Abstract
Despite the relevant therapeutic progresses made in these last 2 decades, the prognosis of acute myeloid leukemia (AML) remains poor. Phorbol esters are used at very low concentrations as differentiating agents in the therapy of myeloid leukemias. Tumor necrosis factor–related apoptosis-inducing ligand (TRAIL), in turn, is a death ligand that spares normal cells and is therefore currently under clinical trials for cancer therapy. Emerging evidence, however, suggests that TRAIL is also involved in nonapoptotic functions, like cell differentiation. PKCϵ is differentially modulated along normal hematopoiesis, and its levels modulate the response of hematopoietic precursors to TRAIL. Here, we investigated the effects of the combination of phorbol esters (phorbol ester 4-β-phorbol-12,13-dibutyrate [PDBu]) and TRAIL in the survival/differentiation of AML cells. We demonstrate here that PDBu sensitizes primary AML cells to both the apoptogenic and the differentiative effects of TRAIL via PKCϵ down-modulation, without affecting TRAIL receptor surface expression. We believe that the use of TRAIL in combination with phorbol esters (or possibly more specific PKCϵ down-modulators) might represent a significative improvement of our therapeutic arsenal against AML.
Introduction
Although these last 2 decades have seen relevant progresses in the therapy of acute myelogenous leukemia (AML) in terms of cytogenetic prognostic factors, bone marrow transplantation, and targeted therapies,1 the 5-year survival rate is still 20% to 30%2 for adult primary AML, and is even worse for AML arising from myeloproliferative disorders or myelodysplastic syndromes (MDSs).3 Moreover, conventional antitumor treatments make a selective pressure for p53-inactivated tumor cells, and the development of drug resistance remains a serious problem. In this context, a special effort is currently dedicated to targeted therapies whose cornerstones—at least in terms of clinical success—have been all-trans retinoic acid (ATRA) for promyelocytic leukemia4,5 and imatinib for chronic myelogenous leukemia,6 while several other compounds—including apoptosis inducers and antiapoptotic inhibitors—are in the pipeline in the future perspective of combinations of multiple targeted therapies for the treatment of AML.
Recombinant, soluble tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) is currently being developed as a promising natural immune molecule for patients with cancer because it selectively induces apoptosis in transformed or stressed cells but not in most normal cells.7 Although primary AML cells are generally resistant to TRAIL-induced apoptosis in patients with cancer, phase 1 and 2 clinical trials using agonistic mAbs that engage the human TRAIL receptors 1 (TRAIL-R1) and 2 (TRAIL-R2) have also provided encouraging results.7 Although hepatotoxicity was observed as side effect using agonistic antibodies against TRAIL-R2,8 the emerging idea is that TRAIL will likely be used as one component of more complex antitumor therapeutic regimens. In this regard, a number of drugs and biological agents have already been shown to sensitize tumor cells to TRAIL.9 Since recombinant TRAIL acts synergistically with chemotherapy and radiotherapy, combinations of these with TRAIL or with cytokines that mobilize effector cells equipped with membrane-bound TRAIL (like natural killer [NK] cells, T cells, monocytes, and neutrophils) might prove effective in patients with cancer.9-15 A deeper understanding of the mechanisms of synergy between TRAIL and other antitumor drugs and biological agents is therefore worthy of further scientific effort. In this regard, in a recent paper, Carter et al16 demonstrated that triptolide, a diterpenoid compound, was able to sensitize AML cells to TRAIL-induced apoptosis via a decrease of antiapoptotic XIAP and a p53-mediated increase of TRAIL-R2. However, although TRAIL-R1 and TRAIL-R2 are commonly referred to as the “death receptors” for their well-characterized activity of transducing the apoptotic signal, mounting experimental evidence suggests that they are also involved in nonapoptotic functions. In fact, TRAIL has been recently described as a promoter/inhibitor of the maturation of erythrocytes, megakaryocytes, and monocytes.17-21
It is well established that several isoforms of PKC have a role in the regulation of TRAIL activity.19,21-23 In general, PKC isoenzymes play central roles in various cellular signaling pathways, participating in a variety of protein phosphorylation cascades that regulate/modulate cellular structure and gene expression.24-29 Specifically, we found that PKCϵ protects late erythroid precursors from the apoptogenic effects of TRAIL, to which are—on the contrary—exposed in their earlier phases of differentiation.19 In parallel, we also demonstrated that the timing and expression levels of PKCϵ act as regulators of the megakaryocytic differentiation of human CD34 cells.21
A variety of compounds with the capacity to modulate PKC activity have been studied.23,24 These differ in selectivity, potency, and isoenzyme preference. Two such agents, bryostatin 1 and phorbol esters, are being evaluated in clinical trials.30-35 Several phorbol ester–induced effects are mediated by the temporal activation, translocation, and/or suppression of selected PKC isoforms.36-38 In general, acute exposure to phorbol esters activate different PKC isoforms, while chronic exposure down-regulates the activity of these enzymes.36,37,39,40 Phorbol esters induce a broad range of cellular effects, including maturation/differentiation of hematopoietic cell lines41,42 and apoptosis in prostate cancer cells.43 In a preclinical study on prostate cancer cells, phorbol esters (TPA) and paclitaxel in combination had a stronger inhibitory effect on the growth of tumor cells both in vitro and in vivo, suggesting the possibility of using TPA alone or in combination with paclitaxel in patients with prostate cancer.44 Moreover, phorbol esters are also administered at very low concentrations in patients with myeloid leukemias.31,34,35 On the contrary, in a phase 1 clinical trial with escalating doses of phorbol esters in patients with relapsed or refractory malignancies, serious adverse events were described at high doses of administration.35
Given this complex background, we studied here the effects of TRAIL in association with the phorbol ester 4-β-phorbol-12,13-dibutyrate (PDBu) on primary AML cells. We found that PDBu sensitizes AML cells to both the apoptogenic and the prodifferentiative effects of TRAIL by modulating intracellular PKCϵ levels and the molecular signaling machinery downstream apoptogenic TRAIL receptors, without affecting surface TRAIL receptors expression.
Methods
Cell cultures and treatment
Approval for this study was obtained from the Institutional Review Boards of the Department of Anatomy, Pharmacology and Forensic Medicine and of the Department of Clinical Sciences at the University of Parma (Parma, Italy).
Peripheral blood (PB) cells from 11 patients with acute myeloid leukemia (AML) were collected after informed consent was obtained in accordance with the Declaration of Helsinki. Aliquots were immediately used for flow cytometry phenotyping (Table 1). Cells were grown in 20% fetal bovine serum (FBS)–enriched RPMI medium, at the density of 2 × 106 cells/mL, in the presence of 20 nM PDBu (Sigma-Aldrich, St Louis, MO) to induce megakaryocyte (MK) differentiation, in the presence or absence of 25 ng/mL recombinant TRAIL.
Clinical characteristics of patients with AML
| Patient no. . | Sample source . | Blasts, % . | CD33, % . | CD34, % . | FAB . |
|---|---|---|---|---|---|
| 1 | PB | 93 | 87 | 0 | M0/M1 |
| 2 | BM | 90 | 99 | 85 | M4 |
| 3 | BM | 85 | 99 | 99 | M1 |
| 4 | BM | 90 | 99 | 30 | M4 |
| 5 | PB | 45 | 80 | 99 | M0 |
| 6 | BM | 65 | 50 | 99 | M0 |
| 7 | BM | 43 | 52 | 50 | M4 |
| 8 | BM | 60 | 99 | 0 | M5 |
| 9 | PB | 90 | 97 | 14 | M4 |
| 10 | BM | 83 | 76 | 86 | M2 |
| 11 | BM | 95 | 92 | 0 | M5 |
| Patient no. . | Sample source . | Blasts, % . | CD33, % . | CD34, % . | FAB . |
|---|---|---|---|---|---|
| 1 | PB | 93 | 87 | 0 | M0/M1 |
| 2 | BM | 90 | 99 | 85 | M4 |
| 3 | BM | 85 | 99 | 99 | M1 |
| 4 | BM | 90 | 99 | 30 | M4 |
| 5 | PB | 45 | 80 | 99 | M0 |
| 6 | BM | 65 | 50 | 99 | M0 |
| 7 | BM | 43 | 52 | 50 | M4 |
| 8 | BM | 60 | 99 | 0 | M5 |
| 9 | PB | 90 | 97 | 14 | M4 |
| 10 | BM | 83 | 76 | 86 | M2 |
| 11 | BM | 95 | 92 | 0 | M5 |
The CD33 and CD34 surface expression was studied by flow cytometry gating on the blast population. The AML classification was made according to the French- American-British classification (FAB).
TF-1, HeL, and K562 cell lines express surface antigens specific for both the erythroid (glycophorin; CD71) and the megakaryocytic (MK; CD41, CD61, and CD42b) cell lineages. TF-1 cells also express CD34 and are cytokine-dependent for their growth, with a more immature phenotype than HeL and K562 cells.
TF-1, HeL, and K562 cells were grown in 10% FBS-enriched RPMI medium at the optimal density of 0.5 × 106 cells/mL. TF-1 cells were maintained in the presence of 3 ng/mL IL-3 (PeproTech, London, United Kingdom). TF-1, HeL, and K562 cells were treated with the phorbol ester PDBu (TF-1: 5 nM; HeL and K562: 20 nM) to induce MK differentiation in the presence or absence of 25 ng/mL recombinant TRAIL (Alexis Biochemical, San Diego, CA).
Cell morphology was analyzed by light microscopy at day 3 of culture. Aliquots of cultured cells were centrifuged with a StatSpin CytoFuge (StatSpin, Norwood, MA) at 20g for 4 minutes. Slides were stained with Giemsa (Sigma-Aldrich) and examined by an Eclipse 80i digital light microscope (Nikon, Tokyo, Japan). The images were captured using the ACT-2U software (Nikon).
Flow cytometric analysis
Aliquots of 0.3 × 106 cells/experimental point were labeled by a panel of anti–TRAIL-Rs mAbs (Alexis Biochemical). Expression of TRAIL-R1, TRAIL-R2, TRAIL-R3, and TRAIL-R4 was analyzed by indirect staining using 1 μg HS101 anti–human TRAIL-R1, HS201 anti–human TRAIL-R2, HS301 anti–human TRAIL-R3, and HS401 anti–human TRAIL-R4 mAbs, followed by R-phycoerythrin (RPE)–conjugated goat anti–mouse IgG (Immunotech, Luminy, France) as a second reagent.
To follow megakaryocytic differentiation, aliquots of 0.3 × 106 cells/experimental point were labeled with RPE-conjugated anti-CD61 (Pharmingen/Becton Dickinson, San Jose, CA), RPE-conjugated anti-CD41 (Chemicon, Temecula, CA), cyanin-5 (Cy5)–conjugated anti-CD42b (Pharmingen/Becton Dickinson), RPE-conjugated anti-CD34 (Immunotech), RPE-conjugated anti-CD33 (Pharmingen/Becton Dickinson), RPE-conjugated anti-CD14 (Chemicon), and RPE-conjugated anti–glycophorin A (DAKO, Glostrup, Denmark). Working dilutions of all reagents were previously optimized by serial dilution tests. Analysis of the samples was performed by an Epics XL flow cytometer (Beckman Coulter, Fullerton, CA) and the Expo ADC software (Beckman Coulter). The instrument was calibrated daily with a set of standardized beads (DAKO). These consisted of a set of beads each with a different known amount of either FITC, PE, or Cy5 expressed in units of MESF (molecules of equivalent soluble fluorescein). Thus, a standard curve was constructed each day by plotting MESF values for the beads against the median channel in which the peak was displayed.45
To quantify the platelet production in culture, fixed volumes of the culture supernatants were collected, incubated with anti-CD41–RPE and calcein AM (Sigma-Aldrich; to exclude fragments) and added to a fixed volume of calibration beads (DAKO) at known concentration. Both the platelets and beads populations were simultaneously identified in flow cytometry on the forward scatter (FS) versus logarithmic side scatter (SS) dot plot. The absolute platelets count was performed on the gated CD41+/calcein AM+ platelet population.
PKCϵ overexpression, siRNA design, and transfection
The green fluorescent protein (GFP)–PKCϵ expression and control plasmid were kindly provided by Prof Peter Parker (Cancer Research UK, London Research Institute, London, United Kingdom). The murine GFP-tagged PKCϵ and the murine GFP-K552M mutated PKCϵ constructs were cloned in the pCDNA3.1 hygro vector fused with GFP.46
PKCϵ expression levels were down-regulated in primary AML cells by transfection of double-stranded siRNAs (dsRNA) designed to target sequences corresponding to nt 223 to 244, 429 to 450, 942 to 963, and 1158 to 1179 on human PKCϵ mRNA (NM005400). The target sequences were as follows: 5′-aagat caaaa tctgc gaggcc-3′, 5′-aagat cgagc tggctg tcttt-3′, 5′-aacta caagg tccct accttc-3′, and 5′-aaaaa gctca ttgct ggtgcc-3′. The respective sense and antisense RNA sequences were synthesized by the Silencer siRNA Construction Kit (Ambion, Austin, TX).47 Nonspecific siRNA duplexes containing the same nucleotides, but in irregular sequence (ie, scrambled PKCϵ siRNA), were prepared according to the manufacturer's protocol and used as controls.
To maximize transfection efficiency, GFP-PKCϵ plasmids (1 μg/transfection) and PKCϵ siRNAs (100 nM each) were delivered using the Amaxa nucleofection technology (Amaxa, Koeln, Germany) according to the manufacturer's protocols.47
Western blot
Cultured cells were counted and 2 × 106 cells were collected at specific time points, washed in phosphate-buffered saline (PBS), and centrifuged at 200g for 10 minutes. Pellets were resuspended in a cell lysis buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 mM Na3VO4, and 1 mM NaF) supplemented with fresh protease inhibitors, and protein concentration was determined using the BCA protein assay kit (Pierce, Rockford, IL). A total of 30 μg of proteins from each sample were then migrated in 5% SDS-acrylamide gels and blotted onto nitrocellulose filters.
Blotted filters were blocked and incubated with specific primary antibodies diluted as described in the maunfacturers' protocols. Specifically, rabbit polyclonal anti-PKCϵ (Upstate, Lake Placid, NY) and anti–caspase-3 (Cell Signaling, Lake Placid, NY) antibodies were used at the concentration of 1 μg/mL and at a dilution of 1:500, respectively. The mAbs anti–caspase-8 (Alexis Biochemical) and anti-FLIP long (Alexis Biochemicals) were diluted 1:500. The mAb anti–β-actin (Sigma-Aldrich) was diluted 1:5000.
Filters were washed and further incubated for 1.5 hours at room temperature with 1:5000 peroxidase-conjugated anti–rabbit or with 1:2000 peroxidase-conjugated anti–mouse IgG (Pierce) in the primary antibody working solution at room temperature. Specific reactions were revealed with the ECL Supersignal West Pico Chemiluminescent Substrate detection system (Pierce).
Assessment of apoptosis
Cell culture viability was assessed by trypan blue exclusion. Apoptotic cells were identified by flow cytometry either as subdiploid peaks generated by DNA fragmentation or by annexin V/propidium iodide (PI) staining. Specifically, cells were either permeabilized by ethanol in the presence of RNAse H buffer and stained with 50 μg/mL PI, or phosphatidylserine was stained by FITC-conjugated annexin V (ACTIPLATE; Valter Occhiena, Torino, Italy) in Ca2+ and PI staining buffer, following the manufacturer's protocol.
Statistical analysis
The variables were compared between the 4 treatment groups using one-way analysis of variance (ANOVA). Pairwise P values were determined using the Dunnett and t tests. All the statistical tests were performed at the .05 P value.
The combination index (CI) was determined by the Chou and Talalay method and was expressed as the average of the CI values obtained at the ED50, ED75, and ED90. All the experiments were performed 3 times, and the results were expressed as means plus or minus SD. A CI of less than, equal to, and more than 1 indicates synergy, additivity, and antagonism, respectively.
Results
Primary AML cells are resistant to TRAIL
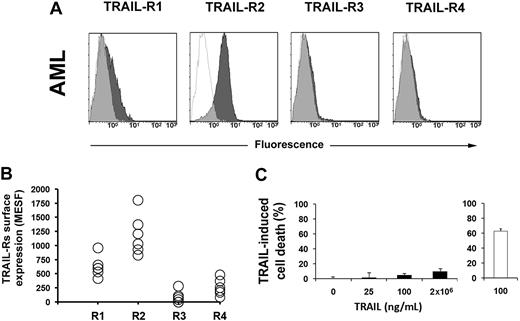
TRAIL is cytotoxic for several tumor cells, including hematological malignancies. To test whether primary AML blasts were sensitive to TRAIL, we first studied their surface expression of TRAIL receptors. Figure 1A and B shows that AML blasts express TRAIL-R2, while the expression level of the other receptors is dim (R1) or hardly detectable (R3 and R4). This was not surprising, since TRAIL-R2 is typically expressed by primary hematopoietic cells. However, soluble TRAIL essentially did not kill leukemic cells (Figure 1C), as previously demonstrated by Riccioni et al.48
TRAIL-Rs expression and sensitivity to TRAIL in AML. (A) Flow cytometric analysis of TRAIL-Rs expression in primary AML cells. Cells were stained with specific mAbs anti–TRAIL-R1, anti–TRAIL-R2, anti–TRAIL-R3, and anti–TRAIL-R4, as described in “Methods.” Specific fluorescence histograms (▩) are superimposed to negative controls (isotype-matched irrelevant mAbs; empty histograms). One representative experiment is shown out of 6. (B) Quantification of TRAIL-Rs expression on the surface of cells from 6 patients with AML. Absolute numbers of surface antigens expressed per cell (MESF). (C) Apoptosis of primary AML cells treated with increasing doses of TRAIL for 48 hours. Jurkat cells were used as control (empty histogram; 100 ng/mL TRAIL for 48 hours). Primary AML cells are resistant to TRAIL-induced apoptosis. Data from 6 independent experiments are expressed as means plus or minus SD.
TRAIL-Rs expression and sensitivity to TRAIL in AML. (A) Flow cytometric analysis of TRAIL-Rs expression in primary AML cells. Cells were stained with specific mAbs anti–TRAIL-R1, anti–TRAIL-R2, anti–TRAIL-R3, and anti–TRAIL-R4, as described in “Methods.” Specific fluorescence histograms (▩) are superimposed to negative controls (isotype-matched irrelevant mAbs; empty histograms). One representative experiment is shown out of 6. (B) Quantification of TRAIL-Rs expression on the surface of cells from 6 patients with AML. Absolute numbers of surface antigens expressed per cell (MESF). (C) Apoptosis of primary AML cells treated with increasing doses of TRAIL for 48 hours. Jurkat cells were used as control (empty histogram; 100 ng/mL TRAIL for 48 hours). Primary AML cells are resistant to TRAIL-induced apoptosis. Data from 6 independent experiments are expressed as means plus or minus SD.
We then tested the sensitivity of TF-1, K562, and HeL hematopoietic cell lines to TRAIL-induced cytotoxicity. All the cell lines expressed TRAIL death receptors (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). At variance with primary AML cells, TRAIL used alone was able to induce apoptosis in the leukemic cell lines (Figure S1B). Cell viability was maximally reduced by 100 ng/mL TRAIL for 24 hours (Figure S1C).
PDBu induces differentiation of myeloid cell lines and primary AML cells
Notwithstanding TRAIL-Rs expressions, primary AML cells—differently from myeloid cell lines—are resistant to TRAIL. Given that the sensitivity of hematopoietic cells to TRAIL largely depends on PKC levels, we asked whether phorbol esters—as general PKC activators used in the therapy of myelogenous leukemia—could sensitize primary AML cells to the apoptogenic effects of TRAIL.
We therefore first performed a dose response set of experiments, looking for the most effective doses of PDBu both in terms of induction of maturation and cytotoxicity of the myeloid cell lines and of the primary AML blasts.
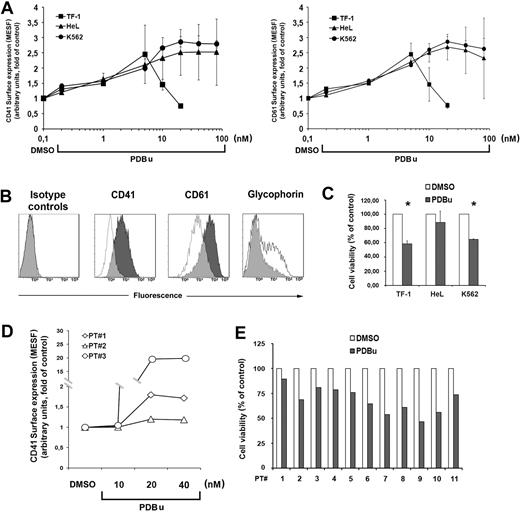
Figure 2A shows the dose-response experiments with increasing doses of PDBu on the cell lines. Differentiation was monitored as CD41, CD61, and glycophorin surface marker expression.
PDBu-induced MK differentiation of leukemic cells. (A) MK differentiation of TF-1, HeL, and K562 cells treated with increasing doses of PDBu for 3 days (left panel, CD41 surface expression; right panel, CD61 surface expression). The maximal cell differentiation was obtained with 5 nM PDBu for TF-1 and 20 nM PDBu for HeL and K562, while higher doses were toxic for the cells. Data from 5 independent experiments are expressed as means plus or minus SD. (B) Immunophenotype of TF-1 cells cultured with 5 nM PDBu for 3 days. DMSO-treated TF-1 cells (empty histograms) as controls. (C) PDBu-related cell death. Cells were treated for 3 days with the optimal doses of PDBu (5 nM for TF-1; 20 nM for HeL and K562). Cell viability is represented as percentage of the control (DMSO). Data from 5 independent experiments are expressed as means plus or minus SD. *P < .05. (D) MK differentiation of primary AML cells treated with increasing doses of PDBu for 3 days. The maximal cell differentiation was obtained with 20 nM PDBu. Data from 3 patients with AML. (E) PDBu-related cell death. Cells were treated with 20 nM PDBu for 3 days. Cell viability is represented as percentage of the control (DMSO). Data are from 11 patients.
PDBu-induced MK differentiation of leukemic cells. (A) MK differentiation of TF-1, HeL, and K562 cells treated with increasing doses of PDBu for 3 days (left panel, CD41 surface expression; right panel, CD61 surface expression). The maximal cell differentiation was obtained with 5 nM PDBu for TF-1 and 20 nM PDBu for HeL and K562, while higher doses were toxic for the cells. Data from 5 independent experiments are expressed as means plus or minus SD. (B) Immunophenotype of TF-1 cells cultured with 5 nM PDBu for 3 days. DMSO-treated TF-1 cells (empty histograms) as controls. (C) PDBu-related cell death. Cells were treated for 3 days with the optimal doses of PDBu (5 nM for TF-1; 20 nM for HeL and K562). Cell viability is represented as percentage of the control (DMSO). Data from 5 independent experiments are expressed as means plus or minus SD. *P < .05. (D) MK differentiation of primary AML cells treated with increasing doses of PDBu for 3 days. The maximal cell differentiation was obtained with 20 nM PDBu. Data from 3 patients with AML. (E) PDBu-related cell death. Cells were treated with 20 nM PDBu for 3 days. Cell viability is represented as percentage of the control (DMSO). Data are from 11 patients.
TF-1 cells showed the highest expression of CD41 and CD61 at 5 nM PDBu for 72 hours: PDBu induces cell death at higher concentrations, while HeL and K562 showed the best response at 20 nM for 72 hours. Glycophorin expression decreased in parallel, as expected (Figure 2B). The large standard deviations observed with high doses of PDBu were due to its relative toxicity, and therefore the lowest concentrations able to increase CD41 and CD61 cells surface expression were chosen. At these concentrations, PDBu induced a variable degree of cell death that, however, did not exceed 40% (Figure 2C). Primary AML cells were induced to the maximal expression of CD41 by 20 nM PDBu for 72 hours with a variable induction of cell death as well (Figure 2D,E).
PDBu sensitizes AML cells to the effects of TRAIL
We then added TRAIL to PDBu-treated AML cells and cell lines. Since it is known that TRAIL can exert both apoptogenic and prodifferentiative effects, we monitored again both phenotypical antigen expression and cell viability.
Figure 3A shows the effects of 25 ng/mL of TRAIL on the PDBu (5 nM)–induced progression of MK differentiation of TF-1 cells. The association of TRAIL to PDBu significantly boosts the expression of MK lineage-related antigens, while glycophorin A expression virtually disappears. Moreover, given that TRAIL per se is apoptogenic in the cell lines, we were not surprised to observe a synergistic effect of TRAIL and PDBu in the induction of cell death, as revealed by the CI calculation at ED50, ED75, and ED90 (Figure 3B).
PDBu sensitizes myeloid cells to TRAIL. (A) Expression of CD41, CD61, and glycophorin antigens on the surface of TF-1 cells cultured with or without 5 nM PDBu in the presence or absence of 25 ng/mL TRAIL for 3 days. Data from 5 experiments are expressed as means plus or minus SD of absolute numbers of surface antigens expressed per cell (MESF). *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (B) Apoptosis of leukemic cell lines treated for 3 days with increasing doses of PDBu (□), TRAIL (○), or both agents (▴). Cell death was determined PI staining. Data from 3 independent experiments are expressed as means plus or minus SD. The combination index (CI) was expressed as the average of the CI values obtained at the ED50, ED75, and ED90 and indicates synergism between TRAIL and PDBu in the induction of cell death. (C) Expression of CD41, CD14, CD33, and CD34 antigens on the surface of AML cells cultured with or without 20 nM PDBu in the presence or absence of 25 ng/mL TRAIL for 3 days. Top panels show the absolute numbers of surface antigens expressed per cell (MESF). Bottom panels show the percentage of positive cells. Tendency lines are calculated as means plus or minus SD. *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (D) Light microscopy of primary AML cells cultured in the presence of TRAIL plus PDBu. Cells progressively acquired an enlarged and multinucleated morphology, typical of differentiating megakaryocytes (Giemsa staining; original magnification, ×40; insets, ×100). (E) Relative number of platelets in the culture of AML cells at day 7 of treatment with TRAIL, PDBu, or their combination. The number of platelets was calculated by flow cytometry on the gated CD41+/calcein AM+ population analyzed in combination with a known number of calibration beads. Data from 3 independent experiments are shown (means ± SD). *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (F) Apoptosis of AML cells treated for 3 days with or without 20 nM PDBu in the presence or absence of 25 ng/mL TRAIL. Cell viability is shown as percentage of the control (DMSO). Data from 11 patients with AML are expressed as means plus or minus SD. *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (G) Cell viability of AML cells cultured with or without 20 nM PDBu in the presence or absence of 25 ng/mL TRAIL. A representative flow cytometric detection of PI incorporation by death cells is shown.
PDBu sensitizes myeloid cells to TRAIL. (A) Expression of CD41, CD61, and glycophorin antigens on the surface of TF-1 cells cultured with or without 5 nM PDBu in the presence or absence of 25 ng/mL TRAIL for 3 days. Data from 5 experiments are expressed as means plus or minus SD of absolute numbers of surface antigens expressed per cell (MESF). *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (B) Apoptosis of leukemic cell lines treated for 3 days with increasing doses of PDBu (□), TRAIL (○), or both agents (▴). Cell death was determined PI staining. Data from 3 independent experiments are expressed as means plus or minus SD. The combination index (CI) was expressed as the average of the CI values obtained at the ED50, ED75, and ED90 and indicates synergism between TRAIL and PDBu in the induction of cell death. (C) Expression of CD41, CD14, CD33, and CD34 antigens on the surface of AML cells cultured with or without 20 nM PDBu in the presence or absence of 25 ng/mL TRAIL for 3 days. Top panels show the absolute numbers of surface antigens expressed per cell (MESF). Bottom panels show the percentage of positive cells. Tendency lines are calculated as means plus or minus SD. *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (D) Light microscopy of primary AML cells cultured in the presence of TRAIL plus PDBu. Cells progressively acquired an enlarged and multinucleated morphology, typical of differentiating megakaryocytes (Giemsa staining; original magnification, ×40; insets, ×100). (E) Relative number of platelets in the culture of AML cells at day 7 of treatment with TRAIL, PDBu, or their combination. The number of platelets was calculated by flow cytometry on the gated CD41+/calcein AM+ population analyzed in combination with a known number of calibration beads. Data from 3 independent experiments are shown (means ± SD). *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (F) Apoptosis of AML cells treated for 3 days with or without 20 nM PDBu in the presence or absence of 25 ng/mL TRAIL. Cell viability is shown as percentage of the control (DMSO). Data from 11 patients with AML are expressed as means plus or minus SD. *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (G) Cell viability of AML cells cultured with or without 20 nM PDBu in the presence or absence of 25 ng/mL TRAIL. A representative flow cytometric detection of PI incorporation by death cells is shown.
More importantly, PDBu sensitizes primary AML cells to TRAIL, promoting both cell differentiation and cell death. The combined treatment of AML cells with PDBu and TRAIL in fact induced a decrease of CD34 and CD33 surface antigen expression, which was accompanied by the increase of CD41 expression, both in terms of percentage of positive cells (Figure 3C bottom panels) and absolute numbers of antigens expressed/cell (Figure 3C top panels). On the contrary, there was no significant modulation of the CD14 expression.
MK differentiation was also studied by morphology and platelet production. Primary AML cells treated with PDBu and TRAIL became larger and multinucleated (Figure 3D) and were able to release increased number of platelets after 7 days of culture (Figure 3E). Moreover, although TRAIL was not cytotoxic to primary AML cells, the association of TRAIL to PDBu significantly enhanced AML cell death (Figure 3F,G).
PDBu down-modulates PKCϵ expression in leukemic cells
Since we have previously demonstrated that the effects of TRAIL in hematopoietic cells largely depend on PKCϵ expression levels, with high PKCϵ levels protective against TRAIL-induced apoptosis, we asked if PDBu could induce a specific modulation of PKCϵ expression.
We therefore assayed the levels of PKCϵ upon PDBu-induced MK differentiation in the presence or absence of 25 ng/mL TRAIL.
As shown in Figure 4, both primary AML cells and cell lines constitutively express PKCϵ at various levels. However, when the cells are treated with PDBu, the PKCϵ levels invariably decrease. TRAIL alone is also able to reduce PKCϵ protein levels, and the association of TRAIL to PDBu always potentiates the reduction (Figure 4B,D).
TRAIL and PDBu down-modulate PKCϵ. (A) Detection of endogenous PKCϵ protein in AML cells by Western blot. PKCϵ was revealed by the specific antibody. β-actin was monitored for protein loading. AML cells were treated for 3 days with DMSO (controls), PDBu (20 nM), and TRAIL (25 ng/mL), alone or in combination. PDBu, TRAIL, and, more evidently, their combination, induce a decrease of PKCϵ. A representative experiment is shown. (B) Levels of PKCϵ protein expression in AML cells treated for 3 days with DMSO (controls), PDBu (20 nM), and TRAIL (25 ng/mL), alone or in combination. Densitometric measurements of Western blots from 6 AML samples (means ± SD). *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone). (C) Detection of endogenous PKCϵ protein in TF-1, HeL, and K562 cells by Western blot. β-actin was monitored for protein loading. TF-1, HeL, and K562 cells were treated for 3 days with DMSO (controls), PDBu (TF-1, 5 nM; HeL and K562, 20 nM), and TRAIL (25 ng/mL), alone or in combination. Although the different cell lines show different basal levels of PKCϵ expression, PDBu, TRAIL and, more evidently, their combination, invariably induce a decrease of PKCϵ. (D) Levels of PKCϵ proteins expression in TF-1, HeL, and K562 cells treated for 3 days with DMSO (controls), PDBu (TF-1, 5 nM; HeL and K562, 20 nM), and TRAIL (25 ng/mL), alone or in combination. Densitometric measurements of Western blots from 5 independent experiments (means ± SD). *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone.
TRAIL and PDBu down-modulate PKCϵ. (A) Detection of endogenous PKCϵ protein in AML cells by Western blot. PKCϵ was revealed by the specific antibody. β-actin was monitored for protein loading. AML cells were treated for 3 days with DMSO (controls), PDBu (20 nM), and TRAIL (25 ng/mL), alone or in combination. PDBu, TRAIL, and, more evidently, their combination, induce a decrease of PKCϵ. A representative experiment is shown. (B) Levels of PKCϵ protein expression in AML cells treated for 3 days with DMSO (controls), PDBu (20 nM), and TRAIL (25 ng/mL), alone or in combination. Densitometric measurements of Western blots from 6 AML samples (means ± SD). *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone). (C) Detection of endogenous PKCϵ protein in TF-1, HeL, and K562 cells by Western blot. β-actin was monitored for protein loading. TF-1, HeL, and K562 cells were treated for 3 days with DMSO (controls), PDBu (TF-1, 5 nM; HeL and K562, 20 nM), and TRAIL (25 ng/mL), alone or in combination. Although the different cell lines show different basal levels of PKCϵ expression, PDBu, TRAIL and, more evidently, their combination, invariably induce a decrease of PKCϵ. (D) Levels of PKCϵ proteins expression in TF-1, HeL, and K562 cells treated for 3 days with DMSO (controls), PDBu (TF-1, 5 nM; HeL and K562, 20 nM), and TRAIL (25 ng/mL), alone or in combination. Densitometric measurements of Western blots from 5 independent experiments (means ± SD). *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone.
PDBu-mediated down-modulation of PKCϵ sensitizes leukemic cells to TRAIL
To formally prove that PDBu-mediated down-modulation of PKCϵ was causal to the sensitization of leukemic cells to TRAIL, we (1) overexpressed PKCϵ or an inactive PKCϵ K522M mutated (PKCϵm) kinase (as negative control)46 in the TF-1 cell line; and (2) down-modulated the expression of PKCϵ by specific siRNA transfection in primary AML cells. Cell viability and differentiation in the presence of PDBu and TRAIL was then evaluated in transfected cells by flow cytometry. Figure 5A shows that TF-1 cells could be efficiently transfected by PKCϵ-GFP constructs, as previously demonstrated.19 PDBu could not sensitize PKCϵ-overexpressing cells to either the cytotoxic effects of TRAIL (Figure 5B) or to its prodifferentiative effects along the megakaryocytic lineage (Figure 5C). On the contrary, the overexpression of the mutated form of PKCϵ was not able to prevent the PDBu-induced sensitization of TF-1 cells to TRAIL.
PKCϵ overexpression or down-modulation. (A) Western blot of PKCϵ in TF-1 cells overexpressing PKCϵ (PKCϵ) or mutated PKCϵ (PKCϵm). Exogenous PKCϵ (PKCϵ-GFP) and endogenous PKCϵ (PKCϵ) proteins are shown in the immunoblot. (B) Cell viability of TF-1 cells overexpressing PKCϵ or PKCϵm. After 24 hours, the cells were treated with or without 5 nM PDBu in the presence or absence of 25 ng/mL TRAIL for 3 days. Data from 3 experiments are expressed as means plus or minus SD. *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (C) PKCϵ overexpression reduces the density of surface MK markers in TF-1 cells. TF-1 cells were transfected with PKCϵ (black histograms, PKCϵ) or mutated PKCϵ (gray histograms, PKCϵm). After 24 hours, the cells were cultured with or without 5 nM PDBu and analyzed 3 days later. Data from 3 experiments are expressed as means plus or minus SD of absolute numbers of surface antigens (MESF) expressed per cell. *P < .05 versus control; #P < .05 PKCϵ versus PKCϵm. (D) Western blot of PKCϵ in primary AML cells transfected with PKCϵ-specific siRNA (PKCϵ siRNA) or control siRNA (Scrambled siRNA). (E) Cell viability of primary AML cells transfected with PKCϵ-siRNA or control siRNA (Scrambled siRNA). After 48 hours, the cells were treated with or without 20 nM PDBu in the presence of 500 ng/mL TRAIL for 3 days. Data from 3 experiments are expressed as means plus or minus SD. *P < .05 versus control; #P < .05 PKCϵ siRNA versus Scrambled siRNA. (F) PKCϵ down-regulation induces the expression of CD41 MK marker in primary AML cells while reducing the density of surface CD33. AML cells were transfected with PKCϵ siRNA or control siRNA (Scrambled siRNA). After 48 hours, the cells were cultured with or without 20 nM PDBu in the presence of 500 ng/mL TRAIL and analyzed 3 days later. Data from 3 experiments are expressed as means plus or minus SD of absolute numbers of surface antigens (MESF) expressed per cell. *P < .05; #P < .05 PKCϵ siRNA versus Scrambled siRNA.
PKCϵ overexpression or down-modulation. (A) Western blot of PKCϵ in TF-1 cells overexpressing PKCϵ (PKCϵ) or mutated PKCϵ (PKCϵm). Exogenous PKCϵ (PKCϵ-GFP) and endogenous PKCϵ (PKCϵ) proteins are shown in the immunoblot. (B) Cell viability of TF-1 cells overexpressing PKCϵ or PKCϵm. After 24 hours, the cells were treated with or without 5 nM PDBu in the presence or absence of 25 ng/mL TRAIL for 3 days. Data from 3 experiments are expressed as means plus or minus SD. *P < .05 versus control; #P < .05 TRAIL plus PDBu versus PDBu alone. (C) PKCϵ overexpression reduces the density of surface MK markers in TF-1 cells. TF-1 cells were transfected with PKCϵ (black histograms, PKCϵ) or mutated PKCϵ (gray histograms, PKCϵm). After 24 hours, the cells were cultured with or without 5 nM PDBu and analyzed 3 days later. Data from 3 experiments are expressed as means plus or minus SD of absolute numbers of surface antigens (MESF) expressed per cell. *P < .05 versus control; #P < .05 PKCϵ versus PKCϵm. (D) Western blot of PKCϵ in primary AML cells transfected with PKCϵ-specific siRNA (PKCϵ siRNA) or control siRNA (Scrambled siRNA). (E) Cell viability of primary AML cells transfected with PKCϵ-siRNA or control siRNA (Scrambled siRNA). After 48 hours, the cells were treated with or without 20 nM PDBu in the presence of 500 ng/mL TRAIL for 3 days. Data from 3 experiments are expressed as means plus or minus SD. *P < .05 versus control; #P < .05 PKCϵ siRNA versus Scrambled siRNA. (F) PKCϵ down-regulation induces the expression of CD41 MK marker in primary AML cells while reducing the density of surface CD33. AML cells were transfected with PKCϵ siRNA or control siRNA (Scrambled siRNA). After 48 hours, the cells were cultured with or without 20 nM PDBu in the presence of 500 ng/mL TRAIL and analyzed 3 days later. Data from 3 experiments are expressed as means plus or minus SD of absolute numbers of surface antigens (MESF) expressed per cell. *P < .05; #P < .05 PKCϵ siRNA versus Scrambled siRNA.
Subsequently, we silenced the PKCϵ expression in AML cells by transfection of specific siRNA and assayed transfected cells for the effects of PDBu and TRAIL. Figure 5D shows that AML cells could be efficiently transfected by PKCϵ-specific siRNA. The down-modulation of PKCϵ not only potentiated the cytotoxic effects of PDBu and TRAIL combination, but also—to a lower extent—made AML cells sensitive to TRAIL alone, as expected (Figure 5E). The MK differentiation of surviving cells was enhanced as well, as shown by the increased expression of surface CD41, while even CD33 expression was decreased (Figure 5F).
Caspase-8, caspase-3, and FLIP in PDBu-treated AML cells
Since PDBu sensitizes AML cells to the effects of TRAIL, we finally studied the expression of major signaling molecules downstream apoptogenic TRAIL receptors.
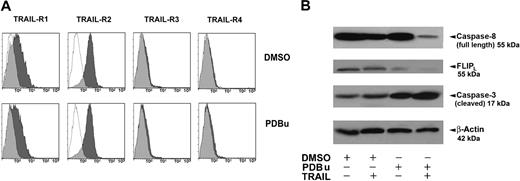
First, PDBu treatment does not modify the surface expression of TRAIL receptors (Figure 6A). Subsequently, we explored the levels of full-length (inactive) caspase-8, FLIPL, and caspase-3, in primary AML cells cultured with PDBu in the presence or absence of 25 ng/mL TRAIL. Primary AML cells expressed high levels of full-length caspase-8, a variable level of FLIP, and low levels of caspase-3 (Figure 6B). Collectively, the data reported in Figure 6B show that (1) PDBu induces a decrease of FLIPL levels, thus predisposing leukemic cells to the apoptogenic effect of TRAIL; and (2) the addition of TRAIL to PDBu induces an activation of the extrinsic apoptotic pathway, as indicated by the marked decrease of procaspase-8 (seemingly due to its cleavage, and thus to its activation) and the clear increase of the cleaved (active) form of caspase-3.
PDBu effects on TRAIL-Rs downstream signaling. (A) Flow cytometric analysis of TRAIL-Rs expression on primary AML cells treated with PDBu or without PDBu (DMSO) 2 nM for 3 days. Specific fluorescence histograms (▩) are superimposed to negative controls (isotype-matched irrelevant mAbs; empty histograms). A representative of 6 independent experiments is shown. (B) Detection of endogenous full-length (inactive) caspase-8, FLIPL, and caspase-3 proteins in AML cells by Western blot. The proteins were revealed by the specific antibodies. β-actin control was used for protein loading. AML cells were treated for 3 days with DMSO (control), PDBu (20 nM), or TRAIL (25 ng/mL), alone or in combination. While TRAIL alone slightly induces caspase-3, PDBu alone also decreases FLIPL levels. However, the combination of PDBu and TRAIL greatly activate caspase-3 and caspase-8 (decrease of full-length, inactive form) at the same time, completely down-modulating FLIPL. A representative of 3 independent experiments is shown.
PDBu effects on TRAIL-Rs downstream signaling. (A) Flow cytometric analysis of TRAIL-Rs expression on primary AML cells treated with PDBu or without PDBu (DMSO) 2 nM for 3 days. Specific fluorescence histograms (▩) are superimposed to negative controls (isotype-matched irrelevant mAbs; empty histograms). A representative of 6 independent experiments is shown. (B) Detection of endogenous full-length (inactive) caspase-8, FLIPL, and caspase-3 proteins in AML cells by Western blot. The proteins were revealed by the specific antibodies. β-actin control was used for protein loading. AML cells were treated for 3 days with DMSO (control), PDBu (20 nM), or TRAIL (25 ng/mL), alone or in combination. While TRAIL alone slightly induces caspase-3, PDBu alone also decreases FLIPL levels. However, the combination of PDBu and TRAIL greatly activate caspase-3 and caspase-8 (decrease of full-length, inactive form) at the same time, completely down-modulating FLIPL. A representative of 3 independent experiments is shown.
Discussion
Besides its well-known apoptogenic effects, TRAIL signaling does not only lead to the activation of effector caspases and apoptosis, but can also induce nonapoptotic pathways, including the activation of mitogen-activated protein (MAP) kinases and NF-κB, leading to cell proliferation and differentiation.49 The differential molecular mechanism(s) underlying the apoptogenic versus the differentiative/proliferative effects of TRAIL are not yet clear, although the NF-κB pathway appears to be relevant.50 However,the expression of antiapoptotic Bcl-2 and Bcl-xL proteins is regulated by PKCϵ levels in hematopoietic cells,19,21 and these proteins have proven to be effective in preventing TRAIL-induced apoptosis in hematopoietic progenitors.
Since (1) induction of differentiation and induction of apoptosis are both useful strategies against cancer in general and leukemia in particular; (2) AML cells are generally resistant to the apoptogenic effects of TRAIL; (3) PKCϵ levels modulate the apoptogenic response of hematopoietic progenitors to TRAIL; and (4) phorbol esters, which induce the translocation of PKC isoforms from the cytosolic to the particulate fraction controlling their access to substrates, induce the maturation of hematopoietic cell lines, and are now used in phase 1 clinical trials against myeloid leukemia, we decided to test the hypothesis that phorbol esters could sensitize AML cells to the effects of TRAIL, an effect that could have an impact on the therapeutic strategies for AML.
Notwithstanding the expression of TRAIL-R2, primary AML cells are resistant to TRAIL, while TRAIL is apoptogenic for the TF-1, K562, and HeL cell lines. Treatment of primary AML cells with 20 nM PDBu for 72 hours induced both cytotoxicity and cell differentiation along the MK lineage. The addition of 25 ng/mL TRAIL to PDBu-treated AML cells synergistically enhanced both cytotoxicity and cell differentiation. Searching for the molecular basis of the sensitization to TRAIL, we immediately focused on PKCϵ, since this isoform of PKC has a key role in the modulation of the effects of TRAIL in normal hematopoiesis. One could imagine that, because phorbol esters are general activators of PKC, induction of terminal MK differentiation of leukemic cells by PDBu would keep PKCϵ at high levels. On the contrary, PKCϵ is down-modulated during PDBu-induced MK differentiation of both primary AML cells and leukemic cell lines, similarly to what happens in normal, TPO-induced, MK differentiation of primary CD34 progenitors.
Overexpression of PKCϵ—but not of a catalytically inactive, mutated form of PKCϵ—abolishes both the cytotoxic and the prodifferentiative effects of TRAIL on PDBu-treated leukemic cells. On the contrary, the down-modulation of PKCϵ levels in AML cells by PKCϵ-specific siRNA enhances both the cytotoxic and the prodifferentiative effects of the association of PDBu and TRAIL. Although other compounds known to sensitize tumor cells to TRAIL, such as the recently described triptolide,16 enhance death receptor expression, this was not the case for PDBu, which did not modify the phenotypical expression of TRAIL-Rs. However, several key molecules of the TRAIL-Rs–dependent apoptotic signaling are modulated by the association of TRAIL to PDBu in AML cells. Specifically, the association of TRAIL and PDBu definitely activate caspase-8 and caspase-3 while decreasing FLIP levels, thus opening the way for a fully apoptogenic signaling in AML cells. Specifically, our results suggest that PDBu predisposes leukemic cells to the apoptogenic effects of TRAIL through a decrease of FLIPL. TRAIL, in conclusion, holds the intuitive advantage of being both cytotoxic and—at the same time—prodifferentiative for the PDBu-treated leukemic cell population. AMLs are characterized by the predominance of immature cells, mainly blasts. Our results suggest that PDBu primarily suppresses the protection of AML blasts from the action of apoptogenic death ligands. In parallel, given the role of PKCs—and specifically PKCϵ—in the MK differentiation, PDBu theoretically creates biologically favorable conditions to allow terminal differentiation of surviving blasts. One limitation of this study is the relative small number of patients studied. Given the differentiative heterogeneity of AML (Table 1), it is, however, tempting to speculate that the combination of PDBu and TRAIL preferentially induces cell death of more immature phenotypes while promoting terminal differentiation of the more mature ones.
Our data are limited to the induction of megakaryocytic differentiation. The analogy of the effects of TRAIL with the normal MK development raises the question of whether this double-faceted effect of TRAIL would also take place when inducing cell differentiation along lineages other than MK. However, it must be noted that a similar prodifferentiative effect of TRAIL has been described in HL-60 cells,17 while ATRA has been demonstrated to induce apoptosis of differentiating promyelocitic cells via a paracrine induction of TRAIL.51
Overall, our data suggest that using TRAIL in combination with phorbol esters (or possibly other, more specific, PKCϵ modulators) could represent a useful strategy to induce both apoptosis and/or terminal differentiation of AML cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Instrumentation Laboratory-Italy for technical support and to Luciana Cerasuolo, Vincenzo Palermo, Domenico Manfredi, and Davide Dallatana for technical support.
This work was supported by Ministero dell'Università e della Ricerca-Programmi di ricerca di Rilevante Interesse Nazional (MUR-PRIN) 2006 grant to G.G., Associazione Italiana Leucemie (AIL) and Programma di Ricerca-Università 2007/2009 grants to M.V., and by a grant from the Associazione Italiana per la Ricerca sul Cancro (AIRC) to A.B.
Authorship
Contribution: G.G. designed and performed research and analyzed data; P.M. designed research and analyzed data; C.C., C. Micheloni, and C. Malinverno performed research; P.L. contributed tools; A.B. analyzed data; and M.V. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marco Vitale, Department of Anatomy, Pharmacology and Forensic Medicine, Human Anatomy Section, University of Parma, Ospedale Maggiore, Via Gramsci 14, I-43100 Parma, Italy; e-mail: marco.vitale@unipr.it.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal