognostic genes previously identified remain relevant in the R-CHOP era, and that loss of immunosurveillance and high proliferation together identify patients with the worst outcome.
Methods
Patient materials
Three- to 5-micron unstained cuts from FFPET blocks were used from 93 cases of DLBCLs treated primarily with CHOP or similar CHOP-like chemotherapy and 116 cases treated with R-CHOP. Cases of transformed lymphomas were excluded. Frozen blocks from the CHOP-alone cases had been analyzed as part of a prior publication.1 As previously reported, these cases had undergone consensus review by a panel of expert hematopathologists and confirmed as DLBCL. The R-CHOP cases were taken from the current case files at the University of Arizona, British Columbia Cancer Agency, and Oregon Health Sciences Center. Of these 116 R-CHOP cases, frozen blocks from 32 were also used in another study and had undergone review by an expert panel.11 All tissues used for this retrospective study came from pretreatment diagnostic biopsies using excess diagnostic tissue under institutional review board–approved protocols. This project has been reviewed and approved by the University of Arizona Human Subjects Institutional Review Board, in accordance with the Declaration of Helsinki.
Assay methods
The performance of the ArrayPlate assay customized for use in DLBCLs has been described previously by our group.5 Additional descriptive details of the assay, schematic figure, and probe sequences are given in Table S1, Figure S1, and Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The complete previous publication is available at the online archives for Laboratory Investigation.5
We used the key genes12 identified as prognostically important in 4 previous papers of DLBCLs, which accounted for 36 genes of interest,1-4 as listed in Table 1 in the order in which they were listed in the original references. The housekeeping gene, TATA Box Binding Protein, was chosen for normalizing the data based on its stable expression at moderate levels in 12 lymphoma cell lines and 80 B- and T-cell lymphoma samples compared with 11 other “housekeeping” genes, using quantitative RT-PCR as well as our own previous experience with this gene in the ArrayPlate assay, which showed it to be moderately expressed with minimal variability in all samples tested to date.5,13
Prognostic genes identified in prior studies of CHOP-treated patients assessed using ArrayPlate
| Accession no. . | Name in original reference . | ArrayPlate name . | Reference (gene order within reference) . |
|---|---|---|---|
| NM_138931 | BCL6 | BCL6 | Rosenwald et al1 (1), Lossos et al4 (6) |
| NM_175739 | IMAGE 1334260 | GCET1 (SERPINA9) | Rosenwald et al1 (2) |
| NM_152785 | IMAGE 814622 | GCET2 | Rosenwald et al1 (3) |
| NM_033554 | HLA-DPα | HLA-DPA1 | Rosenwald et al1 (4) |
| NM_002122 | HLA-DQα | HLA-DQA1 | Rosenwald et al1 (5) |
| NM_019111 | HLA-DRα | HLA-DRA | Rosenwald et al1 (6) |
| NM_002124 | HLA-DRβ | HLA-DRB | Rosenwald et al1 (7) |
| NM_001102 | α-Actinin | ACTN1 | Rosenwald et al1 (8) |
| NM_000090 | Collagen type III α1 | COL3A1 | Rosenwald et al1 (9) |
| NM_001901 | Connective-tissue growth factor | CTGF | Rosenwald et al1 (10) |
| NM_212482 | Fibronectin | FN1 | Rosenwald et al1 (11), Lossos et al4 (5) |
| NM_014745 | KIAA0233 | FAM38A | Rosenwald et al1 (12) |
| NM_002658 | Urokinase plasminogen activator | PLAU | Rosenwald et al1 (13) |
| NM_002467 | MYC | MYC | Rosenwald et al1 (14) |
| NM_019095 | E21G3 (Nucleostemin) | C20orf155 | Rosenwald et al1 (15) |
| NM_006993 | NPM3 | NPM3 | Rosenwald et al1 (16) |
| NM_001718 | BMP6 | BMP6 | Rosenwald et al1 (17) |
| NM_005574 | LM02 | LMO2 | Lossos et al4 (1) |
| NM_000633 | BCL2 | BCL2 | Lossos et al4 (2) |
| NM_002983.1 | SCYA3 | CCL3 | Lossos et al4 (3) |
| NM_001759.2 | CCND2 | CCND2 | Lossos et al4 (4) |
| NM_001939 | DRP2 dystrophin related protein 2 | DRP2 | Shipp et al3 (1) |
| NM_002738 | PRKACB protein kinase C-beta-1 | PRKCB1 | Shipp et al3 (2) |
| NM_014456 | H731 nuclear antigen | PDCD4 | Shipp et al3 (3) |
| NM_005909 | 3′ UTR of unknown protein | MAP1B | Shipp et al3 (4) |
| NM_005077 | Transducin-like enhancer protein 1 | TLE1 | Shipp et al3 (5) |
| NM_014251 | Uncharacterized | SLC25A13 | Shipp et al3 (6) |
| NM_002600 | Phosphodiesterase 4B, cAMP-specific (PDE4B) | PDE4B | Shipp et al3 (7) |
| NM_001497 | UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 1 | B4GALT1 | Shipp et al3 (8) |
| NM_002739 | Protein kinase C, gamma (PRKCG) | PRKCG | Shipp et al3 (9) |
| NM_002557 | Oviductal glycoprotein | OVGP1 | Shipp et al3 (10) |
| NM_173198 | Mitogen induced nuclear orphan receptor (MINOR) | NR4A3 | Shipp et al3 (11) |
| NM_012256 | Zinc-finger protein C2H2-150 | ZNF212 | Shipp et al3 (12) |
| NM_000867 | 5-Hydroxytryptamine 2B receptor | HTR2B | Shipp et al3 (13) |
| NM_001752 | Catalase | CAT | Tome et al2 (1) |
| NM_000636 | Manganese superoxide dismutase | SOD2 | Tome et al2 (2) |
| M34960 | TATA box binding protein | TBP | Lossos et al13 |
| Accession no. . | Name in original reference . | ArrayPlate name . | Reference (gene order within reference) . |
|---|---|---|---|
| NM_138931 | BCL6 | BCL6 | Rosenwald et al1 (1), Lossos et al4 (6) |
| NM_175739 | IMAGE 1334260 | GCET1 (SERPINA9) | Rosenwald et al1 (2) |
| NM_152785 | IMAGE 814622 | GCET2 | Rosenwald et al1 (3) |
| NM_033554 | HLA-DPα | HLA-DPA1 | Rosenwald et al1 (4) |
| NM_002122 | HLA-DQα | HLA-DQA1 | Rosenwald et al1 (5) |
| NM_019111 | HLA-DRα | HLA-DRA | Rosenwald et al1 (6) |
| NM_002124 | HLA-DRβ | HLA-DRB | Rosenwald et al1 (7) |
| NM_001102 | α-Actinin | ACTN1 | Rosenwald et al1 (8) |
| NM_000090 | Collagen type III α1 | COL3A1 | Rosenwald et al1 (9) |
| NM_001901 | Connective-tissue growth factor | CTGF | Rosenwald et al1 (10) |
| NM_212482 | Fibronectin | FN1 | Rosenwald et al1 (11), Lossos et al4 (5) |
| NM_014745 | KIAA0233 | FAM38A | Rosenwald et al1 (12) |
| NM_002658 | Urokinase plasminogen activator | PLAU | Rosenwald et al1 (13) |
| NM_002467 | MYC | MYC | Rosenwald et al1 (14) |
| NM_019095 | E21G3 (Nucleostemin) | C20orf155 | Rosenwald et al1 (15) |
| NM_006993 | NPM3 | NPM3 | Rosenwald et al1 (16) |
| NM_001718 | BMP6 | BMP6 | Rosenwald et al1 (17) |
| NM_005574 | LM02 | LMO2 | Lossos et al4 (1) |
| NM_000633 | BCL2 | BCL2 | Lossos et al4 (2) |
| NM_002983.1 | SCYA3 | CCL3 | Lossos et al4 (3) |
| NM_001759.2 | CCND2 | CCND2 | Lossos et al4 (4) |
| NM_001939 | DRP2 dystrophin related protein 2 | DRP2 | Shipp et al3 (1) |
| NM_002738 | PRKACB protein kinase C-beta-1 | PRKCB1 | Shipp et al3 (2) |
| NM_014456 | H731 nuclear antigen | PDCD4 | Shipp et al3 (3) |
| NM_005909 | 3′ UTR of unknown protein | MAP1B | Shipp et al3 (4) |
| NM_005077 | Transducin-like enhancer protein 1 | TLE1 | Shipp et al3 (5) |
| NM_014251 | Uncharacterized | SLC25A13 | Shipp et al3 (6) |
| NM_002600 | Phosphodiesterase 4B, cAMP-specific (PDE4B) | PDE4B | Shipp et al3 (7) |
| NM_001497 | UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 1 | B4GALT1 | Shipp et al3 (8) |
| NM_002739 | Protein kinase C, gamma (PRKCG) | PRKCG | Shipp et al3 (9) |
| NM_002557 | Oviductal glycoprotein | OVGP1 | Shipp et al3 (10) |
| NM_173198 | Mitogen induced nuclear orphan receptor (MINOR) | NR4A3 | Shipp et al3 (11) |
| NM_012256 | Zinc-finger protein C2H2-150 | ZNF212 | Shipp et al3 (12) |
| NM_000867 | 5-Hydroxytryptamine 2B receptor | HTR2B | Shipp et al3 (13) |
| NM_001752 | Catalase | CAT | Tome et al2 (1) |
| NM_000636 | Manganese superoxide dismutase | SOD2 | Tome et al2 (2) |
| M34960 | TATA box binding protein | TBP | Lossos et al13 |
Statistical analysis
General.
Statistical analyses of the association of gene expression, as measured by Array Plate qNPA technology, with survival were performed on the 116 cases treated with R-CHOP and the 93 cases treated with CHOP or CHOP-like regimens alone. The logarithms of gene-expression values were standardized to have standard deviation equal to 1.
Initial evaluation of ArrayPlate results related to patient survival (univariate analysis, comparison between CHOP- and R-CHOP-treated case results).
Hazard ratios, 95% confidence intervals, and P values for the univariate associations between standardized log gene expression levels and patient overall survival were obtained using Cox proportional hazard regression.14 To account for the relatively large number of test statistics, the overall statistical significance of the set of hypothesis tests against a global null hypothesis of no association was calculated, by permutation resampling, based on the “tail strength” (TS) statistic.15 A test of the overall statistical interaction of gene expression and treatment type was considered in a similar fashion.
Multivariate analysis.
In an exploratory analysis of parsimonious multivariate models, we used best subset selection, which determines the “best” model based on the global score χ2 statistic.16 Candidate genes used in the model building process were those achieving nominal P values less than .05. The top 3 models for each of 1, 2, 3, and 4 variable models were derived. For presentation purposes, for each factor included, the overall best identified model, patients were categorized by high versus low gene expression (above or below the median value). Patients were then grouped according to the number of adverse risk factors, and survival was examined.
Adjustment of 2-gene model for clinical International Prognostic Index (IPI) score.
Finally, we assessed whether the AP gene-risk model retained significance of the biologic aspects of the malignant cells after adjusting for the clinically based IPI index.17
Variable cutoff point analysis on 2 key genes.
Separately, cutoff point analysis was performed on the factors identified in multivariate modeling to optimize identification of expression levels of highest risk. Permutation resampling was used to adjust significance levels of the proportional hazards score tests among all evaluated cutoff points.18 In addition, to control statistical variability of the cutoff point analysis, a minimum possible group size of 10% of total patients was set for the analysis.
Results
Performance of assay in FFPET blocks
Of 209 cases attempted, there was only one that did not result in adequately detectable signal. In situ hybridization using a polyDT probe (Ventana Medical Systems, Tucson, AZ) demonstrated that the mRNA was degraded (data not shown). This failure was therefore attributed to sample inadequacy rather than a technical failure of the assay itself. TATA-binding protein was moderately and consistently expressed in all samples, as in our previous work, and again used as the control gene for normalization of the data.5
Overall rationale and sequence of statistical analyses
Initial evaluation of HTG results included univariate analysis of individual gene levels with respect to patient survival in both treatment groups, using the logarithm of the gene expression measurements. To further explore potentially important genes, we calculated hazard ratios of death and assessed whether they trended in the direction predicted by the previously reported literature. For each of the treatment groups, we assessed the overall significance of the panel of genes using the tail strength statistic and permutation resampling. Any gene that was significantly associated (P < .05) with overall survival in univariate modeling was assessed for potential inclusion in a multivariate risk model using Cox regression analysis.14 To determine the best model, we used best subset selection, which determines the “best” model based on the global score χ2 statistic, and adjusted this model for clinical IPI score.16 Lastly, a variable cutoff point analysis was performed on the 2 key genes to determine whether there were more relevant cutoff points, rather than the preselected 50th percentile, which might have biologic implications. Permutation sampling was used to adjust for multiple comparisons in the cutoff point optimization.
CHOP results
For chemotherapy-alone (mainly CHOP) treated cases, gene expression levels were significantly correlated with overall survival at P less than .05 for 15 of 36 prognostic genes, including the major histocompatibility complex (MHC) class II genes HLA-DRA and HLA-DPA1, germinal center–associated genes BCL6, GCET1 (SERPINA9), stromal associated genes (ACTN1, COL3A1, CTGF, FN1), proliferation genes MYC, CCND2, PRKCB1, as well as PDCD4, TLE, B4GALT1, and BCL-2. These genes represented all 4 prognostic signatures from Rosenwald et al,1 4 of 13 genes reported by Shipp et al,3 and 3 of 6 genes from Lossos et al.4 An additional gene, CCL3, was borderline significant at a P value of .062.
R-CHOP results
For the R-CHOP–treated patients, 11 of the 36 genes analyzed were significantly associated with survival at the P less than .05 cutoff level. These genes were GCET1 (SERPINA9), HLA-DQA1, HLA-DRB, ACTN1, COL3A1, PLAU, MYC, BCL6, LMO2, PDCD4, and SOD2. An additional gene, FN1, was marginally significant at a P value of .078. Results of univariate analyses compared for the 2 treatment eras are shown side by side in Table 2. To emphasize the genes with recurrent significance, the P values at .05 or less are footnoted. Average 2-year overall survival for each gene cut at above and below the median expression level are also summarized in Table 2. We note that survival rates at 2 years were chosen as simple descriptive summary statistics. Similar results were seen with 3- and 4-year rates, although estimates were more unstable because of more censored cases. The P values presented in the tables are based on Cox score tests using the continuous logarithm of gene expression and therefore do not depend on the choice of summary survival estimates presented.
Results of univariate analyses of gene expression with overall survival
| Gene . | CHOP . | CHOP plus R . | ||||
|---|---|---|---|---|---|---|
| HR . | P . | 2-yr OS*, % . | HR . | P . | 2-yr OS*, % . | |
| BCL6 | 0.65 | .008† | 69, 52 | 0.62 | .007† | 82, 69 |
| GCET1 (SERPINA9) | 0.75 | .01† | 69, 53 | 0.62 | .013† | 83, 68 |
| GCET2 | 0.93 | .608 | 59, 66 | 0.93 | .608 | 77, 74 |
| HLA-DPA1 | 0.71 | .036† | 63, 58 | 0.77 | .115 | 84, 68 |
| HLA-DQA1 | 1.14 | .35 | 63, 62 | 0.65 | .020 | 83, 68 |
| HLA-DRA | 0.72 | .02† | 63, 58 | 0.91 | .580 | 84, 68 |
| HLA-DRB | 0.98 | .921 | 58, 66 | 0.71 | .030† | 89, 62 |
| ACTN1 | 0.66 | .03† | 73, 49 | 0.62 | .011† | 85, 66 |
| COL3A1 | 0.78 | .016† | 71, 50 | 0.67 | .029† | 81, 69 |
| CTGF | 0.79 | .026† | 75, 45 | 0.80 | .211 | 84, 67 |
| FN1 | 0.77 | .01† | 67, 54 | 0.73 | .078 | 78, 73 |
| FAM38A | 1.16 | .462 | 61, 60 | 0.85 | .426 | 80, 71 |
| PLAU | 0.73 | .122 | 68, 54 | 0.56 | .001† | 84, 67 |
| MYC | 1.40 | .047† | 54, 67 | 1.64 | .007† | 65, 86 |
| C20ORF155 | 1.32 | .369 | 52, 70 | 1.03 | .851 | 72, 79 |
| NPM3 | 1.27 | .25 | 58, 64 | 1.22 | .303 | 74, 76 |
| BMP6 | 1.26 | .216 | 64, 60 | 0.86 | .402 | 77, 74 |
| LMO2 | 1.03 | .832 | 57, 64 | 0.62 | .011† | 82, 69 |
| BCL2 | 1.44 | .018† | 57, 66 | 1.11 | .569 | 71, 80 |
| CCL3 | 1.40 | .062 | 48, 73 | 0.82 | .296 | 84, 67 |
| CCND2 | 1.45 | .002† | 53, 69 | 1.23 | .271 | 76, 75 |
| DRP2 | 1.02 | .878 | 66, 60 | 0.94 | .719 | 75, 76 |
| PRKCB1 | 1.47 | .028† | 51, 71 | 0.99 | .951 | 79, 73 |
| PDCD4 | 1.89 | .001† | 50, 73 | 1.53 | .023† | 67, 84 |
| MAP1B | 1.05 | .772 | 63, 64 | 0.94 | .717 | 78, 72 |
| TLE1 | 1.60 | .001† | 42, 80 | 1.16 | .428 | 70, 81 |
| SLC25A13 | 1.15 | .676 | 58, 63 | 0.89 | .540 | 78, 73 |
| PDE4B | 1.19 | .423 | 56, 64 | 1.17 | .402 | 75, 76 |
| B4GALT1 | 1.87 | .001† | 49, 71 | 0.82 | .258 | 80, 72 |
| PRKCG | 1.05 | .701 | 49, 72 | 1.02 | .924 | 77, 71 |
| OVGP1 | 1.25 | .279 | 52, 73 | 1.02 | .924 | 77, 71 |
| NR4A3 | 1.30 | .151 | 49, 71 | 0.80 | .227 | 72, 80 |
| ZNF212 | 0.99 | .965 | 58, 64 | 1.04 | .810 | 73, 78 |
| HTR2B | 1.04 | .834 | 48, 68 | 0.76 | .210 | 81, 64 |
| CAT | 1.24 | .50 | 54, 67 | 0.99 | .962 | 80, 71 |
| SOD2 | 1.10 | .573 | 60, 61 | 0.64 | .014† | 87, 64 |
| Gene . | CHOP . | CHOP plus R . | ||||
|---|---|---|---|---|---|---|
| HR . | P . | 2-yr OS*, % . | HR . | P . | 2-yr OS*, % . | |
| BCL6 | 0.65 | .008† | 69, 52 | 0.62 | .007† | 82, 69 |
| GCET1 (SERPINA9) | 0.75 | .01† | 69, 53 | 0.62 | .013† | 83, 68 |
| GCET2 | 0.93 | .608 | 59, 66 | 0.93 | .608 | 77, 74 |
| HLA-DPA1 | 0.71 | .036† | 63, 58 | 0.77 | .115 | 84, 68 |
| HLA-DQA1 | 1.14 | .35 | 63, 62 | 0.65 | .020 | 83, 68 |
| HLA-DRA | 0.72 | .02† | 63, 58 | 0.91 | .580 | 84, 68 |
| HLA-DRB | 0.98 | .921 | 58, 66 | 0.71 | .030† | 89, 62 |
| ACTN1 | 0.66 | .03† | 73, 49 | 0.62 | .011† | 85, 66 |
| COL3A1 | 0.78 | .016† | 71, 50 | 0.67 | .029† | 81, 69 |
| CTGF | 0.79 | .026† | 75, 45 | 0.80 | .211 | 84, 67 |
| FN1 | 0.77 | .01† | 67, 54 | 0.73 | .078 | 78, 73 |
| FAM38A | 1.16 | .462 | 61, 60 | 0.85 | .426 | 80, 71 |
| PLAU | 0.73 | .122 | 68, 54 | 0.56 | .001† | 84, 67 |
| MYC | 1.40 | .047† | 54, 67 | 1.64 | .007† | 65, 86 |
| C20ORF155 | 1.32 | .369 | 52, 70 | 1.03 | .851 | 72, 79 |
| NPM3 | 1.27 | .25 | 58, 64 | 1.22 | .303 | 74, 76 |
| BMP6 | 1.26 | .216 | 64, 60 | 0.86 | .402 | 77, 74 |
| LMO2 | 1.03 | .832 | 57, 64 | 0.62 | .011† | 82, 69 |
| BCL2 | 1.44 | .018† | 57, 66 | 1.11 | .569 | 71, 80 |
| CCL3 | 1.40 | .062 | 48, 73 | 0.82 | .296 | 84, 67 |
| CCND2 | 1.45 | .002† | 53, 69 | 1.23 | .271 | 76, 75 |
| DRP2 | 1.02 | .878 | 66, 60 | 0.94 | .719 | 75, 76 |
| PRKCB1 | 1.47 | .028† | 51, 71 | 0.99 | .951 | 79, 73 |
| PDCD4 | 1.89 | .001† | 50, 73 | 1.53 | .023† | 67, 84 |
| MAP1B | 1.05 | .772 | 63, 64 | 0.94 | .717 | 78, 72 |
| TLE1 | 1.60 | .001† | 42, 80 | 1.16 | .428 | 70, 81 |
| SLC25A13 | 1.15 | .676 | 58, 63 | 0.89 | .540 | 78, 73 |
| PDE4B | 1.19 | .423 | 56, 64 | 1.17 | .402 | 75, 76 |
| B4GALT1 | 1.87 | .001† | 49, 71 | 0.82 | .258 | 80, 72 |
| PRKCG | 1.05 | .701 | 49, 72 | 1.02 | .924 | 77, 71 |
| OVGP1 | 1.25 | .279 | 52, 73 | 1.02 | .924 | 77, 71 |
| NR4A3 | 1.30 | .151 | 49, 71 | 0.80 | .227 | 72, 80 |
| ZNF212 | 0.99 | .965 | 58, 64 | 1.04 | .810 | 73, 78 |
| HTR2B | 1.04 | .834 | 48, 68 | 0.76 | .210 | 81, 64 |
| CAT | 1.24 | .50 | 54, 67 | 0.99 | .962 | 80, 71 |
| SOD2 | 1.10 | .573 | 60, 61 | 0.64 | .014† | 87, 64 |
The 2-year OS percentage is split at less than the median and greater than or equal to the median.
P value .05 or less.
Comparative overall survival curves in the different treatment eras for HLA-DRB (an MHC class II gene), BCL6, and MYC are demonstrated in Figure 1. These examples demonstrate the ability of the ArrayPlate assay to generate meaningful quantitative data that can be related to patient outcome. The results also demonstrate that, for these well-known prognostic genes, there is continued evidence of prognostic relevance in R-CHOP–treated patients.
Overall survival in years for 3 representative genes showing patients treated with CHOP versus R-CHOP according to gene expression levels.HLA-DRB is cut above and below 25%; BCL6 and MYC are cut at median. (A) CHOP-treated cases, all IPI scores: (Ai) HLA-DRB; (Aii) BCL6; (Aiii) MYC (N = 93). (B) R-CHOP cases, all IPI scores, HLA-DR, BCL6, and MYC (N = 116).
Overall survival in years for 3 representative genes showing patients treated with CHOP versus R-CHOP according to gene expression levels.HLA-DRB is cut above and below 25%; BCL6 and MYC are cut at median. (A) CHOP-treated cases, all IPI scores: (Ai) HLA-DRB; (Aii) BCL6; (Aiii) MYC (N = 93). (B) R-CHOP cases, all IPI scores, HLA-DR, BCL6, and MYC (N = 116).
For most genes in both treatment groups, the estimated hazard ratios of death trended in the direction predicted by the original studies (Table 3). Hazard ratios correspond to a change in 1 standard deviation in log expression levels and a hazard ratio more than 1 indicates an association between high expression (above the median) with poorer outcome, whereas hazard ratios less than 1 indicate an association between high expression with better outcome. Therefore, an estimated hazard ratio that is very small in magnitude (eg, close to 0) corresponds to a gene with strong association between higher expression and longer survival.
Hazard ratios of overall survival in R-CHOP–treated patients and agreement with original study regarding predictive capacity
| Gene* . | HR (95% CI)† . | Agree with trend in original study . |
|---|---|---|
| BCL6 | 0.62 (0.44-0.87) | Yes |
| GCET1 (SERPINA9) | 0.62 (0.41-0.92) | Yes |
| GCET2 | 0.93 (0.66-1.31) | Yes |
| HLA-DPA1 | 0.77 (0.56-1.05) | Yes |
| HLA-DQA1 | 0.65 (0.45-0.93) | Yes |
| HLA-DRA | 0.91 (0.67-1.24) | Yes |
| HLA-DRB | 0.71 (0.53-0.95) | Yes |
| ACTN1 | 0.62 (0.43-0.89) | Yes |
| COL3A1 | 0.67 (0.47-0.97) | Yes |
| CTGF | 0.80 (0.56-1.14) | Yes |
| FN1 | 0.73 (0.51-1.04) | Yes |
| FAM38A | 0.85 (0.58-1.26) | Yes |
| PLAU | 0.56 (0.40-0.79) | Yes |
| MYC | 1.64 (1.16-2.31) | Yes |
| C20ORF155 | 1.03 (0.73-1.47) | No |
| NPM3 | 1.22 (0.84-1.76) | Yes |
| BMP6 | 0.86 (0.59-1.23) | No |
| LMO2 | 0.62 (0.43-0.90) | Yes |
| BCL2 | 1.11 (0.77-1.60) | Yes |
| CCL3 | 0.82 (0.58-1.18) | No |
| CCND2 | 1.23 (0.85-1.77) | Yes |
| DRP2 | 0.94 (0.65-1.35) | Yes |
| PRKCB1 | 0.99 (0.69-1.42) | No |
| PDCD4 | 1.53 (1.07-2.21) | Yes |
| MAP1B | 0.94 (0.66-1.33) | Yes |
| TLE1 | 1.16 (0.81-1.65) | Yes |
| SLC25A13 | 0.90 (0.63-1.28) | Yes |
| PDE4B | 1.17 (0.81-1.68) | Yes |
| B4GALT1 | 0.82 (0.57-1.16) | No |
| PRKCG | 1.02 (0.69-1.51) | Yes |
| OVGP1 | 1.03 (0.73-1.47) | Yes |
| NR4A3 | 0.80 (0.55-1.15) | Yes |
| ZNF212 | 1.04 (0.74-1.48) | Yes |
| HTR2B | 0.76 (0.49-1.18) | Yes |
| CAT | 0.99 (0.70-1.41) | No |
| SOD2 | 0.64 (0.45-0.92) | Yes |
| Gene* . | HR (95% CI)† . | Agree with trend in original study . |
|---|---|---|
| BCL6 | 0.62 (0.44-0.87) | Yes |
| GCET1 (SERPINA9) | 0.62 (0.41-0.92) | Yes |
| GCET2 | 0.93 (0.66-1.31) | Yes |
| HLA-DPA1 | 0.77 (0.56-1.05) | Yes |
| HLA-DQA1 | 0.65 (0.45-0.93) | Yes |
| HLA-DRA | 0.91 (0.67-1.24) | Yes |
| HLA-DRB | 0.71 (0.53-0.95) | Yes |
| ACTN1 | 0.62 (0.43-0.89) | Yes |
| COL3A1 | 0.67 (0.47-0.97) | Yes |
| CTGF | 0.80 (0.56-1.14) | Yes |
| FN1 | 0.73 (0.51-1.04) | Yes |
| FAM38A | 0.85 (0.58-1.26) | Yes |
| PLAU | 0.56 (0.40-0.79) | Yes |
| MYC | 1.64 (1.16-2.31) | Yes |
| C20ORF155 | 1.03 (0.73-1.47) | No |
| NPM3 | 1.22 (0.84-1.76) | Yes |
| BMP6 | 0.86 (0.59-1.23) | No |
| LMO2 | 0.62 (0.43-0.90) | Yes |
| BCL2 | 1.11 (0.77-1.60) | Yes |
| CCL3 | 0.82 (0.58-1.18) | No |
| CCND2 | 1.23 (0.85-1.77) | Yes |
| DRP2 | 0.94 (0.65-1.35) | Yes |
| PRKCB1 | 0.99 (0.69-1.42) | No |
| PDCD4 | 1.53 (1.07-2.21) | Yes |
| MAP1B | 0.94 (0.66-1.33) | Yes |
| TLE1 | 1.16 (0.81-1.65) | Yes |
| SLC25A13 | 0.90 (0.63-1.28) | Yes |
| PDE4B | 1.17 (0.81-1.68) | Yes |
| B4GALT1 | 0.82 (0.57-1.16) | No |
| PRKCG | 1.02 (0.69-1.51) | Yes |
| OVGP1 | 1.03 (0.73-1.47) | Yes |
| NR4A3 | 0.80 (0.55-1.15) | Yes |
| ZNF212 | 1.04 (0.74-1.48) | Yes |
| HTR2B | 0.76 (0.49-1.18) | Yes |
| CAT | 0.99 (0.70-1.41) | No |
| SOD2 | 0.64 (0.45-0.92) | Yes |
HR indicates hazard ratio; and CI, confidence interval.
Standardized to normal (0, 1) distribution.
HR of 1 indicates no effect on risk; HR between 0 and 1, good risk; HR greater than 1, poor risk.
Comparison of CHOP and R-CHOP data
To address the testing of the multiple genes in the panel, an overall test of the 36 P values was performed using the TS statistic and permutation resampling. There is evidence of association between the overall 36-gene panel and outcome in both CHOP-treated patients (TS, P = .007) and R-CHOP patients (TS, P = .013).15 We also considered an overall test of differences by treatment group in the association between each gene and survival (statistical interaction). Although power for interaction testing is limited, there was no evidence of a differential effect of the overall 36-gene expression panel between the 2 treatment types (TS, P = .250).
As an overall assessment of important prognostic features, the IPI distribution was assessed among patients in the 2 treatment types. In the CHOP-alone patients, 41% had IPI of 0 to 1, 48% had IPI of 2 to 3, and 11% had IPI of 4 to 5. In the CHOP-R-treated patients, 40% had IPI of 0 to 1, 56% had IPI of 2 to 3, and 4% had IPI of 4 to 5. There was no evidence of a difference between the 2 treatment groups (P = .18). Table 4 details the distribution of the individual factors of the IPI score between patients in the 2 treatment eras.
Distribution of factors in the International Prognostic Index (IPI) between patients in the 2 treatment eras
| IPI factor . | CHOP, % . | R-CHOP, % . |
|---|---|---|
| Age older than 60 y | 47 | 49 |
| LDH over the upper limit of normal | 54 | 60 |
| More than stage II | 48 | 60 |
| More than 1 extranodal site | 17 | 17 |
| Performance status more than 1 | 16 | 29 |
| IPI factor . | CHOP, % . | R-CHOP, % . |
|---|---|---|
| Age older than 60 y | 47 | 49 |
| LDH over the upper limit of normal | 54 | 60 |
| More than stage II | 48 | 60 |
| More than 1 extranodal site | 17 | 17 |
| Performance status more than 1 | 16 | 29 |
Prognostic model
As an exploratory analysis of multivariable prognostic models, best 1, 2, 3, and 4 variable models, as determined by best subsets analysis, were calculated (data not shown). The best 2-variable model was the combination of MYC and HLA-DRB, with a model χ2 of 16.6. However, we note that other 2-variable models, including MYC with HLA-DQA1 or PLAU, had modestly smaller model χ2 statistics. Given the relatively small number of events in this study, conclusive statements about the overall best model are not possible. There was no evidence that 3 variable models yielded any statistical improvement in model fit.
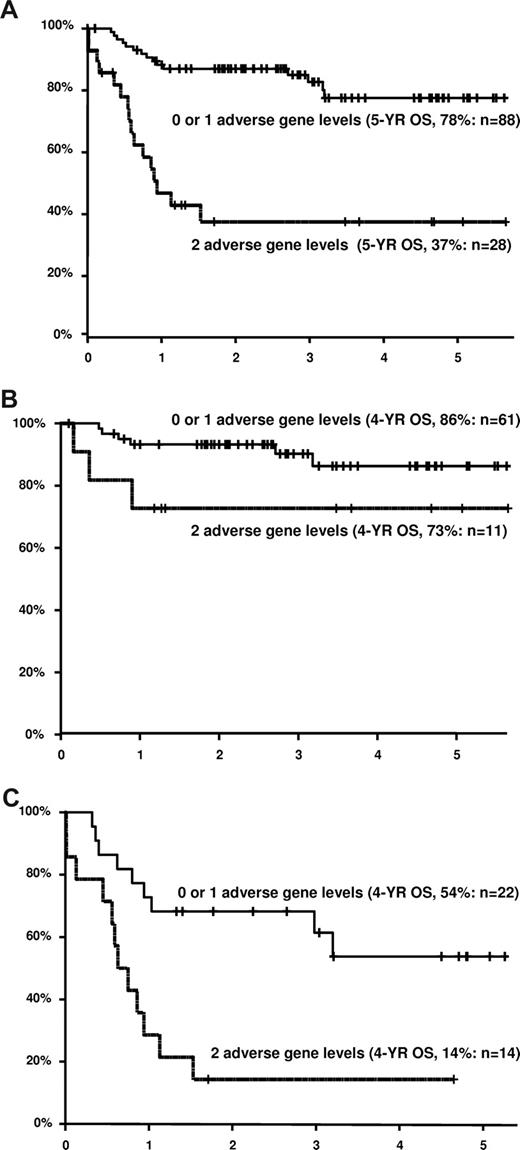
Patients were defined as having high or low levels of MYC and HLA-DRB. Twenty-eight patients (24%) had both adverse gene levels. These patients had much worse survival than patients with 0 or 1 adverse gene level (2-year overall survival, 38% vs 87%), as shown in Figure 2A. Differences are presented for both the high and low IPI subgroups (Figures 2B,C). The survival disadvantage for patients with both adverse gene levels appears particularly pronounced in patients with high IPI (2-year estimate, 14% vs 68%), although there was no evidence of an interaction between number of adverse gene levels and IPI group (P = .88).
Overall survival in years for patients treated with R-CHOP according to IPI score and expression levels of HLA-DRB and/or MYC. Cutoff point levels are above and below the median for both genes. Adverse gene level for HLA-DR is for expression below the median, whereas adverse gene level for MYC is for expression above the median (A) All IPI groups (N = 116). (B) Low IPI group (scores 0-2, N = 72). (C) High IPI group (scores 3-5, N = 36). The combined number of cases in panels B and C are fewer than in panel A because of several cases with missing IPI information.
Overall survival in years for patients treated with R-CHOP according to IPI score and expression levels of HLA-DRB and/or MYC. Cutoff point levels are above and below the median for both genes. Adverse gene level for HLA-DR is for expression below the median, whereas adverse gene level for MYC is for expression above the median (A) All IPI groups (N = 116). (B) Low IPI group (scores 0-2, N = 72). (C) High IPI group (scores 3-5, N = 36). The combined number of cases in panels B and C are fewer than in panel A because of several cases with missing IPI information.
Both CHOP and R-CHOP data were combined to further explore the nature of the association of expression of these 2 genes with survival using cutoff point analysis. For HLA-DRB, the highest χ2 value indicating the most significant cutoff point was at the 20th percentile of gene expression (P = .01 based on permutation resampling to account for the multiple testing). For MYC, the most significant cutoff point (P = .01) was at the 80th percentile of expression (Figure 3). Given the adaptive nature of cutoff point selection, any multivariate model based on cut-off point levels identified in this analysis would be best validated independently. We emphasize that, although the 80th percentile was the optimal cutoff point for MYC (corresponding to a χ2 value of > 15 and a nominal P < .001), there were a wide range of cutoff point values that were also nominally significant (P < .025). This indicates that other cutoff points may lead to interesting prognostic models.
Variable cutoff point analysis for HLA-DRB and MYC genes. Gene expression level on x-axis; log rank score on y-axis; permutation P value indicated. (A) HLA-DRB. (B) MYC. The peaks in the log rank scores indicate the most significant cut-off points in the data yielding the largest differences in overall survival.
Variable cutoff point analysis for HLA-DRB and MYC genes. Gene expression level on x-axis; log rank score on y-axis; permutation P value indicated. (A) HLA-DRB. (B) MYC. The peaks in the log rank scores indicate the most significant cut-off points in the data yielding the largest differences in overall survival.
Discussion
GEP studies in cancer have relied on snap-frozen tissues, which are often not obtained at diagnosis and are becoming decreasingly available in the current era of small biopsies. To overcome this obstacle, in this paper we describe the use of a quantitative S1 nuclease protection assay to measure gene expression levels in FFPET blocks of DLBCLs, related to results to patient outcome in 2 different treatment eras, and used multivariate and variable cutoff point analysis to identify molecules related to antigen presentation and cell cycle/proliferation as key indicators of patient outcome.
The ArrayPlate qNPA multiplexed mRNA assay used for this project was originally designed to support genomics-driven drug discovery efforts.19 The standardized protocol is automated and optimized for processing large numbers of samples. The S1 nuclease digestion that the qNPA incorporates is a well-established approach that predates more widely used PCR-based procedures for quantifying gene expression.20 For a 12-gene signature predictive of phospholipidosis in human liver cells to examine treatments with 80 different compounds, mRNA scores correlated significantly (R2 = 0.95) between the ArrayPlate and real-time PCR assays.21 Unlike RT-PCR, however, the ArrayPlate qNPA did not require RNA isolation, reverse transcription, or amplification. Instead, target mRNA-specific 50-mer qNPA probes were retained in a stoichiometric fashion. The direct probe-to-target hybridization on which the assay relies was critical for the successful GEP of FFPET blocks of material.5 We demonstrated that the assay measured mRNA, whether it was released from FFPET sections or cross-linked in situ. The use of relatively short target 50-mers in the sample meant that robust assay results were obtained even for archived specimens nearly 20 years old in which RNA degradation was presumably substantial, resulting in an extremely low failure rate of only 1 in 209 cases (0.5%). With most comparable techniques, failure rates of all attempted cases are seldom reported and anecdotally high.
In our series of CHOP-treated cases, qNPA results for many genes were significantly associated with overall survival when using a standard cutoff point at the median level of expression. These genes represented key expression signatures previously identified as prognostically important in large discovery-oriented GEP experiments. Because these cases had been a part of the original case series by Rosenwald et al, the results showing significant correlations in 10 of the 17 outcome predictor score genes from that paper served to demonstrate the ability of the qNPA to generate data from FFPET blocks that can be correlated to patient survival.1 We also concluded that many of the genes identified by the different research groups each do represent important aspects of DLBCL biology, which deserve further exploration. We wish to emphasize that the biologic importance of these findings lies in the functional groups of genes rather than specific genes themselves because, in any one relatively small dataset, one or the other of a set of genes may reach statistical significance.
In the R-CHOP–treated patients, again many genes were significantly associated with overall survival using a cutoff at the median level of expression. For the genes that did not reach statistical significance, the trends in the hazard ratios largely agreed with those from the original publications indicating that many of the “nonsignificant” genes may become significant if analyzed in a larger dataset. Nonetheless, our findings indicate that representatives from each of the previous functional groups of genes remain relevant in the R-CHOP era and that the key biologic aspects of DLBCLs are not selectively affected by addition of monoclonal antibody to CHOP chemotherapy. This interpretation is supported by the global test of interaction between the 2 treatment groups, which did not show any significant difference in gene levels as related to outcome overall.
In agreement with our findings of the continued importance of expression levels of germinal center associated genes (BCL6, GCET1 in the CHOP era; and BCL6, GCET1, and LM02 in the R-CHOP era), a recent paper demonstrated that LM02 immunohistochemical staining was a significant predictor of survival in R-CHOP–treated patients similar to chemotherapy alone.22 Recent reports using quantitative RT-PCR and GEP have described similar findings of the continued importance of most genes.11,23 A possible exception is the BCL2 gene, which was significant in the CHOP era but not in our series of R-CHOP–treated cases (although the hazard ratio trended in the correct direction with higher BCL2 levels representing a higher risk). Whether this represents the real impact of adding rituximab to the chemotherapy regimen as reported by Mounier et al using immunohistochemistry needs to be clarified in a larger cohort of patients.10 Small changes in the significance of genes in CHOP- versus R-CHOP–treated patients would be difficult to find without large numbers of cases because most patients are simultaneously treated with rituximab and CHOP, which may obfuscate the survival changes from rituximab. Application of the variable cutoff point analysis (applied here to HLA-DRB and MYC only) to all of the genes in this study may be an interesting exercise to determine whether the use of different cutoff points allows the impact of gene expression to be seen more readily. However, we are not attempting to call out specific genes but rather to emphasize that the same groups of genes remain relevant in the current treatment era.
We and others have previously demonstrated that, with decreasing MHC class II expression, patient outcome worsens with those in the lowest 10% having an extremely poor outcome.24-26 In this paper, we again demonstrated the significance of the MHC class II antigen gene expression levels (when cut at the median in our screening analysis). In addition, we show the strong significance of very low levels of MHC class II using the variable cutoff point analysis, which identified the most significant impact on survival at the 20% expression level consistent with our previous work. Although the MHC class II signature effect demonstrates similar trends with CHOP and R-CHOP, there is nonetheless a diminished MHC class II effect in the R-CHOP group, which could reflect some partial reversal of the negative prognostic value of MHC class II. A large study may be needed to further investigate this issue. The importance of lost MHC class II appears to be related to a loss of tumor immunosurveillance, as evidenced by decreased numbers of tumor-infiltrating T cells as reported by us and others.24,27-29 Continued significance of these genes in the R-CHOP era is not surprising given the strong influence of tumor immunosurveillance on lymphomagenesis and outcome. This class of molecules therefore constitutes a therapeutic target in a subset of DLBCL.
Previous studies have identified high levels of MYC as being prognostically important in DLBCL in the CHOP era. It is therefore of interest that we were able to confirm the importance of MYC in the R-CHOP era and show, by using a quantitative method and variable cutoff point analysis, that the highest gene expression levels were most significant, and to narrow the collection of genes down to a useful signature of MYC and one other gene that identified a subgroup with only a 14% overall 2-year survival. This subgroup actually had approximately 40% of the observed deaths occurring within the first 2 years. MYC has a complicated story in aggressive B-cell lymphomas. In studies of DLBCLs, the presence of dual translocation of both BCL2, such as in t(14;18) as well as MYC as in t(8;14), have been identified as having poor outcome.30 It is possible that dual translocations, alternative MYC translocations, amplifications, or mutations could all result in increased gene expression levels in these cases. Some of these cases could be DLBCLs that harbor a MYC translocation as a primary pathogenetic event, for which the impact on outcome is unknown. This is an excellent topic for future study.
The best selected 2-variable model predicting overall survival in DLBCL was the combination of MYC and HLA-DRB. It is striking that MYC-based proliferation and MHC antigen expression are important in R-CHOP–treated DLBCLs and are also the defining attributes of Burkitt's lymphoma suggesting that lack of immunosurveillance (through loss of either major histocompatibility class I or class II expression) and high proliferation are the key features defining the most aggressive B-cell lymphomas.1,31,32 Down-regulation of immune response has been identified as a key feature of Burkitt lymphoma and poor-outcome DLBCL previously.24,28,29,31,33-36 Highly proliferative DLBCL has also been identified as a key feature of poor-outcome DLBCL.1,4,37 This tentative 2-gene model therefore is biologically plausible based on previous literature. Although these genes were identified using the median cutoff, the variable cutoff point analysis went on to demonstrate that those patients in either the lowest 20% of MHC class II expression or highest 20% of MYC expression have the poorest outcome. In addition, we note that the use of both high-risk IPI and the 2 genes identifies a subpopulation of 14 patients in which only 2 survive and 12 die during the first 2 years, yielding a 2-year overall survival rate of 14%. The remaining patients, with neither of these factors, appear to represent the curable population of DLBCL, whereas those with one or more of these factors should be the focus of future therapeutic development.
More broadly, the ability to use unstained cuts from fixed paraffin-embedded tissue blocks for multigene quantification has the potential to change the practice of pathology. In particular, the ability to interrogate routinely handled tissues, using minimal amounts of material, without RNA extraction, allows application of newly discovered gene expression biomarkers to patients uniformly treated on clinical trials (because fixed tissue is usually the only type of tissue easily available in such a context). Thus, this portable technology solves the major practical matter of obtaining and shipping snap-frozen tissue biopsies for analysis, allowing for the necessary validation studies required for biomarker application to clinical practice.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the entire Leukemia and Lymphoma Molecular Profiling Project research group for producing the large well-annotated datasets that provided the data on the original group of CHOP-treated patients.
R.D.G. and J.M.C. are supported by the NCIC Terry Fox Program Project (grant 016003). L.R.'s laboratory is supported by the American Cancer Society (grant RSG0605501L1B).
Authorship
Contribution: L.M.R. designed research, contributed cases, analyzed data, and wrote the paper; M.L.L. designed research, analyzed data, and wrote the paper; J.M.U. analyzed data and wrote the paper; T.P.M. and D.O.P. designed research and edited the paper; T.M.G. designed research and wrote parts of the paper; C.M.S. performed the research and analyzed data; R.R.M. contributed new analytical tools and edited the paper; B.S. supported new analytical tool development; R.M.B., E.C., A.R., L.H.S., J.M.C., and N.J. submitted case materials and reviewed the paper; and R.D.G. submitted cases, designed the research, and reviewed the paper.
Conflict-of-interest disclosure: R.R.M., C.M.S., and B.S. are employees of High Throughput Genomics. R.R.M. and B.S. hold stock and stock options in High Throughput Genomics. C.M.S. holds stock options in High Throughput Genomics. The remaining authors declare no competing financial interests.
Correspondence: Lisa M. Rimsza, Department of Pathology, University of Arizona, 1501 N. Campbell Avenue, PO Box 245043, Tucson, AZ 85724-5043; e-mail: lrimsza@email.arizona.edu.