Previous research has shown that glycolytic enzymes (GEs) exist as multienzyme complexes on the inner surface of human erythrocyte membranes. Because GE binding sites have been mapped to sequences on the membrane protein, band 3, that are not conserved in other mammalian homologs, the question arose whether GEs can organize into complexes on other mammalian erythrocyte membranes. To address this, murine erythrocytes were stained with antibodies to glyceraldehyde-3-phosphate dehydrogenase, aldolase, phosphofructokinase, lactate dehydrogenase, and pyruvate kinase and analyzed by confocal microscopy. GEs were found to localize to the membrane in oxygenated erythrocytes but redistributed to the cytoplasm upon deoxygenation, as seen in human erythrocytes. To identify membrane proteins involved in GE assembly, erythrocytes from mice lacking each of the major erythrocyte membrane proteins were examined for GE localization. GEs from band 3 knockout mice were not membrane associated but distributed throughout the cytoplasm, regardless of erythrocyte oxygenation state. In contrast, erythrocytes from mice lacking α-spectrin, ankyrin, protein 4.2, protein 4.1, β-adducin, or dematin headpiece exhibited GEs bound to the membrane. These data suggest that oxygenation-dependent assembly of GEs on the membrane could be a general phenomenon of mammalian erythrocytes and that stability of these interactions depends primarily on band 3.
Introduction
The glycolytic enzymes (GEs), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), aldolase, phosphofructokinase (PFK), lactate dehydrogenase (LDH), and pyruvate kinase (PK), have been shown to organize into multienzyme complexes on the inner surface of the human erythrocyte membrane.1 Because assembly of these complexes is sensitive to physiologic stimuli, such as band 3 phosphorylation and hemoglobin (Hb) deoxygenation, it has been suggested that regulation of GE assembly may serve an important biologic function, perhaps in regulation of glucose metabolism. Indeed, association of the complexes on the membrane has been observed to correlate with a shift in glucose metabolism from glycolysis to the pentose phosphate pathway.2,3
The specific docking sites of GEs on band 3 have been mapped to tandem similar sequences (6-DDYED-10 and 19-EEYED-23) near the NH2-terminus of the polypeptide.4,–6 However, these NH2-terminal sequences are not well conserved in other mammalian species, including the mouse (Figure 1), raising the question of whether GE assembly into complexes is a feature of all mammalian red cells or a unique characteristic of human erythrocytes. Preliminary evidence in favor of GE binding to nonhuman erythrocyte membranes has come from staining data of Ercolani et al7 and Weber et al8 who show that antibodies to GAPDH label the membrane in both rat and mouse erythrocytes, respectively. However, no membrane staining of any other GEs has ever been reported in nonhuman erythrocytes, and the tyrosines with flanking acidic residues (Y8 and Y21) that are phosphorylated in human band 3 and regulate GE association with the membrane are absent in murine band 3.
Alignment of the human, dog, mouse, rat, and cow NH2-terminus of band 3. Alignment of the first 65 amino acids of human, dog, mouse, rat, and cow AE1 using the CLUSTALW program (http://align.genome.jp//). Asterisks (*) indicate complete sequence conservation; double and single dots, less conservation; (^) symbols, the 2 tyrosine phosphorylation sites at the NH2-terminus of human band 3.
Alignment of the human, dog, mouse, rat, and cow NH2-terminus of band 3. Alignment of the first 65 amino acids of human, dog, mouse, rat, and cow AE1 using the CLUSTALW program (http://align.genome.jp//). Asterisks (*) indicate complete sequence conservation; double and single dots, less conservation; (^) symbols, the 2 tyrosine phosphorylation sites at the NH2-terminus of human band 3.
To determine the generality of GE assembly on erythrocyte membranes, we undertook to characterize the membrane binding sites, if any, of GAPDH, aldolase, PFK, LDH, and PK on murine erythrocyte membranes. In the present study, we demonstrate that (1) GEs are indeed localized to the membrane in oxygenated murine erythrocytes, (2) GEs release from the membrane upon deoxygenation of the mouse red cells, (3) GEs are cytosolic in both oxygenated and deoxygenated erythrocytes from band 3 knockout mice,9 (4) GEs are displaced from the membrane by antibodies to the cytoplasmic domain of band 3 and by recombinant cytoplasmic domain of band 3 itself, and (5) GEs are largely membrane localized in mature oxygenated cells from mice deficient in protein 4.2,10 β-adducin,11 protein 4.1,12 dematin headpiece domain,13 ankyrin (nb/nb),14 and α-spectrin (sph/sph).15 These data suggest that GE associations with erythrocyte membranes could represent a widespread phenomenon and that the stability of these interactions depends primarily on the presence of band 3.
Methods
Reagents
Poly-L-lysine and cold fish skin gelatin solutions were acquired from Sigma-Aldrich (St Louis, MO). Acrolein was purchased from Sigma-Aldrich. Goat anti–rabbit muscle aldolase, PFK, LDH, and PK were from Polysciences (Warrington, PA). Polyclonal anti-GAPDH and anticytoplasmic domain of band 3 (anti-cdb3) were generated in our own laboratory by immunization of rabbits against the purified human proteins according to standard procedures. Secondary antibodies Rhodamine Red X and Cy2-labeled anti–rabbit and Rhodamine Red X, Cy2 and Cy5 antigoat IgG were from Jackson ImmunoResearch Laboratories (West Grove, PA). Syto RNASelect green fluorescent cell stain was from Invitrogen (Carlsbad, CA) and used as indicated for fixed cells by the manufacturer. The cytoplasmic domain of murine band 3 (cdb3, residues 1-398) was a recombinant protein expressed and purified in our laboratory and is described in “Preparation of murine cdb3.”
Preparation of murine cdb3
A sense strand primer (5′-TACATATGGGGGACATGCGGGACCAC-3′) and an antisense primer (5′-AAGCTTTCAAAAGATCCGGCCTGTGCG-3′) were used both to amplify cDNA encoding murine cdb3 (residues 1-398) from murine full-length band 3 cDNA (Invitrogen) and to introduce a NdeI site and a HindIII restriction enzyme site. Using these restriction sites, murine cdb3 cDNA was inserted into a pT7-7 expression vector and protein was expressed in BL21(DE3) pLys S cells (Invitrogen). Recombinant cdb3 was then purified by the procedure used to purify recombinant human cdb3.6
Preparation of cells
Mouse blood was collected either by cardiac puncture or retro-orbital sinus bleeding into heparinized tubes, shipped overnight on ice to Purdue University, and processed immediately on arrival (no more than 24 hours from the time of blood withdrawal). Samples were always accompanied by a travel control to assure that any possible stresses associated with shipping did not alter the results. Erythrocytes were washed twice in 330 mOsm phosphate-buffered saline (PBS), pH 7.4, containing 5 mM glucose. Processing for indirect immunofluorescence and evaluation of the effects of red blood cell deoxygenation were performed as previously described.1 Deoxygenation with argon was achieved by gently passing humidified argon across the erythrocyte suspension for 60 minutes. This deoxygenation caused the anticipated color change in the erythrocyte suspension; but because the argon was humidified, no change in suspension volume or hemolysis occurred. Oxygenation was achieved by simply washing the cells in oxygenated buffers.
Deoxygenation with sodium dithionite was accomplished by evacuating 3 mL PBS-glucose solution in a 25-mL vacuum flask for 10 minutes and then flowing humidified argon across the solution for 10 minutes; 10 mM dry sodium dithionite was then added to the deoxygenated solution, followed by addition of washed red blood cells to a final hematocrit of 10%. Finally, argon was again gently blown across the sealed suspension for 3 minutes, and paraformaldehyde fixation was performed as previously described.1 Fixation of the deoxygenated cells before exposure to air was required to prevent rebinding of the enzymes to the membrane, which is a rapidly reversible process. Paraformaldehyde was used for initial fixation because acrolein cannot fix cells in the absence of O2. This study received Institutional Animal Care and Use Committee approval for the use of mice.
Confocal microscopy
Mouse erythrocytes were fixed, permeabilized, and stained as previously described.1 Cells were subsequently attached to poly-L lysine-coated coverslips (no. 1.5 VWR) in humidified chambers for 15 to 20 minutes. Before observation, cells were mounted on microscope slides using M800 Aqua Mount (Fisher Scientific, Pittsburgh, PA) and sealed with nail polish. Images were acquired using an MRC-1024 UV confocal system (Bio-Rad, Hemel Hempstead, United Kingdom) on a Diaphot 300 inverted microscope (Nikon, Tokyo, Japan) using a 60×/1.4 NA oil-immersion lens. The acquisition software used was Lasersharp 2000 version 5.0 (Bio-Rad). Images were converted from Bio-Rad's pic format to .tif format using ThumbsPlus version 6.0 (Cerious Software, Charlotte, NC) and assembled into figures using Photoshop version 7.0 (Adobe Systems, San Jose, CA).
Results
Glycolytic enzymes are membrane localized in unperturbed oxygenated mouse erythrocytes
We have demonstrated a reversible association of GEs with the cytoplasmic domain of band 3 (cdb3) on the human erythrocyte membrane, and we have shown that this association is regulated by erythrocyte oxygenation and band 3 phosphorylation.1 The sequences of human band 3 shown to be involved in this interaction,6 however, are not highly conserved in the mouse or in other mammalian species (Figure 1), raising the possibility that membrane association of GEs might be a late evolutionary development. To explore whether GEs from murine erythrocytes are associated with the membrane, we fixed, permeabilized, and stained fresh murine erythrocytes with antibodies to GAPDH, aldolase, PFK, LDH, and PK, and evaluated the GE distributions in stained whole cells. As seen in Figure 2, all 5 GEs were found highly localized to the membrane in oxygenated murine erythrocytes.
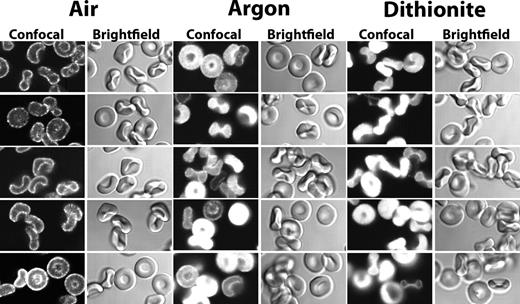
GE staining in oxygenated and in deoxygenated mouse erythrocytes. Confocal images of mouse erythrocytes after their staining for glycolytic enzymes under oxygenated (Air), deoxygenated with humidified argon (Argon), or deoxygenated with 10 mM of sodium dithionite (Dithionite) followed by humidified argon (“Preparation of cells”).
GE staining in oxygenated and in deoxygenated mouse erythrocytes. Confocal images of mouse erythrocytes after their staining for glycolytic enzymes under oxygenated (Air), deoxygenated with humidified argon (Argon), or deoxygenated with 10 mM of sodium dithionite (Dithionite) followed by humidified argon (“Preparation of cells”).
Enzyme association is largely reversed on erythrocyte deoxygenation
Previous studies have demonstrated that deoxygenated hemoglobin and GEs compete for overlapping binding sites at the NH2-terminus of band 3 in human erythrocytes, resulting in GE displacement from the membrane during red cell deoxygenation.16,17 To determine whether murine GEs are similarly responsive to Hb deoxygenation, fresh murine erythrocytes were depleted of O2 before fixation and staining and examined by confocal microscopy. As shown in Figure 2, regardless of the method of deoxygenation, GEs translocate from the membrane to the cytoplasm on oxygen removal. It should be noted that, although the rate and mechanism of oxygen depletion are different for the dithionite and for the argon methods (dithionite consumes O2 by reacting with it, whereas argon depletes O2 from the atmosphere above the cell suspension, allowing the reversible dissociation of O2 from the cells in suspension18 ), neither method was seen to induce cell shape change or cell lysis (see bright field images of cells), suggesting that the GE translocations are not the result of damage to the erythrocyte membrane. Taken together, these findings imply that oxygenation-dependent assembly of GEs on the erythrocyte membrane occurs in mice as well as humans, despite significant divergence in their band 3 sequences.
Band 3 is the center of enzyme organization on the erythrocyte membrane
Although GAPDH, aldolase, PFK, LDH, and PK all translocate to the cytoplasm on erythrocyte deoxygenation, only GAPDH, aldolase, and PFK have been shown to bind human band 3.6 Indeed, in a previous study, we were unable to document a direct interaction between band 3 and either PK or LDH.6 Because band 3 constitutes the only known binding site for deoxyHb on the membrane, displacement of PK and LDH during red cell deoxygenation was hypothesized to derive from their binding to proteins closely associated with band 3.1 However, the proximal membrane docking sites of LDH and PK have yet to be identified.
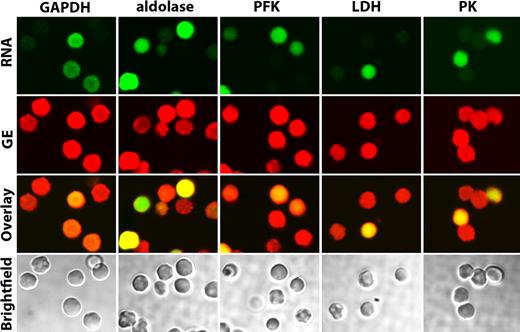
The availability of mice deficient in each of the major erythrocyte membrane proteins together with the knowledge that murine erythrocyte membranes bind GEs similar to human membranes now enables us to try to identify these putative proximal membrane binding sites. For this purpose, erythrocytes from band 3 knockout mice9 were gently washed in oxygenated PBS and fixed with acrolein as described in “Preparation of cells.” After permeabilization, the fixed cells were stained as described in “Confocal microscopy” and examined by confocal microscopy for both GE distribution and the presence of RNA (indicative of a reticulocyte). As seen in Figure 3, none of the enzymes tested was found to localize on the membrane of band 3 knockout mice. Although the majority of cells were admittedly reticulocytes (Syto RNA Green stained panel in Figure 3), even the few mature red cells that lacked RNA (lack of green staining) displayed no GE enrichment on the membrane. These data therefore suggest that band 3 is essential for proper assembly of GEs on the mature erythrocyte membrane.
Localization of GEs in oxygenated band 3 knockout erythrocytes. Confocal images of band 3 null erythrocytes stained for both RNA (to identify reticulocytes, green stain) and GEs (red stain), as indicated. A merger of both stains is shown in the third row (overlay), and bright field images are displayed in the bottom row. Note that even the very few mature erythrocytes present (negative for SytoRNA stain, green color) do not show enzyme localization at the membrane.
Localization of GEs in oxygenated band 3 knockout erythrocytes. Confocal images of band 3 null erythrocytes stained for both RNA (to identify reticulocytes, green stain) and GEs (red stain), as indicated. A merger of both stains is shown in the third row (overlay), and bright field images are displayed in the bottom row. Note that even the very few mature erythrocytes present (negative for SytoRNA stain, green color) do not show enzyme localization at the membrane.
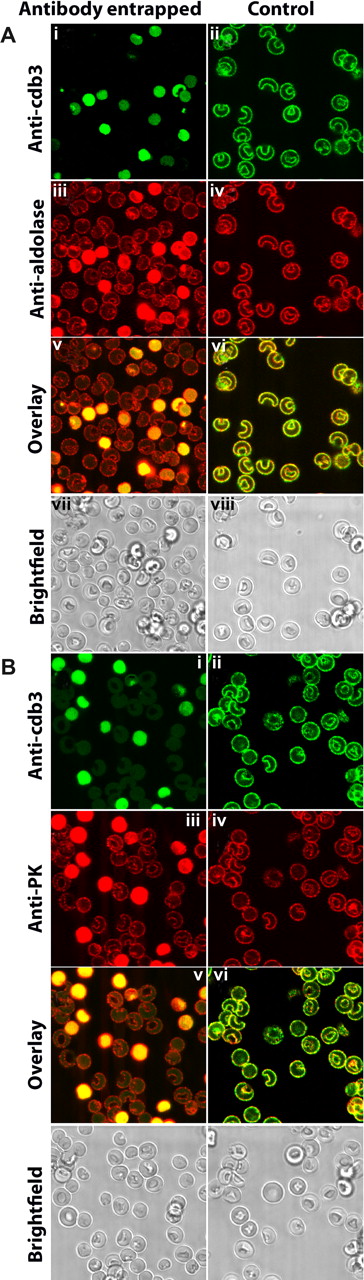
To explore this hypothesis in greater detail, murine erythrocytes were resealed with an antibody to cdb3 and examined for displacement of GE from the membrane. Importantly, erythrocytes that were found to stain positive for entrapment of anti-cdb3 also revealed displacement of GE from the membrane. This is shown clearly in Figure 4 where red cells that contain anti-cdb3 antibody in their cytosols (left panel, green staining in cytoplasm) are also seen to display strong anti-aldolase and anti-pyruvate kinase staining throughout their cytoplasms (left panel, red staining), suggesting that anti-cdb3 can block binding of GE to the membrane.
Localization of glycolytic enzymes in murine erythrocytes resealed in the presence or absence of an antibody to the cytoplasmic domain of band 3. Freshly drawn murine erythrocytes were washed 3 times to remove serum and buffy coat, and divided into 2 aliquots. One suspension of erythrocytes was lysed and resealed in the presence of rabbit polyclonal antibody raised against cdb3.1 The other suspension (control) were similarly lysed and resealed in the absence of any antibody. After resealing, both erythrocyte preparations were similarly fixed with acrolein, permeabilized with Triton X-100 (“Preparation of cells”), and stained with the desired antibodies. (i) Antirabbit antibody conjugated to Cy2 (492 510) was used to identify cells containing entrapped anti-cdb3 antibody. (ii) Cells (lacking entrapped antibody) were first stained with the same rabbit anti-cdb3 antibody and then labeled with Cy2-conjugated antirabbit IgG. Membrane staining in this panel demonstrates the specificity of the antibody for band 3. (iii,iv) Stained first with goat antibodies specific for either aldolase (A) or pyruvate kinase (B) and then with antigoat antibody conjugated to Cy5 (650 670). (v,vi) Overlays of subpanels i and iii, and ii and iv, respectively. (vii,viii) Bright field images of subpanels i, iii, v, and ii, iv, vi, respectively.
Localization of glycolytic enzymes in murine erythrocytes resealed in the presence or absence of an antibody to the cytoplasmic domain of band 3. Freshly drawn murine erythrocytes were washed 3 times to remove serum and buffy coat, and divided into 2 aliquots. One suspension of erythrocytes was lysed and resealed in the presence of rabbit polyclonal antibody raised against cdb3.1 The other suspension (control) were similarly lysed and resealed in the absence of any antibody. After resealing, both erythrocyte preparations were similarly fixed with acrolein, permeabilized with Triton X-100 (“Preparation of cells”), and stained with the desired antibodies. (i) Antirabbit antibody conjugated to Cy2 (492 510) was used to identify cells containing entrapped anti-cdb3 antibody. (ii) Cells (lacking entrapped antibody) were first stained with the same rabbit anti-cdb3 antibody and then labeled with Cy2-conjugated antirabbit IgG. Membrane staining in this panel demonstrates the specificity of the antibody for band 3. (iii,iv) Stained first with goat antibodies specific for either aldolase (A) or pyruvate kinase (B) and then with antigoat antibody conjugated to Cy5 (650 670). (v,vi) Overlays of subpanels i and iii, and ii and iv, respectively. (vii,viii) Bright field images of subpanels i, iii, v, and ii, iv, vi, respectively.
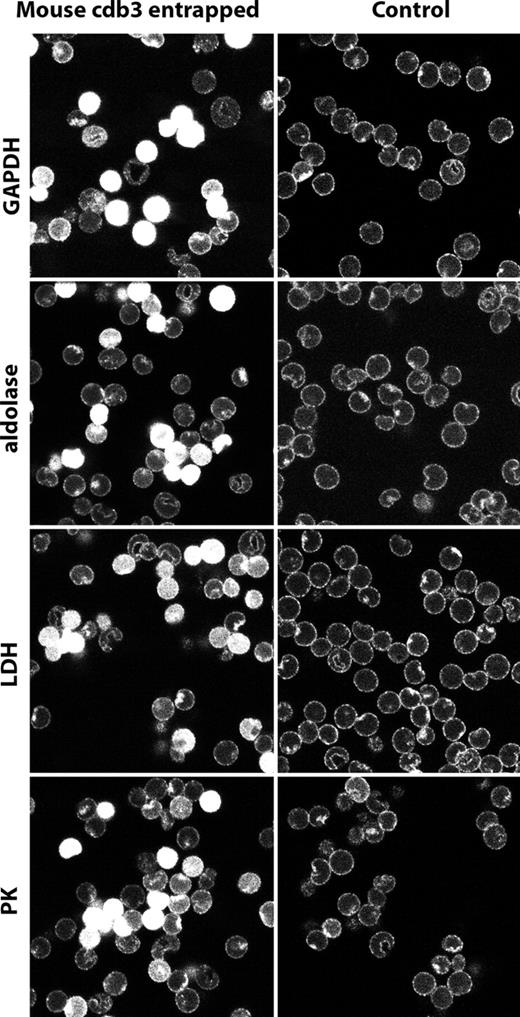
To corroborate this result further, fresh murine erythrocytes were resealed with a cloned cytoplasmic domain of murine band 3 (residues 1-398), and after fixation and permeabilization, similarly stained with antibodies to GAPDH, aldolase, LDH, and PK. As shown in Figure 5, cells that had entrapped the cloned mouse cdb3 display GEs distributed uniformly throughout their cytoplasms, indicating that free cdb3 can compete with endogenous cdb3 for assembling GEs. Finally, the same cloned mouse wild-type cdb3 was tested for its in vitro capacity to inhibit GAPDH activity, an assay commonly used to evaluate its binding affinity for cdb3.5,6 As shown in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article), cdb3 inhibits the activity of the enzyme by approximately 70%. Taken together, we conclude that band 3 provides an important site for organization of GEs on murine red cells, just like it does on human red cells.1,4,5,6
Localization of glycolytic enzymes in murine erythrocytes resealed in the presence of the cytoplasmic domain of murine band 3 (residues 1-398). Murine wild-type cdb3 was dialyzed against 5 mM of potassium phosphate containing 160 mM of sodium chloride, pH 7.4, and concentrated to 6.8 mg/mL using a centricon-30 (Millipore, Billerica, MA). Freshly drawn mouse erythrocytes were washed in the above buffer and resuspended at 50% hematocrit in the same buffer containing mouse cdb3. The suspension was introduced into mini dialysis units, and lysed and resealed as described by Campanella et al.1 After resealing, erythrocytes were fixed, permeabilized, and stained for GAPDH, aldolase, LDH, and PK and visualized with secondary antibody conjugated with Cy2 (492 510). Control erythrocytes were lysed and resealed in the absence of cdb3, but otherwise treated identically. Glycolytic enzymes are displaced from the membrane in cells containing resealed murine cdb3.
Localization of glycolytic enzymes in murine erythrocytes resealed in the presence of the cytoplasmic domain of murine band 3 (residues 1-398). Murine wild-type cdb3 was dialyzed against 5 mM of potassium phosphate containing 160 mM of sodium chloride, pH 7.4, and concentrated to 6.8 mg/mL using a centricon-30 (Millipore, Billerica, MA). Freshly drawn mouse erythrocytes were washed in the above buffer and resuspended at 50% hematocrit in the same buffer containing mouse cdb3. The suspension was introduced into mini dialysis units, and lysed and resealed as described by Campanella et al.1 After resealing, erythrocytes were fixed, permeabilized, and stained for GAPDH, aldolase, LDH, and PK and visualized with secondary antibody conjugated with Cy2 (492 510). Control erythrocytes were lysed and resealed in the absence of cdb3, but otherwise treated identically. Glycolytic enzymes are displaced from the membrane in cells containing resealed murine cdb3.
Requirement of other membrane proteins for assembly of GEs on the membrane.
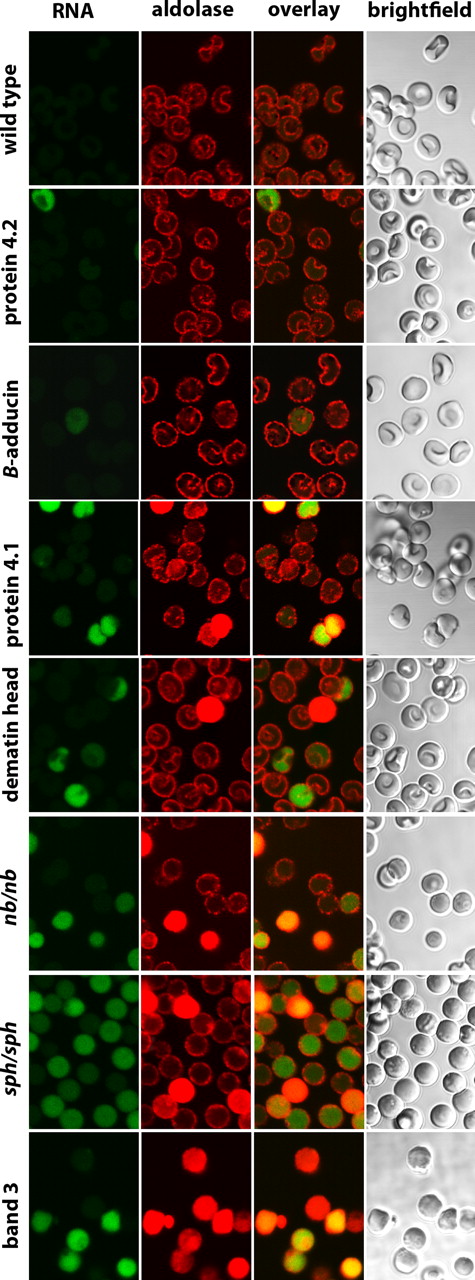
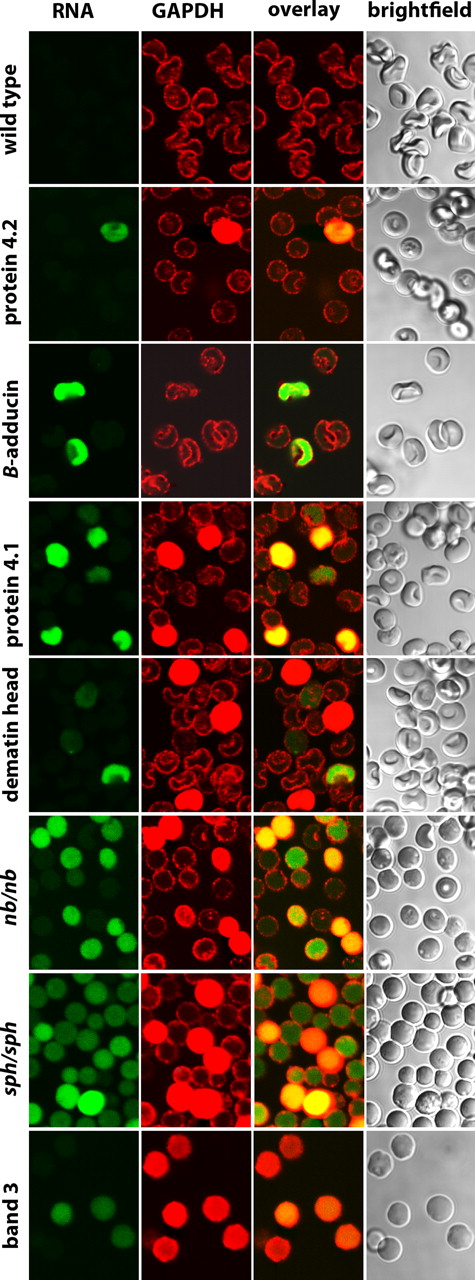
To determine whether other membrane proteins might be similarly essential for GE organization on the membrane, erythrocytes from knockout mice for protein 4.2,10 β-adducin,11 protein 4.1,12 and dematin,13 as well as red cells from mice containing spontaneous hypomorphic mutations leading to nearly quantitative deficiencies in ankyrin (nb/nb mice)14 and α-spectrin (sph/sph mice)15 were also examined. In contrast to membranes lacking band 3, GEs in these other defective erythrocytes still exhibit various extents of membrane localization, suggesting that the essential components of their membrane binding sites remain intact (Figures 6, 7, S2). Nevertheless, membrane localization is neither quantitative nor equivalent among all of the deficient membranes. Thus, membranes lacking protein 4.2, β-adducin, and protein 4.1 show little to moderate perturbation of GE binding, whereas membranes lacking ankyrin, dematin, and α-spectrin exhibit a moderate to substantial fraction of cells with aberrant GE distributions (Figures 6, 7, S2). Importantly, most cells (but clearly not all cells) with cytosolic GEs also stain green with Syto RNA stain, suggesting many of these cells are reticulocytes rather than mature erythrocytes. Because reticulocytes still run the Krebs cycle and maintain much higher rates of glycolysis than mature erythrocytes,19 a different organization of GEs may be optimal.
Distribution of aldolase in erythrocytes from mice deficient in major erythrocyte membrane proteins. Intact erythrocytes from wild-type mice and mice deficient in protein 4.2, β-adducin, protein 4.1, dematin headpiece domain, ankyrin (nb/nb), α-spectrin (sph/sph), and band 3 were fixed, permeabilized, and stained for aldolase and RNA, as described in “Methods.”
Distribution of aldolase in erythrocytes from mice deficient in major erythrocyte membrane proteins. Intact erythrocytes from wild-type mice and mice deficient in protein 4.2, β-adducin, protein 4.1, dematin headpiece domain, ankyrin (nb/nb), α-spectrin (sph/sph), and band 3 were fixed, permeabilized, and stained for aldolase and RNA, as described in “Methods.”
Distribution of GAPDH in erythrocytes from mice deficient in major erythrocyte membrane proteins. All conditions are identical to those in Figure 6, except that anti-GAPDH was used to stain the cells rather than antialdolase.
Distribution of GAPDH in erythrocytes from mice deficient in major erythrocyte membrane proteins. All conditions are identical to those in Figure 6, except that anti-GAPDH was used to stain the cells rather than antialdolase.
Finally, it should not go unnoticed that the staining intensities of most of the free cytosolic GEs are invariably much greater than the staining intensities of their membrane-bound counterparts. Indeed, in most cases, the fluorescence intensities of displaced enzymes were found to saturate the detector under the same conditions where the fluorescent signal from the membrane-bound enzymes was minimal. This differential in fluorescence intensity suggests that the epitopes recognized by anti-GE antibodies are largely occluded on membrane-bound GEs, but fully accessible on displacement of the GEs into the cytoplasm. One possible hypothesis to explain this consistent observation is that GEs are so tightly associated into complexes on the membrane that accessibility of their epitopes is limited.
Discussion
Nucleated cells have highly compartmentalized interiors with enzyme pathways that are largely confined to organelles and cytosolic structures, such as mitochondria, microtubules, and microfilaments. The mammalian erythrocyte, in contrast, has none of these cytosolic components, raising the question of whether organized metabolic pathways can be assembled in mammalian erythrocytes. Our recent observation that GEs are membrane bound in human erythrocytes1 may partially address this question; however, the weak homology between GE binding sites on human and murine homologs of band 3 has since raised concern that GE organization might be solely a late evolutionary development. We have now demonstrated that GEs are also membrane bound in oxygenated, healthy, murine erythrocytes and that these associations are disassembled during normal deoxygenation in a manner similar to that seen in human erythrocytes. The observation that enzyme epitopes are largely inaccessible in membrane-bound GEs further suggests that interactions among GEs and other membrane components are tight, at least to the extent that they exclude antibodies. Indeed, data by Hoffman and others20,21 demonstrate that at least one type of GE complex on the erythrocyte membrane can sequester ATP and channel it directly to the membrane's Na+/K+-ATPase. The GE complexes characterized by Hoffman are also sufficiently tightly assembled that they can be shown to concentrate fluorescent ATP analogs on the membrane.22 Whether the glycolytic compartments characterized by these workers23,24 are similar to those observed in our study cannot be determined from the data.
Although many previous studies have concluded that band 3 constitutes a binding site for GEs on the membrane,4,5 no experiments have evaluated the possible participation of other membrane proteins in organization of GE assemblies. Using hypomorphic mutants and knockout mice, we have demonstrated that only the absence of band 3 leads to quantitative displacement of GEs from the membrane. Whereas at least half of all erythrocytes in the sph/sph α-spectrin-deficient mice displayed cytosolic GEs, the relatively few cells with little or no mRNA showed membrane binding of their GEs. Although other membrane proteins may still participate in membrane localization of GEs, these data argue that band 3 is the only membrane protein that is completely essential for GE assembly.
Finally, recent observations suggest that band 3 may exist in 2 distinct populations on the erythrocyte membrane. One population, associated with ankyrin, links β-spectrin to the membrane near the spectrin dimer↔tetramer association site.25 The other population, attached to adducin, anchors the junctional complex to the membrane.26 To fully understand the architecture of the erythrocyte membrane, it will be important to determine whether one or both of these band 3 populations can organize GEs on the membrane.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Athar Chishti for the generous gift of mouse blood from (dematin head domain and band 3 null).
This work was supported by National Institutes of Health grants GM24417-29 (P.S.L.) and HL075714 (L.L.P.).
National Institutes of Health
Authorship
Contribution: M.E.C. performed experiments, analyzed data, and wrote the paper; H.C. performed experiments and helped analyze data; N.J.W., L.L.P., D.M.G, and N.M. provided mouse blood and edited the manuscript; and P.S.L. designed studies, analyzed data, and helped write the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Philip S. Low, Department of Chemistry, Purdue University, 560 Oval Drive, West Lafayette, IN 47907; e-mail: plow@purdue.edu.