Abstract
Mantle cell lymphoma (MCL) is genetically characterized by the t(11;14)(q13;q32) translocation and a high number of secondary chromosomal alterations. However, only a limited number of target genes have been identified. We have studied 10 MCL cell lines and 28 primary tumors with a combination of a high-density single-nucleotide polymorphism array and gene expression profiling. We detected highly altered genomes in the majority of the samples with a high number of partial uniparental disomies (UPDs). The UPD at 17p was one of the most common, and it was associated with TP53 gene inactivation. Homozygous deletions targeted 4 known tumor suppressor genes (CDKN2C, BCL2L11, CDKN2A, and RB1) and 6 new genes (FAF1, MAP2, SP100, MOBKL2B, ZNF280A, and PRAME). Gene amplification coupled with overexpression was identified in 35 different regions. The most recurrent amplified regions were 11q13.3-q13.5, 13q31.3, and 18q21.33, which targeted CCND1, C13orf25, and BCL2, respectively. Interestingly, the breakpoints flanking all the genomic alterations, including UPDs, were significantly associated with genomic regions enriched in copy number variants and segmental duplications, suggesting that the recombination at these regions may play a role in the genomic instability of MCL. This integrative genomic analysis has revealed target genes that may be potentially relevant in MCL pathogenesis.
Introduction
Mantle-cell lymphoma (MCL) is an aggressive B-cell neoplasm genetically characterized by the t(11;14)(q13;q32) translocation and cyclin D1 overexpression. Experimental studies have shown that cyclin D1 can function as an oncogene, but its tumorigenic and transforming properties require the cooperation of other mechanisms.1-3 The need for additional oncogenic events in the progression of MCL has also been supported by clinical observations. In particular, low numbers of cells carrying the t(11;14) translocation have been found in the blood of 1% to 2% of healthy people.4 On the other hand, the genomic complexity of MCL and certain secondary chromosomal aberrations are associated with the prognosis of the patients, suggesting that these additional genetic events influence the behavior of this tumor.5-8
Molecular studies using comparative genomic hybridization (CGH),5,6,9,10 multicolor fluorescent in situ hybridization (M-FISH),11 bacterial artificial chromosome (BAC) arrays,7,8,12-14 and single-nucleotide polymorphism (SNP) arrays15 have revealed a large number of chromosomal alterations in MCL. Recurrent aberrations include losses, gains, and amplifications of certain chromosomal regions that may contain crucial genes implicated in relevant tumoral pathways. Molecular studies have identified a number of these genes; and notably, most of them are involved in cell-cycle regulation and DNA damage response pathways.3,16,17 The recent identification of a homozygous deletion of the proapoptotic gene BCL2L11 in MCL cell lines suggests that elements of the cell-survival pathways may be also genetically altered in these lymphomas.13,18 Nevertheless, the targets of most recurrent chromosomal alterations are still unknown. In this sense, BAC and 10K-SNP array studies have narrowed down some of the minimal regions involved in chromosomal alterations.7,8,12-15 However, the potential relevant genes remain elusive, probably resulting from low resolution of some areas or the absence of information on the expression levels of the genes included in these regions.
MCL is one of the lymphoid neoplasms with the highest levels of genomic instability,19 but the mechanisms involved in this phenomenon are not well understood. A new class of genetic variation in the human genome, named structural variation, has recently been recognized and composes approximately 12% of the human genome sequence.20 Structural variations are presented mainly as copy number variants (CNVs)20-22 and segmental duplications (SDs).23 SDs are highly homologous DNA duplicated sequences that occur at more than one site of the genome and define hotspots of chromosomal instability by predisposing these regions to rearrangements by nonallelic homologous recombination. Interestingly, SDs are frequently found at the breakpoints of disease-associated rearrangements.24 CNVs have been recently identified in healthy populations20-22 and consist of deletions and duplications that contribute to genomic variability and potentially to disease susceptibility. Of note, CNVs frequently overlap with coding genes and SDs,21,25 suggesting that these may be inherently unstable genomic regions that can trigger genomic rearrangements.24,26,27 The potential mediation and/or stimulation of chromosomal alterations in tumor samples driven by structural variants is not well known, and their possible relationship in lymphoid neoplasms has not been previously assessed.
In this study, we have performed a comprehensive high-resolution integrative analysis of the recurrent chromosomal alterations by high-density SNP array and mRNA expression profiling of a series of MCL cell lines and primary tumors to identify new genetic alterations and potential target genes that may be relevant in the pathogenesis of these tumors. The relatively precise mapping of the breakpoints of the chromosomal alterations has allowed us the identification of their possible association with structural variations in the human genome and the potential role of CNVs and SDs in the pathogenesis of MCL genomic instability.
Methods
Samples
Ten MCL cell lines (HBL2, UPN1, MINO, REC1, GRANTA519, NCEB1, MAVER1, Z138, JEKO1, and JVM2) and 28 primary MCL were analyzed (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). To assure a high tumor-cell content in the primary tumors, mononuclear cells were isolated from the peripheral blood of 16 leukemic MCL patients by gradient centrifugation, and the tumor cells were purified using anti-CD19 magnetic microbeads (Miltenyi Biotec, Auburn, CA). Tumor-cell purity greater than 98% was obtained in all samples, as determined by flow cytometry. Matched DNA from normal samples was available in 5 patients. In addition, we selected 7 highly leukemic primary MCLs with more than 85% of tumor-cell content and 5 samples from frozen tissues with high tumor content (∼> 85%). According to our dilution experiments (see details in Document S1), a high tumor content of more than 85% minimizes the potential interference of contaminating normal cells in the array analysis. All samples were collected from the Tumor Bank of the Pathology Department, Hospital Clinic (Barcelona, Spain) and the Institute of Pathology (Würzburg, Germany). All cases carried the t(11;14) translocation and/or overexpressed cyclin D1 mRNA or protein. Total RNA and DNA were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). TP53 mutational analysis was performed from exons 4 to 11 as previously described.28 A panel of 24 additional nonpurified primary MCL samples was used for quantitative real-time polymerase chain reaction (qPCR) validation studies. The study was approved by the Hospital Clinic Review Board, and informed consent was obtained in accordance with the Declaration of Helsinki.
SNP arrays and data analysis
The simultaneous genome-wide detection of DNA copy number alterations and loss of heterozygosity (LOH) were investigated using the standard GeneChip Mapping-100K assay protocol (Affymetrix, Santa Clara, CA). Briefly, 2 aliquots of 250 ng of genomic DNA were digested with XbaI and HindIII enzymes, ligated to the adaptors, and amplified by PCR. PCR products were concentrated and fragmented, and then end-labeled with biotin, denatured, and hybridized to the 2 arrays of the GeneChip Mapping-100K array set (Affymetrix). The arrays were processed using the Fluidics Station 450, GeneChip Scanner 3000, and GCOS workstation version 1.4 (Affymetrix). The evaluation was done using GDAS and CNAT V3.0 (Affymetrix). To minimize background noise, a smoothing window of 0.5 Mb was applied. The determination of the working criteria used to define copy number alterations and LOH was based on previously known alterations in diploid cell lines (Document S1). Uniparental disomy (UPD) was defined as copy number neutral LOH and uniparental trisomy (UPT) as LOH in a gained or amplified region. Genomic abnormalities were examined by 2 independent observers.
Expression arrays and integrative analysis
Expression profiling was performed with the HG-U133 Plus 2.0 arrays (Affymetrix), following the Affymetrix standard protocols as previously published.29 Gene expression differential analysis was performed to identify deregulated target genes within altered regions. Cell lines and purified and nonpurified MCL cases were treated separately for these integrative analyses. Probe sets considered “nonpresent” (with detection call “absent” or “marginal”) in more than 90% of samples were excluded. For the determination of target tumor suppressor genes included in the regions of homozygous deletions, we compared the expression levels of probe sets in the different groups of cell lines or primary MCL cases: those with potential homozygous deletions, those with loss, and those with no copy number change or gain/amplification. Only probe sets called “nonpresent” in the group of homozygous deletion cases and called “present” in the remaining samples were considered as potential target genes. To identify putative overexpressed oncogenes in amplified regions, we compared the expression levels of the genes included in the overlapping region in amplified cases and in cases with no copy number changes. Cases with gains, UPD/UPT, or loss of the same chromosomal region were excluded for the comparisons. Only probe sets with at least a 2-fold change were considered overexpressed.
Quantitative real-time PCR
Relative DNA copy number changes and mRNA expression of different genes were determined by qPCR using the ABI Prism 7700 Sequence Detector System (Applied Biosystems, Foster City, CA). One microgram of total RNA was reverse-transcribed with SuperScript First-Strand kit (Invitrogen). TaqMan Gene Expression Assays (Applied Biosystems) were used for DNA and RNA validation, respectively. Detailed sequences of all primers and assays are available on request. The comparative threshold cycle method (2−ΔΔCt) was applied using ALB and GUSB genes as endogenous references for copy number and expression analysis, respectively. Based on previous experiments30 and dilution experiments with cell lines, we established the following cutoffs: less than 0.7 and less than 0.3 for single loss and homozygous deletions, respectively, and less than 0.6 and less than 0.2 for low and absent expression, respectively.
FISH
FISH analysis was performed on cultured cells following the manufacturer's recommendations using the dual-color dual-fusion probe LSI IGH/BCL2, locus-specific LSI CCND1 orange probe, and CEP11 green probe (Vysis-Abott, Downers Grove, IL). The images were captured by an Olympus bx-60 microscope (Olympus, Melville, NY), at 100×. Digital image acquisition, processing, and evaluation were performed using ISIS digital image analysis system version 5.0 (Metasystems, Altlussheim, Germany).
Copy number variation and segmental duplication enrichment in chromosomal breakpoints
To investigate whether breakpoints of MCL alterations represent random events or may reflect a susceptibility of these regions to DNA rearrangement resulting from particular genome architecture (ie, presence of CNVs and SDs), we evaluated the presence of these structural variants in the 100-kb and 50-kb regions flanking each chromosomal breakpoint. In addition, we performed the analysis considering separately the different types of chromosomal alterations (UPD/UPT, homozygous deletion, loss, gain, and amplification). Computational simulations were performed to assess whether the observed number of breakpoints associated with CNVs and SDs could be the result of chance or represented a significant enrichment. Thus, we randomly assigned 1000 times the position of the breakpoints along the chromosomes and calculated the expected number, as well as expected median, mean, maximum, and minimum number of breakpoints containing CNVs or SDs. The P value was calculated as the number of times that the observed number equaled or exceeded the expected value divided by the total number of permutations plus 1 (observed ≥ expected)/(permutations + 1).31 P values less than .05 were considered significant. Analyses were done using R (R Project for Statistical Computing; Vienna University of Economics and Business Administration, Vienna, Austria).
Results
Copy number changes and LOH in MCL cell lines
We studied 10 MCL cell lines with the GeneChip Mapping-100K SNP array. The SNP call rate in all experiments was more than 94.2%. All cell lines showed copy number alterations, with a total of 179 losses, 133 gains, and 25 high-level DNA amplifications (Table S1). The altered regions observed in at least 4 cell lines were gains of 8q24.21, 11q13.3-q14.2, 12q13.3-q14.1, and 18q21.31-q22.1, and losses of 6q22.32-q24.3, 9pter-p21.3, and 17pter-p13 regions. Some of the target genes included in these regions were MYC, CCND1, CDK4/MDM2, BCL2, CDKN2A, and TP53. The genotyping platform used in this study allowed the sensitive detection of LOH associated with no DNA copy number alteration or even with a copy number increase that correspond to the so-called UPD and UPT, respectively. Each MCL cell line showed at least one whole chromosome or partial UPD/UPT. Overall, 69 regions of UPD/UPT were identified with a mean of 6.9 per cell line (range, 1-26; Figure 1A; Table S1). HBL2 had an extremely large number of UPD/UPT regions (19 UPDs and 7 UPTs) present in almost all chromosomes. The minimal regions of overlapping UPD/UPT in at least 4 cell lines were 2q31.1-q32.1, 2q37.1-qter, and 9q (Figure 1B) and in 3 cell lines, 2q11.2-q12.2, 2q31.1-q32.1, and 16q23.1-q24.1.
Uniparental disomy and uniparental trisomy in MCL. (A) Ideogram of the distribution of the UPD/UPT regions detected by SNP arrays. UPD/UPTs are represented on the left side of each chromosome; thin bars represent UPD regions, whereas thick bars represent gained/amplified regions with LOH (UPT). UPD/UPTs detected in the 10 cell lines are indicated in green, whereas UPD/UPTs detected in the primary MCL samples are indicated in blue. Acquired UPDs detected only in tumor cells were indicated in orange. The more recurrent overlapping regions were shaded in gray. (B) Chromosome 9 total/partial UPDs and homozygous deletions detected in MCL cell lines. Chromosome 9 profile is represented from pter (left) to qter (right). For each cell line, the top panel represents copy number alteration (genome smooth analysis [GSA], P value) and the bottom panel represents LOH (−log10 LOH P value). Regions with genomic losses and concomitant LOH are underlined with thick red bars, homozygous deletions (HD) are indicated with dark red squares, and regions with UPD are underlined with thick black bars. MINO showed a whole chromosome 9 UPD, whereas the 3 remaining cell lines showed a 9q UPD and concomitant loss of 9pter-p21. REC1 and MAVER1 showed homozygous deletions of CDKN2A gene at 9p21.3, and MAVER1 had 2 additional homozygous deletions (indicated by asterisks; Table 1).
Uniparental disomy and uniparental trisomy in MCL. (A) Ideogram of the distribution of the UPD/UPT regions detected by SNP arrays. UPD/UPTs are represented on the left side of each chromosome; thin bars represent UPD regions, whereas thick bars represent gained/amplified regions with LOH (UPT). UPD/UPTs detected in the 10 cell lines are indicated in green, whereas UPD/UPTs detected in the primary MCL samples are indicated in blue. Acquired UPDs detected only in tumor cells were indicated in orange. The more recurrent overlapping regions were shaded in gray. (B) Chromosome 9 total/partial UPDs and homozygous deletions detected in MCL cell lines. Chromosome 9 profile is represented from pter (left) to qter (right). For each cell line, the top panel represents copy number alteration (genome smooth analysis [GSA], P value) and the bottom panel represents LOH (−log10 LOH P value). Regions with genomic losses and concomitant LOH are underlined with thick red bars, homozygous deletions (HD) are indicated with dark red squares, and regions with UPD are underlined with thick black bars. MINO showed a whole chromosome 9 UPD, whereas the 3 remaining cell lines showed a 9q UPD and concomitant loss of 9pter-p21. REC1 and MAVER1 showed homozygous deletions of CDKN2A gene at 9p21.3, and MAVER1 had 2 additional homozygous deletions (indicated by asterisks; Table 1).
A UPD at 17pter-p13.1 was observed only in the MINO cell line, which was associated with a homozygous mutation of TP53 (Table S4). Five additional cell lines had TP53 inactivating mutations associated with the loss of the remaining allele.
Copy number changes and LOH in primary MCL
Twenty-eight primary MCLs were analyzed with the GeneChip Mapping-100K SNP arrays. The SNP call rate in all experiments was more than 93%. Copy number alterations were observed in 86% (24 of 28) of the cases, with a total of 135 losses, 60 gains, and 24 amplifications (Table S1). The most frequent overlapping gains were found at 3q26.1-qter (39%) and 8q24-qter (32%), whereas frequent overlapping losses were found at 9p21, 11q22-q23 (36% each), 1p21.3-p22.1 (32%), 8pter-p21 (25%), 6q23.3 (21%), and 10pter-p12.31 (18%). Interestingly, a complex pattern of losses was found in chromosome 13: 9 cases showed discontinuous losses (range, 2-6 losses per case), whereas 2 cases (MCL2 and MCL24) showed a single loss and 2 patients (MCL22 and MCL27) showed a monosomy 13. Overall, the 13q13.3-q31.1 and 13q33.3-qter regions were lost in 36% of the patients.
Partial UPDs were detected in 13 of the 28 MCLs (46%) with a mean number of 0.8 per case (range, 0-5; Figure 1A). A total of 22 chromosomal regions showed UPDs, and one locus had a UPT (amplification of 18q21.33-q22.1). Germinal DNA was studied in 5 patients. Four of the 7 UPDs detected in the tumors were also present in the germline of 3 patients, whereas 3 UPDs (2 at 20q and one at 17pter-p12) were acquired in the tumor. Interestingly, the acquired UPDs were recurrent in tumor samples, whereas the germ line UPDs were small and nonrecurrent (Figure 1A). All gains and losses were acquired in the tumors. Recurrent overlapping regions of UPD were observed at 17pter-p13.1 and 20q11.22-q12 (3 cases each) and 5p14-p12 and 16q23.1-q23.3 (2 cases each).
Interestingly, 2 tumors with a UPD at 17pter-p13.1 (MCL3 and MCL14) had an inactivating mutation of TP53. Although we did not find a mutation in exons 4 to 11 of TP53 in the remaining MCL with a 17p UPD (MCL1), this tumor had a total lack of TP53 mRNA expression by microarray analysis. In addition, a mutation of the TP53 gene was found in 4 other cases: 2 with loss of 17p and 2 with no 17p abnormalities (Table S4).
Detection and validation of homozygous deletions in MCL
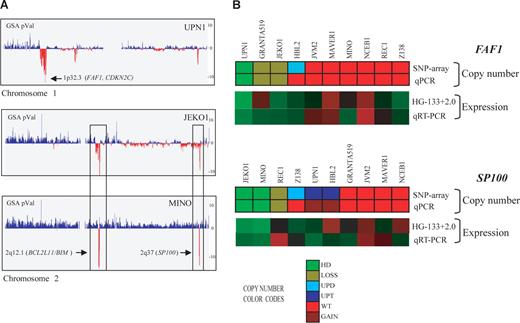
We detected 23 homozygous deletions in 8 of 10 (80%) cell lines and 4 of 28 (14%) primary MCLs. These homozygous deletions involved 11 different regions with a mean size of 807.6 kb and targeted a total of 12 genes that lacked mRNA expression (Table 1). These genes were FAF1 and CDKN2C at 1p32.3 (UPN1) (Figure 2A), ACOXL and BCL2L11 at 2q12.1 (MINO, JEKO1, and Z138), MAP2 at 2q34 (UPN1), SP100 at 2q37 (MINO and JEKO1) (Figure 2A), MOBKL2B at 9p21.2 (MAVER1), MTAP and CDKN2A at 9p21.3 (MAVER1, GRANTA519, HBL2, REC1, and Z138), RB1 at 13q14.2 (UPN1), and ZNF280A and PRAME at 22q11.22 (GRANTA519 and HBL2) (Table 1). In addition, we found 3 homozygous deletions; one of them (9p21.1) did not include any known gene, and in the remaining 2 (13q33.1 and 18q22.1), the probe sets for the respective genes (FGF14 and DSEL) in the expression array were not evaluable.
Genomic loci with homozygous deletions and target genes in MCL
| Cytoband/sample . | Start (kb) . | End (kb) . | Size (kb) . | No. of SNPs . | No. of genes . | Genes with no expression . |
|---|---|---|---|---|---|---|
| 1p32.3 | ||||||
| UPN1 | 50 465.6 | 52 103.6 | 1638.0 | 29 | 7 | FAF1, CDKN2C |
| 2q12.1 | ||||||
| MINO | 111 333.1 | 112 182.9 | 849.8 | 29 | 2 | ACOXL, BCL2L11 |
| JEKO1 | 110 286.5 | 112 155.1 | 1868.5 | 31 | 7 | ACOXL, BCL2L11 |
| Z138 | 111 517.6 | 111 623.9 | 106.3 | 5 | 2 | ACOXL, BCL2L11 |
| 2q34 | ||||||
| UPN1 | 210 048.3 | 210 472.0 | 423.6 | 36 | 1 | MAP2 |
| 2q37 | ||||||
| MINO | 230 936.6 | 231 248.6 | 312.1 | 26 | 1 | SP100 |
| JEKO1 | 231 008.8 | 231 248.6 | 239.8 | 18 | 1 | SP100 |
| 9p21.1 | ||||||
| MAVER1 | 28 761.5 | 30 316.1 | 1554.6 | 163 | 0* | — |
| 9p21.2 | ||||||
| MAVER1 | 27 316.8 | 27 758.4 | 441.6 | 41 | 3 | MOBKL2B |
| 9p21.3 | ||||||
| MAVER1 | 21 762.3 | 22 102.6 | 340.3 | 21 | 3 | MTAP, CDKN2A |
| GRANTA519 | 21 204.9 | 22 519.4 | 1314.5 | 68 | 12 | MTAP, CDKN2A |
| HBL2 | 21 948.5 | 22 102.6 | 154.1 | 12 | 2 | CDKN2A |
| REC1 | 21 948.5 | 22 404.6 | 456.1 | 16 | 2 | CDKN2A |
| Z138 | 21 844.2 | 22 449.0 | 604.8 | 26 | 4 | CDKN2A |
| MCL9 | 21 204.9 | 24 209.7 | 3004.8 | 183 | 14 | MTAP, CDKN2A |
| MCL17 | 20 835.8 | 23 113.2 | 2277.4 | 133 | 22 | ND |
| 13q14.2 | ||||||
| UPN1 | 47 812.8 | 47 817.9 | 5.1 | 6 | 2 | RB1 |
| 13q33.1 | ||||||
| JEKO1 | 101 593.7 | 101 930.8 | 337.1 | 29 | 1† | — |
| 18q22.1 | ||||||
| UPN1 | 63 097.6 | 63 530.9 | 433.3 | 33 | 1† | — |
| 22q11.22 | ||||||
| GRANTA519 | 20 831.6 | 21 479.1 | 647.5 | 21 | 5 | ZNF280A, PRAME |
| HBL2 | 20 831.6 | 21 479.1 | 647.5 | 21 | 5 | ZNF280A, PRAME |
| MCL11 | 21 000.1 | 21 479.1 | 479.1 | 19 | 4 | ZNF280A, PRAME |
| MCL20 | 20 830.9 | 21 270.4 | 439.6 | 18 | 4 | ND |
| Cytoband/sample . | Start (kb) . | End (kb) . | Size (kb) . | No. of SNPs . | No. of genes . | Genes with no expression . |
|---|---|---|---|---|---|---|
| 1p32.3 | ||||||
| UPN1 | 50 465.6 | 52 103.6 | 1638.0 | 29 | 7 | FAF1, CDKN2C |
| 2q12.1 | ||||||
| MINO | 111 333.1 | 112 182.9 | 849.8 | 29 | 2 | ACOXL, BCL2L11 |
| JEKO1 | 110 286.5 | 112 155.1 | 1868.5 | 31 | 7 | ACOXL, BCL2L11 |
| Z138 | 111 517.6 | 111 623.9 | 106.3 | 5 | 2 | ACOXL, BCL2L11 |
| 2q34 | ||||||
| UPN1 | 210 048.3 | 210 472.0 | 423.6 | 36 | 1 | MAP2 |
| 2q37 | ||||||
| MINO | 230 936.6 | 231 248.6 | 312.1 | 26 | 1 | SP100 |
| JEKO1 | 231 008.8 | 231 248.6 | 239.8 | 18 | 1 | SP100 |
| 9p21.1 | ||||||
| MAVER1 | 28 761.5 | 30 316.1 | 1554.6 | 163 | 0* | — |
| 9p21.2 | ||||||
| MAVER1 | 27 316.8 | 27 758.4 | 441.6 | 41 | 3 | MOBKL2B |
| 9p21.3 | ||||||
| MAVER1 | 21 762.3 | 22 102.6 | 340.3 | 21 | 3 | MTAP, CDKN2A |
| GRANTA519 | 21 204.9 | 22 519.4 | 1314.5 | 68 | 12 | MTAP, CDKN2A |
| HBL2 | 21 948.5 | 22 102.6 | 154.1 | 12 | 2 | CDKN2A |
| REC1 | 21 948.5 | 22 404.6 | 456.1 | 16 | 2 | CDKN2A |
| Z138 | 21 844.2 | 22 449.0 | 604.8 | 26 | 4 | CDKN2A |
| MCL9 | 21 204.9 | 24 209.7 | 3004.8 | 183 | 14 | MTAP, CDKN2A |
| MCL17 | 20 835.8 | 23 113.2 | 2277.4 | 133 | 22 | ND |
| 13q14.2 | ||||||
| UPN1 | 47 812.8 | 47 817.9 | 5.1 | 6 | 2 | RB1 |
| 13q33.1 | ||||||
| JEKO1 | 101 593.7 | 101 930.8 | 337.1 | 29 | 1† | — |
| 18q22.1 | ||||||
| UPN1 | 63 097.6 | 63 530.9 | 433.3 | 33 | 1† | — |
| 22q11.22 | ||||||
| GRANTA519 | 20 831.6 | 21 479.1 | 647.5 | 21 | 5 | ZNF280A, PRAME |
| HBL2 | 20 831.6 | 21 479.1 | 647.5 | 21 | 5 | ZNF280A, PRAME |
| MCL11 | 21 000.1 | 21 479.1 | 479.1 | 19 | 4 | ZNF280A, PRAME |
| MCL20 | 20 830.9 | 21 270.4 | 439.6 | 18 | 4 | ND |
ND indicates not done; and —, not applicable.
The deleted 9p21.1 region did not include known genes.
The homozygous deletions at the 13q33.1 and 18q22.1 regions included only one gene each, but the probe sets in the expression array were not evaluable.
Homozygous deletions in MCL cell lines detected by SNP array and qPCR. (A) Profiles of chromosomes 1 and 2 detected by the SNP array in 3 MCL cell lines, represented from pter (left) to qter (right). GSA P values represent copy number alterations. The lowest values (vertical red lines) pointed by arrows highlight regions with homozygous deletions and candidate genes. UPN1 showed a novel homozygous deletion including FAF1 gene at 1p32.3, whereas JEKO1 and MINO showed 2 concomitant homozygous deletions at 2q12.1 and 2q37, targeting BCL2L11 and SP100 genes, respectively. In addition, JEKO1 showed 2p gain and MINO gain of whole chromosome 2. (B) Representation of FAF1 and SP100 gene qPCR validation. For each gene, the copy number alteration (upper panel SNP array and lower panel qPCR) and expression values (top panel expression array and bottom panel qRT-PCR) are shown. The color codes for the DNA analysis represent the copy number changes and show a high concordance between the results of the respective techniques. Similarly, the relative mRNA expression levels are represented in a color scale (from green to red, indicating low to high expression, respectively) and also show a high concordance of the results. The genes included in the homozygous deleted regions (FAF1 in UPN1 and SP100 in JEKO1 and MINO) had undetectable mRNA levels supporting their inactivation in these MCL cell lines.
Homozygous deletions in MCL cell lines detected by SNP array and qPCR. (A) Profiles of chromosomes 1 and 2 detected by the SNP array in 3 MCL cell lines, represented from pter (left) to qter (right). GSA P values represent copy number alterations. The lowest values (vertical red lines) pointed by arrows highlight regions with homozygous deletions and candidate genes. UPN1 showed a novel homozygous deletion including FAF1 gene at 1p32.3, whereas JEKO1 and MINO showed 2 concomitant homozygous deletions at 2q12.1 and 2q37, targeting BCL2L11 and SP100 genes, respectively. In addition, JEKO1 showed 2p gain and MINO gain of whole chromosome 2. (B) Representation of FAF1 and SP100 gene qPCR validation. For each gene, the copy number alteration (upper panel SNP array and lower panel qPCR) and expression values (top panel expression array and bottom panel qRT-PCR) are shown. The color codes for the DNA analysis represent the copy number changes and show a high concordance between the results of the respective techniques. Similarly, the relative mRNA expression levels are represented in a color scale (from green to red, indicating low to high expression, respectively) and also show a high concordance of the results. The genes included in the homozygous deleted regions (FAF1 in UPN1 and SP100 in JEKO1 and MINO) had undetectable mRNA levels supporting their inactivation in these MCL cell lines.
Only 4 homozygous deletions associated with total lack of gene expression were detected in the 28 primary MCLs: 2 at 9p21.3 targeting MTAP and CDKN2A (MCL9 and 17) and 2 at 22q11.22 (MCL11 and 20) including ZNF280A and PRAME.
To validate the homozygous deletions of the deleted genes not previously described in MCL, we performed qPCR analysis of FAF1, MAP2, SP100, MOBKL2B, and PRAME genes in the 10 cell lines and in an independent set of 24 primary MCLs (Table 2; Figure 2B). The homozygous deletions of the 5 genes were confirmed by qPCR (Table 2). In addition, we confirmed by qPCR the 9 heterozygous deletions that had been detected by the array (FAF1 gene in GRANTA519 and JEKO1, SP100 gene in REC1, MOBKL2B gene in GRANTA519, JEKO1, NCEB1 and UPN1 and PRAME gene in MINO and NCEB1). In the independent set of 24 primary MCLs, we found homozygous deletions of PRAME in 2 cases and monoallelic losses in 2 additional tumors. Monoallelic losses of FAF1 (3 cases), SP100 (7 cases), and MOBKL2B (5 cases) were also observed. The lack of mRNA expression in cases with homozygous deletions was also confirmed for 3 selected genes (FAF1, SP100, and MOBKL2B; Figures 2B, S1).
Quantitative real-time PCR validation of homozygous deletions in MCL
| Sample . | FAF1 (1p32.3)* . | MAP2 (2q34)* . | SP100(2q37)* . | MOBKL2B (9p21.2)* . | PRAME (22q11.22)* . |
|---|---|---|---|---|---|
| GRANTA519 | 0.6 | 1.1 | 1.2 | 0.6 | 0† |
| HBL2 | 0.9 | 1 | 1.4 | 1.0 | 0† |
| JEKO1 | 0.5 | 0.5 | 0.1† | 0.5 | 1.0 |
| JVM2 | 1.2 | 0.9 | 1.5 | 1.6 | 0.9 |
| MAVER1 | 1.0 | 0.9 | 1.2 | 0† | 1.2 |
| MINO | 1.1 | 1.7 | 0† | 1.3 | 0.5 |
| NCEB1 | 1.2 | 1.2 | 1.0 | 0.3 | 0.5 |
| REC1 | 1.1 | 0.6 | 0.5 | 1.8 | 0.9 |
| UPN1 | 0† | 0† | 1.9 | 0.5 | 0.8 |
| Z138 | 1.2 | 1.1 | 1.2 | 1.0 | 1.0 |
| MCL validation set | 3/21 | ND | 7/21 | 5/21 | 4‡/21 |
| Sample . | FAF1 (1p32.3)* . | MAP2 (2q34)* . | SP100(2q37)* . | MOBKL2B (9p21.2)* . | PRAME (22q11.22)* . |
|---|---|---|---|---|---|
| GRANTA519 | 0.6 | 1.1 | 1.2 | 0.6 | 0† |
| HBL2 | 0.9 | 1 | 1.4 | 1.0 | 0† |
| JEKO1 | 0.5 | 0.5 | 0.1† | 0.5 | 1.0 |
| JVM2 | 1.2 | 0.9 | 1.5 | 1.6 | 0.9 |
| MAVER1 | 1.0 | 0.9 | 1.2 | 0† | 1.2 |
| MINO | 1.1 | 1.7 | 0† | 1.3 | 0.5 |
| NCEB1 | 1.2 | 1.2 | 1.0 | 0.3 | 0.5 |
| REC1 | 1.1 | 0.6 | 0.5 | 1.8 | 0.9 |
| UPN1 | 0† | 0† | 1.9 | 0.5 | 0.8 |
| Z138 | 1.2 | 1.1 | 1.2 | 1.0 | 1.0 |
| MCL validation set | 3/21 | ND | 7/21 | 5/21 | 4‡/21 |
ND indicates not done.
In the cell lines, the number refers to qPCR result of gene dosage, whereas in the MCL primary cases, the number refers to number of cases with monoallelic loss/no. of cases analyzed.
Homozygous deletions.
Two cases showed homozygous deletions of the PRAME gene.
PRAME and ZNF280A genes at 22q11.22 are mapped in the intronic region of the V segments of the lambda chain cluster, between V7-35 and the more telomeric V2-34. Given the location of these 2 genes, we examined the relationship between PRAME/ZNF280A deletions and the expression of the light chains. Interestingly, the 4 cell lines (HBL2, GRANTA 519, MINO, and NCEB1) and the 2 primary cases (MCL11 and MCL20) with homozygous or monoallelic 22q11.22 deletions expressed lambda chain. On the contrary, none of the 21 samples with kappa chain expression had 22q11.22 deletions. These findings suggest that the loss of PRAME/ZNF280A may be a consequence of the lambda gene rearrangements between the J and V regions located centromeric to V7-35.
Amplifications and target genes in MCL: C13orf25, BCL2, and CCND1
We detected 53 high-level DNA amplifications in 8 of 10 (80%) cell lines and 10 of 28 (36%) primary MCL patients involving 35 different chromosomal regions with a mean size of 7 Mb (range, 0.45-71.3 Mb; Tables 3, S3). Amplifications were more frequent in cell lines (mean, 2.5; range, 0-7) than in primary tumors (mean, 0.9; range, 0-6, Table S1). Six recurrent overlapping regions of amplification with highly overexpressed genes were identified (Table 3). Amplification of 13q31.3 and 18q21.33 regions was detected in 6 samples each, whereas the other 4 amplified regions were observed in only 2 cases each (Table 3).
Recurrent high-level DNA amplifications and focal gains in mantle-cell lymphoma
| Cytoband . | Sample . | Start (kb) . | End (kb) . | Size (kb) . | No. of genes . | Overlapping region (no. of genes) . | Overexpressed genes in the overlapping region* . | Putative microRNA in the overlapping region . |
|---|---|---|---|---|---|---|---|---|
| 8q21.11-q24.3 | MCL26 | 74 752.6 | 146 052.2 | 71 299.5 | > 500 | 8q24.21 (2) | CCNE2, E2F5,†EDD1, EIF2C2, BXL6, FBXO43, FZD6,† (MYC),‡NIBP and several zinc finger proteins§ | — |
| 8q24.21 | JEKO1 | 127 321.4 | 128 809.0 | 1 487.5 | 2 | (MYC)‡ | ||
| 9pter-p24.1 | Z138 | 239.4 | 6 759.2 | 6 519.8 | 30 | 9p24.2-p24.1 (14) | AK3, C9orf38,†C9orf46, CDC37L1, JAK2,†PPAPDC2, RANBP6, RCL1, RFX3,†TPD52L3 | hsa mir 101–2 |
| 9p24.2-p24.1 | MCL21 | 2 332.2 | 5 415.1 | 3 082.9 | 14 | ND | ||
| 10p12.33-p12.1 | MCL19 | 19 237.3 | 26 503.1 | 7 265.9 | 15 | 10p12.31-p12.1 (11) | ND | hsa mir 603 |
| 10p12.31-p12.1 | MCL20 | 22 316.8 | 25 365.9 | 3 049.1 | 11 | ND | ||
| 11q13.3-q13.5 | HBL2 | 68 979.0 | 76 176.0 | 7 197.0 | 77 | 11q13.3-q13.5 (77) | ATG16L2, C11orf30, C11orf51, CCND1,†CENTD2, CHCHD8, CLPB, DHCR7,†FADD,†FCHSDs2, INPPL1, LRRC51, MRPL48, NADSYN1,†NUMA1, ORAOV1,†PGM2L1, POLD3, PPFIA1,†PPME1, RAB6A, RNF169, SPCS2, TNFRSF19L, UCP2,†UCP3, PAAF1 | hsa mirs 139/326 |
| 11q13.3-q14.1 | MAVER1 | 69 059.8 | 80 083.5 | 11 023.7 | 101 | |||
| 11q13.1-q14.3‖ | MCL14 | 64 457.5 | 92 492.5 | 28 034.9 | > 200 | CCND1,†DGAT2,†PGM2L1,†PPME1,†PAAF1† | ||
| 11q13.3-q23.3‖ | MCL1 | 69 041.4 | 119 717.6 | 50 441.9 | > 200 | |||
| 11q13.2-q14.1‖ | MCL26 | 68 435.5 | 81 720.1 | 13 284.5 | > 100 | ATG16L2, C11orf30, SLCO2B1 | ||
| 11q13.2-q21‖ | MCL18 | 68 986.2 | 95 676.4 | 26 690.2 | > 150 | ND | ||
| 13q31.1-q31.2 | REC1 | 85 593.0 | 91 208.3 | 5 615.3 | 2 | 13q31.3 (1) | C13orf25† | hsa mirs 17/18a/19a/19b1/20a/92a1/622 |
| 13q31.2-q32.1 | MCL20 | 86 090.6 | 94 386.2 | 8 295.6 | 7 | |||
| 13q31.2-q31.3 | MCL18 | 88 696.3 | 92 595.3 | 3 899.0 | 1 | |||
| 13q31.3 | HBL2 | 89 815.1 | 90 996.6 | 1 181.5 | 1 | |||
| 13q31.3 | JEKO1 | 89 815.1 | 92 166.6 | 2 351.5 | 1 | |||
| 13q31.3 | MCL17 | 90 738.1 | 91 700.9 | 962.8 | 1 | |||
| 18q12.2-q22.1 | MINO | 31 201.2 | 61 451.2 | 30 250.0 | 89 | 18q21.33 (10) | BCL2,†FVT1, PHLPP, VPS4B | hsa mir 122 |
| 18q21.2-q22.2¶ | HBL2 | 49 877.1 | 65 507.0 | 15 629.9 | 57 | |||
| 18q21.31-q22.2 | Z138 | 52 434.9 | 66 564.3 | 14 129.4 | 48 | |||
| 18q21.32-q22.3 | GRANTA519 | 55 604.1 | 69 248.9 | 13 644.8 | 33 | |||
| 18q21.33-q22.1¶ | MCL21 | 57 749.3 | 63 438.4 | 5 689.2 | 21 | |||
| 18q21.33 | MAVER1 | 58 464.6 | 59 544.8 | 1 080.2 | 10 |
| Cytoband . | Sample . | Start (kb) . | End (kb) . | Size (kb) . | No. of genes . | Overlapping region (no. of genes) . | Overexpressed genes in the overlapping region* . | Putative microRNA in the overlapping region . |
|---|---|---|---|---|---|---|---|---|
| 8q21.11-q24.3 | MCL26 | 74 752.6 | 146 052.2 | 71 299.5 | > 500 | 8q24.21 (2) | CCNE2, E2F5,†EDD1, EIF2C2, BXL6, FBXO43, FZD6,† (MYC),‡NIBP and several zinc finger proteins§ | — |
| 8q24.21 | JEKO1 | 127 321.4 | 128 809.0 | 1 487.5 | 2 | (MYC)‡ | ||
| 9pter-p24.1 | Z138 | 239.4 | 6 759.2 | 6 519.8 | 30 | 9p24.2-p24.1 (14) | AK3, C9orf38,†C9orf46, CDC37L1, JAK2,†PPAPDC2, RANBP6, RCL1, RFX3,†TPD52L3 | hsa mir 101–2 |
| 9p24.2-p24.1 | MCL21 | 2 332.2 | 5 415.1 | 3 082.9 | 14 | ND | ||
| 10p12.33-p12.1 | MCL19 | 19 237.3 | 26 503.1 | 7 265.9 | 15 | 10p12.31-p12.1 (11) | ND | hsa mir 603 |
| 10p12.31-p12.1 | MCL20 | 22 316.8 | 25 365.9 | 3 049.1 | 11 | ND | ||
| 11q13.3-q13.5 | HBL2 | 68 979.0 | 76 176.0 | 7 197.0 | 77 | 11q13.3-q13.5 (77) | ATG16L2, C11orf30, C11orf51, CCND1,†CENTD2, CHCHD8, CLPB, DHCR7,†FADD,†FCHSDs2, INPPL1, LRRC51, MRPL48, NADSYN1,†NUMA1, ORAOV1,†PGM2L1, POLD3, PPFIA1,†PPME1, RAB6A, RNF169, SPCS2, TNFRSF19L, UCP2,†UCP3, PAAF1 | hsa mirs 139/326 |
| 11q13.3-q14.1 | MAVER1 | 69 059.8 | 80 083.5 | 11 023.7 | 101 | |||
| 11q13.1-q14.3‖ | MCL14 | 64 457.5 | 92 492.5 | 28 034.9 | > 200 | CCND1,†DGAT2,†PGM2L1,†PPME1,†PAAF1† | ||
| 11q13.3-q23.3‖ | MCL1 | 69 041.4 | 119 717.6 | 50 441.9 | > 200 | |||
| 11q13.2-q14.1‖ | MCL26 | 68 435.5 | 81 720.1 | 13 284.5 | > 100 | ATG16L2, C11orf30, SLCO2B1 | ||
| 11q13.2-q21‖ | MCL18 | 68 986.2 | 95 676.4 | 26 690.2 | > 150 | ND | ||
| 13q31.1-q31.2 | REC1 | 85 593.0 | 91 208.3 | 5 615.3 | 2 | 13q31.3 (1) | C13orf25† | hsa mirs 17/18a/19a/19b1/20a/92a1/622 |
| 13q31.2-q32.1 | MCL20 | 86 090.6 | 94 386.2 | 8 295.6 | 7 | |||
| 13q31.2-q31.3 | MCL18 | 88 696.3 | 92 595.3 | 3 899.0 | 1 | |||
| 13q31.3 | HBL2 | 89 815.1 | 90 996.6 | 1 181.5 | 1 | |||
| 13q31.3 | JEKO1 | 89 815.1 | 92 166.6 | 2 351.5 | 1 | |||
| 13q31.3 | MCL17 | 90 738.1 | 91 700.9 | 962.8 | 1 | |||
| 18q12.2-q22.1 | MINO | 31 201.2 | 61 451.2 | 30 250.0 | 89 | 18q21.33 (10) | BCL2,†FVT1, PHLPP, VPS4B | hsa mir 122 |
| 18q21.2-q22.2¶ | HBL2 | 49 877.1 | 65 507.0 | 15 629.9 | 57 | |||
| 18q21.31-q22.2 | Z138 | 52 434.9 | 66 564.3 | 14 129.4 | 48 | |||
| 18q21.32-q22.3 | GRANTA519 | 55 604.1 | 69 248.9 | 13 644.8 | 33 | |||
| 18q21.33-q22.1¶ | MCL21 | 57 749.3 | 63 438.4 | 5 689.2 | 21 | |||
| 18q21.33 | MAVER1 | 58 464.6 | 59 544.8 | 1 080.2 | 10 |
ND indicates not done; and —, not applicable.
Target overexpressed genes (> 2-fold) are listed in alphabetical order.
Genes overexpressed 4-fold more than the nonamplified samples, C13orf25 was overexpressed 10 times more than the nonamplified samples.
MYC probe sets were not evaluable.
Only a selection of cancer-related genes is shown for the large 8q region.
11q13 focal gain.
18q21.33 amplification in a region of uniparental disomy.
The 13q31.3 amplification was detected in 3 primary tumors and 3 cell lines (Table 3). The minimal region targeted only C13orf25, which encodes an oncogenic microRNA polycistron, and its RNA was overexpressed more than 10 times in the cell lines with the amplification of the locus.
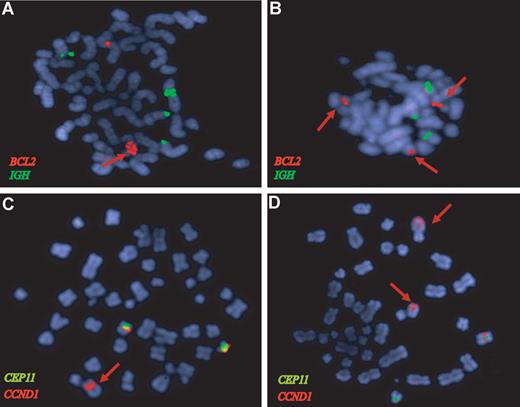
One primary MCL (MCL21) and 5 cell lines had amplifications of the 18q21.33 region with 4 genes (BCL2, FVT1, PHLPP, and VPS4B) showing higher mRNA expression levels than the nonamplified cell lines. BCL2 was the gene with the highest expression levels of the 4 (2- to 3-fold higher than the other amplified genes). Amplification of the BCL2 in MAVER1, HBL2, and one primary case was confirmed by FISH (Figure 3A,B). The UPN1 cell line carried an allelic loss of the 18q21.33 region that was confirmed by FISH (data not shown).
BCL2 and CCND1 gene amplifications in MCL confirmed by FISH analysis. FISH of representative metaphases with BCL2 (red), IGH (green), CCND1 (orange), and CEP11 (green) specific probes. (A) The HBL2 cell line shows a tandem high-copy number BCL2 amplification (red arrow). (B) The BCL2 locus is amplified in a primary MCL (red signals) from the validation set that also showed a 18q21 amplification by CGH. (C) The HBL2 cell line has an amplified CCND1-IGH rearrangement (red arrow) and 2 nontranslocated chromosomes 11 with CCND1 signal (red) below the centromere of chromosome 11 (green). The CCND1-IGH amplified fusion was confirmed by a dual-color dual-fusion probe (data not shown). (D) The MAVER1 cell line has 2 high-level tandem amplifications of CCND1 (red arrows) in 2 different chromosomes: one corresponds to the complex t(11;14) translocation in chromosome 6.11 The chromosome carrying the second amplified region has not been identified. A nontranslocated chromosome 11 with the red CCND1 signal associated with the green centromere 11 probe is also observed.
BCL2 and CCND1 gene amplifications in MCL confirmed by FISH analysis. FISH of representative metaphases with BCL2 (red), IGH (green), CCND1 (orange), and CEP11 (green) specific probes. (A) The HBL2 cell line shows a tandem high-copy number BCL2 amplification (red arrow). (B) The BCL2 locus is amplified in a primary MCL (red signals) from the validation set that also showed a 18q21 amplification by CGH. (C) The HBL2 cell line has an amplified CCND1-IGH rearrangement (red arrow) and 2 nontranslocated chromosomes 11 with CCND1 signal (red) below the centromere of chromosome 11 (green). The CCND1-IGH amplified fusion was confirmed by a dual-color dual-fusion probe (data not shown). (D) The MAVER1 cell line has 2 high-level tandem amplifications of CCND1 (red arrows) in 2 different chromosomes: one corresponds to the complex t(11;14) translocation in chromosome 6.11 The chromosome carrying the second amplified region has not been identified. A nontranslocated chromosome 11 with the red CCND1 signal associated with the green centromere 11 probe is also observed.
Amplification of the 11q13.3 region was found in HBL2 and MAVER1, and it was associated with very high levels of the cyclin D1 long transcript (probe set 208712_at). These CCND1 amplifications corresponded to the translocated allele and were confirmed by FISH (Figure 3C,D). In addition, 4 primary tumors had a focal gain of 11q13 involving the CCND1 locus, and 3 of them had also very high levels of cyclin D1 mRNA.
CNVs and segmental duplications flanking DNA breakpoints
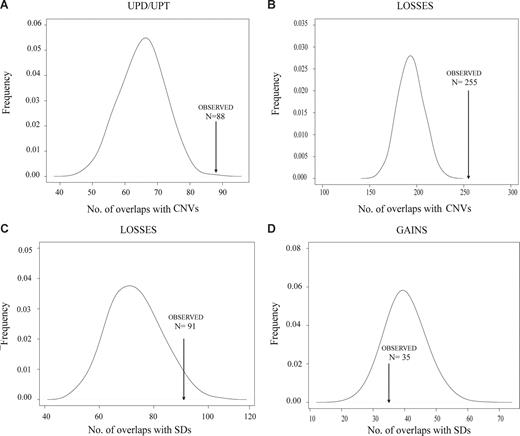
CNVs and SDs are closely related structures that have been considered substrates for nonallelic homologous recombination that may facilitate the generation of chromosomal alterations, similar to fragile sites.27 To determine whether the recurrent DNA breakpoints observed in MCLs could be related to these DNA regions, we performed computational simulation experiments to compare the observed number of chromosomal breakpoints associated with the number of CNVs or SDs in MCL expected by chance. Given the high resolution of the GeneChip Mapping-100K SNP array, we were able to map precisely the breakpoints and evaluate the association of these coordinates with the physical position of CNVs and SDs deposited in the Database of Genomic Variants and in the University of California Santa Cruz tables, respectively.21 We considered the SDs and CNVs overlapping the 100-kb regions flanking each breakpoint. Overall, in the present series of MCLs, we detected 1200 breakpoints of chromosomal alterations. A total of 181 of the 1200 breakpoints (15%) were associated with SDs, a proportion slightly higher than that of the simulated data (median, 152, P = .071). Interestingly, 552 of 1200 breakpoints (46%) overlapped with CNVs, a proportion significantly higher than that of the simulated data (median, 412, P < .001; Table S3). This significant enrichment of CNVs in MCL breakpoints suggests that CNVs may contribute to the generation of genomic imbalances in MCLs. According to the type of alteration (UPD/UPT, homozygous deletion, loss, gain, and amplification), we found a significant enrichment of CNVs with UPD/UPT, homozygous deletions, losses, and gains, whereas a significant enrichment of SDs was found for homozygous deletions and losses (Figure 4; Table S3). Similar significant associations were found when considering the CNVs and SDs at 50-kb flanking each breakpoint (data not shown).
Representative examples of the enrichment of CNVs and segmental duplications in MCL breakpoints. Association of MCL chromosomal breakpoints in CNV/SDs loci based on computational simulations that compared the expected number of breakpoints containing CNV/SDs with the observed number in MCL cell lines and primary tumors. A significant enrichment of breakpoints in CNV loci was found for UPDs/UPTs (A) and chromosomal losses (B). Similarly, a significant enrichment of breakpoints in SD loci was found for losses (C), whereas breakpoints of chromosomal gains have no association with SDs (D).
Representative examples of the enrichment of CNVs and segmental duplications in MCL breakpoints. Association of MCL chromosomal breakpoints in CNV/SDs loci based on computational simulations that compared the expected number of breakpoints containing CNV/SDs with the observed number in MCL cell lines and primary tumors. A significant enrichment of breakpoints in CNV loci was found for UPDs/UPTs (A) and chromosomal losses (B). Similarly, a significant enrichment of breakpoints in SD loci was found for losses (C), whereas breakpoints of chromosomal gains have no association with SDs (D).
Discussion
The high-density 100K SNP array platform is a rapid and robust assay that allows the simultaneous screening of DNA copy number alterations and LOH of the entire genome. Integrating this information with expression array data provides a valuable tool for the identification of pathobiologically relevant cancer genes. We have used this comprehensive approach in a panel of 10 MCL cell lines and 28 primary tumors with a high tumor-cell content. The results of the study expand the view of the genomic complexity of MCL tumors with the identification of numerous and complex UPDs/UPTs and the recognition of new homozygous deletions and amplifications that led to altered expression of putative tumor suppressor genes, oncogenes, and oncogenic microRNAs.
The global profile of genomic alterations observed with the SNP arrays confirms and refines the most common regions previously described in MCL.5-10,12-15 As expected, MCL cell lines showed a 5-fold increased number of alterations compared with MCL primary tumors. However, the most recurrent alterations were also found in MCL primary cases and were associated with expression changes. In addition, we identified, for the first time, the presence of a high number of complex UPDs/UPTs in primary MCLs. UPDs have been recently recognized in different hematologic neoplasms, including acute leukemias,32,33 multiple myeloma,34 chronic lymphocytic leukemia,35 follicular lymphoma,36 T-prolymphocytic leukemia,37 and 5 MCL cell lines.38 The UPD regions in MCL usually target small and widespread chromosomal regions. This type of UPD was also observed in multiple myeloma,34 chronic lymphocytic leukemia,35 and T-prolymphocytic leukemia37 and probably represent a phenomenon originated by multiple mitotic recombination events. In contrast, the UPDs found in follicular lymphoma36 and acute myeloid leukemia32 involve large chromosomal regions reaching the telomere, suggesting that they probably originated from a single mitotic recombination event. Moreover, the UPDs/UPTs detected in most MCLs were found in chromosomal arms concomitantly affected by losses, gains, and other regions of UPD/UPT. The high number and complexity of this phenomenon in MCL genomes have no parallel in other hematologic neoplasms, and it may reflect the genomic instability of these tumors. Interestingly, only T-prolymphocytic leukemia seems to carry also a relatively complex UPD distribution.37
UPDs in certain chromosomal regions may play an important role in the biology of some tumors by selecting mutated/methylated genes that contribute to their pathogenesis, that is, FLT3, WT1, and CEBPA genes in acute myeloid leukemia,39,40 JAK2 in myeloproliferative disorders,41 and IGF2 and H19 genes in hepatoblastoma.42 In our study, the 4 UPDs at 17p detected in 3 primary tumors and one cell line were associated with TP53 gene inactivation. A similar situation has been also observed in 2 chronic lymphocytic leukemia patients.43 The possible significance of the UPDs in other chromosomal regions is not well known because only a limited number of UPDs are located in regions with known tumor suppressor genes. UPTs have not been observed in other hematologic neoplasms. In our study, 7 MCL cell lines and one primary tumor showed UPT of different chromosomal regions. Interestingly, the UPT at 18q21.33 contained a high-level DNA amplification of the BCL2 gene that was also associated with high levels of expression. Interestingly, the parallel analysis of normal DNA of some patients demonstrated that nonrecurrent and small UPDs were already present in the germ line of the patients, but 2 recurrent UPDs were acquired in the tumor sample.
Our integrative genomic and expression analysis has allowed us to identify and validate a high number of homozygous deletions and potential target genes in MCL. Previous studies using BAC arrays had observed homozygous deletions, including different tumor suppressor genes (CDKN2C, BCL2L11, CDKN2A, and ATM), immunoglobulin kappa and lambda loci (2p11.2 and 22q11.22), and regions without known target genes (11p12-p14 and 13q32.3) in a limited number of cell lines and primary tumors.8,13,14,18,29 In the present study, we have found homozygous deletions in 11 different regions, 6 of them not previously identified, and we have confirmed the previously described homozygous deletions of CDKN2C, BCL2L11, CDKN2A, and 22q11.22 in the same cell lines that were originally observed and also expanded the homozygous deletions of BCL2L11, CDKN2A, and 22q11.22 in other cell lines (MINO, MAVER1, and HBL2, respectively). The CDKN2C gene had been reported as the target of the homozygous deletions of 1p32.3. However, we have now observed that this deletion also includes FAF1, an inhibitor of the NF-κB pathway that suppresses the IKK activity and also independently retains NF-κB p65 in the cytoplasm.44
The novel homozygous deletions detected by our approach target MAP2 (2q34), SP100 (2q37), and MOBKL2B (9p21.2) genes. MAP2 is a major regulator of the microtubule dynamics in neurons and has been recently implicated in the migration of epithelial cancer cells.45 SP100 is a constitutive member of nuclear bodies, together with PML, TP53, and DAXX proteins.46 Finally, MOBKL2B was found homozygously deleted in MAVER1, whereas single losses were found in other cell lines. Absence or low levels of mRNA expression of this gene were found also in other samples with no genomic deletion (Figure S1). MOBKL2B is highly homologous to Mob1 gene, a member of the Hippo pathway, which has been involved in cancer development through regulation of proliferation and mitotic checkpoint regulation.47
Only few recurrent amplifications and candidate oncogenes have been identified in MCLs, being the most common 8q24 (MYC), 10p12.2 (BMI1), and 12q13 (CDK4).7,8,15 In the present study, we detected 35 different amplified regions. The most recurrent were at 13q31.3 and 18q21.33, each of them observed in 6 cell lines and primary tumors. Interestingly, C13orf25 was the only gene included in the small 13q31.3 amplicon, and its amplification was associated with expression levels 10-fold higher than in the nonamplified samples, indicating that this gene might be the target of these amplifications in MCL. 13q31-q32 amplifications have been observed in different lymphoid neoplasms, and C13orf25 was cloned as the most probable candidate gene for this amplicon.48 This gene encodes the miR-17-92 polycistronic cluster, which seems to cooperate with MYC to transform mouse B cells and decrease apoptosis.49 Amplifications and overexpression of C13orf25 have been observed occasionally in Burkitt lymphoma, diffuse large B-cell lymphoma, and in the JEKO1 MCL cell line.48,50 13q31-q32 amplification has been previously reported in 10% of MCLs.6 Our finding of C13orf25 as the target gene of these amplifications suggests that this alteration may play a pathogenic role in these tumors.
18q21 is a region frequently gained and amplified in MCL,6,7,15 and higher expression of BCL2 gene had been observed in blastoid variants.51 In this study, we have observed amplifications of this region in 5 cell lines and 2 primary tumors. The minimal overlapping region of this amplicon included 10 genes, and only 4 of them were up-regulated. The BCL2 gene showed the highest mRNA overexpression level, suggesting that it may be the target of these amplifications. The BCL2 amplifications and overexpression may contribute to the pathogenesis of MCL by promoting cell survival, facilitating the genomic instability of the cells,52 and increasing the resistance of the tumor cells to new therapeutic drugs targeting the members of the BCL2 family (ie, BH-3 mimetic drugs).53
CCND1 gene was amplified in 2 cell lines and was also included in the focal 11q13 gains of 4 primary cases. These alterations were associated with very high levels of cyclin D1. The FISH analysis of 2 cell lines (HBL2 and MAVER1) confirmed that the amplification corresponded to the translocated allele. These amplifications may be an additional mechanism to increase the levels of cyclin D1, similar to the effect of the truncations and mutations in the 3′UTR region of the mRNA, which are associated with very high levels of expression of the short 1.5-kb transcript.54 Interestingly, these 2 cell lines only expressed the long mRNA cyclin D1 transcript, suggesting that they do not carry alterations in the 3′UTR mRNA region. The amplification of CCND1/IGH fusion gene has been only described in an unusual leukemic MCL with high cyclin D1 overexpression, very complex karyotype, TP53 gene inactivation, and a very aggressive clinical course.55
In addition to these highly recurrent genetic alterations with important impact on mRNA expression, we detected a high number of less frequent aberrations. The biologic and clinical significance of these 2 types of changes may be different. Whereas the recurrent events targeting potent tumor suppressor genes and oncogenes may have an important pathogenetic role and clinical impact, the less common aberrations may correspond to passenger events reflecting the high level of chromosomal instability of MCLs.3,19
The mechanisms involved in the genomic instability of MCL are not well understood. CNVs and SDs are DNA break-prone regions that may contribute to the generation of chromosomal alterations by facilitating nonallelic homologous recombinations.24,26,27 Indeed, an enrichment of SDs and CNVs has been found at tumor-break regions in some chromosomes of neuroblastoma,56 familial pancreatic cancer,57 and carcinoma cell lines.58 Furthermore, a genome-wide association of chromosomal breakpoints with CNVs and SDs has been reported recently for colon cancer.59 In this study, we have found, for the first time, a significant enrichment of CNVs and SDs in the breakpoints of chromosomal alterations in MCL. We have found as many as 46% of all breakpoints colocalizing with CNV regions, a number similar to the 41% found in colon cancer.59 Particularly, in MCL, CNV regions were frequently associated with UPDs/UPTs, homozygous deletions, losses, and gains but not amplifications, whereas SDs were more frequently related to homozygous deletions and losses. These findings are concordant with previous studies in healthy subjects suggesting that SDs are involved in chromosomal losses but not in gains.25 Moreover, it is the first time that an association of such structural variation is found with UPD/UPT regions. These findings in MCL, a tumor with high levels of genomic instability and alterations in DNA damage repair pathways, may suggest that a number of the chromosomal alterations could arise from the aberrant repair of the nonallelic homologous recombination driven by SDs and CNVs.
In conclusion, our study integrating high-resolution genomic and expression profiling of MCL has identified a high number of complex and genome interspersed UPDs that expands the perspective of the genomic instability in these tumors. In addition, we have delineated and confirmed several homozygous deletions and amplifications that target known and new candidate genes that may be relevant in the pathogenesis of these lymphomas. Finally, we have described a significant enrichment of CNVs and SDs in the breakpoints flanking the chromosomal alterations of MCL, suggesting that these structures may contribute to the genomic instability of the tumors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Melissa Visser, Laura Pla, Montse Sanchez, and Iracema Nayach for excellent technical support.
This work was supported by the Instituto de Salud Carlos III, Fondo Investigaciones Sanitarias (FIS06/0150; S.B.), Comisión Interministerial de Ciencia y Tecnología Española (SAF05/5855; E.C.), Instituto de Salud Carlos III Red Temática de Investigación Cooperativa de Cancer (2006RET2039; E.C.), National Institutes of Health SPECS (grant 5 U01 CA114778-03; E.C., A.R.), and Lymphoma Research Foundation (LRFMCLI-05-023, E.C., A.R.; and LRF07168, P.J.). E.M.H and A.R. are supported by the Interdisciplinary Center for Clinical Research, University of Würzburg, Germany. L.H. is a researcher from Institut d'Investigacions Biomèdiques August Pi i Sunyer and supported by FIS and Programa d'estabilització d'investigadors de la Direcció d'Estrategia i Coordinació del Departament de Salut (Generalitat de Catalunya). Research of X.E.'s laboratory is supported by the Generalitat de Catalunya (Departments of Health and Universities and Innovation).
National Institutes of Health
Authorship
Contribution: S.B. designed and performed research, collected and analyzed data, and wrote the manuscript; L.A. performed simulation experiments and analysis; I.S., M.P., V.F., E.M.H., P.J., and A.N. performed research; V.A. and L.H. discussed results; G.O. and A.R. provided and revised patient samples; X.E. supervised simulation experiments and analysis; E.C. designed the study, revised patient samples, supervised the research, and wrote the manuscript; and all authors have critically reviewed the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elias Campo, Hematopathology Unit, Hospital Clinic, Villarroel 170, 08036-Barcelona, Spain; e-mail: ecampo@clinic.ub.es.

![Figure 1. Uniparental disomy and uniparental trisomy in MCL. (A) Ideogram of the distribution of the UPD/UPT regions detected by SNP arrays. UPD/UPTs are represented on the left side of each chromosome; thin bars represent UPD regions, whereas thick bars represent gained/amplified regions with LOH (UPT). UPD/UPTs detected in the 10 cell lines are indicated in green, whereas UPD/UPTs detected in the primary MCL samples are indicated in blue. Acquired UPDs detected only in tumor cells were indicated in orange. The more recurrent overlapping regions were shaded in gray. (B) Chromosome 9 total/partial UPDs and homozygous deletions detected in MCL cell lines. Chromosome 9 profile is represented from pter (left) to qter (right). For each cell line, the top panel represents copy number alteration (genome smooth analysis [GSA], P value) and the bottom panel represents LOH (−log10 LOH P value). Regions with genomic losses and concomitant LOH are underlined with thick red bars, homozygous deletions (HD) are indicated with dark red squares, and regions with UPD are underlined with thick black bars. MINO showed a whole chromosome 9 UPD, whereas the 3 remaining cell lines showed a 9q UPD and concomitant loss of 9pter-p21. REC1 and MAVER1 showed homozygous deletions of CDKN2A gene at 9p21.3, and MAVER1 had 2 additional homozygous deletions (indicated by asterisks; Table 1).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/113/13/10.1182_blood-2008-07-170183/4/m_zh80040930120001.jpeg?Expires=1765893138&Signature=Mjm3DyN81SI5tcDqB1MeEeHalAuu67Wt12zwz1aBMOS6TXxtcgauvGc3w6IdHE1iKub~fjHup3ZkQtkNKmn9X7kAiwKBs0Zy5VOL8pAGrR57L~nupHWPUBlenskKKQJpBCUd5-NZWqpEXVtFU2tNA-wC8jLHewsI~rjkkal33sNV5XFQd~RzD3OzVZpg3x5Mjigcp4HAkgjt-9sbDr9V9wtcUaaFHXvqe-TYhhDxp7PmgvGFYN4to1dB8JiW5j2ajtJnqeEkkhOw7z8sDqry85CIh-3u3B8JAmQHp7pElYDDuPpY24A-sZ6Q9XkbxEMlHzTyp9-ApJymqIY3kxancg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal