Abstract
Eosinophil chemotaxis and survival within tissues are key components in the development of tissue eosinophilia and subsequent effector responses. In this study, we demonstrate a novel mechanism of eosinophil autoregulation affecting migration and survival mediated through Notch signaling. We show for the first time that human blood eosinophils express Notch receptors and Notch ligands, expressions of which are influenced by the presence of eosinophil-activating granulocyte-macrophage colony-stimulating factor (GM-CSF). Evidence of Notch receptor activation and subsequent transcription of the Notch-responsive gene HES1 were observed in GM-CSF–stimulated eosinophils, confirming functionality of eosinophil-expressed Notch-signaling components. Moreover, by inhibiting Notch signaling with γ-secretase inhibitors or Notch receptor–specific neutralizing antibodies, we demonstrate that autocrine Notch signaling enhances stimulus-mediated actin rearrangement and eosinophil chemokinesis, and impairs eosinophil viability. Taken together, these data suggest autocrine Notch signaling, enhanced in response to tissue- or inflammatory-derived signals, influences eosinophil activity and longevity, which may ultimately contribute to the development of tissue eosinophilia and exacerbation or remediation of eosinophil effector functions.
Introduction
Eosinophils are innate immune leukocytes implicated in the pathogeneses of multiple inflammatory responses, including parasitic helminth infections, RNA viral infections, and allergic diseases (reviewed in Rothenberg and Hogan1 ). Eosinophils are recruited from the circulation by chemotactic factors, including interleukin (IL)–5 and eotaxin-1 (CCL11), and activated within tissues in response to inflammatory-derived mediators.2,3 In chronic airway inflammatory conditions, such as asthma, activated eosinophils within tissues modulate immune responses and elicit effector functions through differential secretion of cytokine, lipid, and cationic protein mediators.1 In contrast, tissue eosinophilia may be protective in the face of some parasitic helminth infections,4,5 and eosinophil-derived RNases are effective combatants against infection with RNA viruses, such as respiratory syncytial virus (RSV).6,7 Delineation of mechanisms mediating chemotaxis, activation, and survival of eosinophils is thus an attractive goal for development of therapies both to alleviate eosinophil-mediated tissue destruction and, conversely, to promote protective functions of eosinophils.
Eosinophil chemotaxis and survival within tissues are enhanced through exposure to specific cytokines, chemokines, and other proinflammatory molecules. Included among the eosinophilopoietins are IL-3, IL-5, and granulocyte-macrophage colony-stimulating factor (GM-CSF), which play critical roles in promoting the generation of eosinophils in the bone marrow, enhancing chemotaxis into tissues, and delaying eosinophil apoptosis within tissues.8 Of note, IL-3, IL-5, and GM-CSF are cytokines also synthesized and released from eosinophils, providing for potential autocrine regulation.1
Notch signaling is a strongly evolutionarily conserved pathway first noted for its principal role in cell-to-cell communications, dictating critical cell fate decisions during development. Signaling is mediated through 4 mammalian Notch receptors (1, 2, 3, and 4) and at least 5 identified Notch ligands, subdivided into 2 families: Jagged 1 (J1) and 2 (J2), and Delta-like 1 (DL1), DL3, and DL4. Binding of Notch ligands to Notch receptors on neighboring cells initiates sequential α- and γ-secretase–mediated proteolytic cleavage events, releasing the intracellular portion of the Notch receptor (NICD) from the plasma membrane of target cells, initiating downstream effects of Notch activation (reviewed in Maillard et al9 ). In addition to critical roles throughout development, recent studies have revealed novel effects of Notch signaling in mature cells, including T and B lymphocytes.9-12 Moreover, Notch signaling is implicated in multiple diverse processes from immune modulation to fibrosis13,14 and oncogenesis.15-17 Currently, inhibition of γ-secretase, which also cleaves the amyloid β precursor protein to produce amyloid β peptide, is in trials as a treatment for Alzheimer disease.18
Notch signaling inhibits eosinophilopoiesis, as demonstrated by enhanced in vitro eosinophil development in the presence of a γ-secretase inhibitor.19 Our studies show, for the first time, that mature human blood eosinophils express Notch receptors and Notch ligands. Eosinophil-expressed Notch ligands are functional, and their expression is enhanced by the eosinophil-activating cytokine GM-CSF. Notch signaling enhances GM-CSF–induced eosinophil polarization and chemokinesis, and diminishes eosinophil viability, demonstrating autoregulatory functions for eosinophil-expressed Notch ligands. This work reveals Notch ligand and receptor expression by circulating human eosinophils, and identifies Notch signaling–mediated autoregulatory mechanisms in eosinophils influencing eosinophil activities and viability in response to tissue- or inflammatory-derived signals. Thus, Notch signaling pathways may provide novel therapeutic targets for treatment of eosinophil-associated pathologies.
Methods
Cell isolation and stimulation
Eosinophils were purified from donor blood by negative selection, as previously described,20 with the exception that hypotonic red blood cell (RBC) lysis was omitted to avoid any potential for RBC lysis to affect eosinophil function. Written informed consent was obtained from donors in accordance with the Declaration of Helsinki, and Institutional Review Board (IRB) approval was obtained from the Beth Israel Deaconess Medical Center. Both mildly atopic and healthy donors were included, with total eosinophils recovered from 320 mL of blood ranging from 8 to 50 million. Briefly, venous blood was collected into a 6% dextran saline solution (Baxter, Deerfield, IL), and RBCs were allowed to sediment. Buffy coat was centrifuged over Ficoll to separate eosinophil-containing granulocyte pellets from peripheral blood mononuclear cells (PBMCs). Eosinophils were isolated from granulocyte pellets by incubation with a depletion antibody (Ab) cocktail (containing Abs against CD2, CD14, CD16, CD19, CD56, and glycophorin A, StemSep; StemCell Technologies, Vancouver, BC), followed by passage over columns (Miltenyi Biotec, Auburn, CA) under magnetic force. Most RBCs present in the granulocyte pellet were retained in the cell separation column during the magnetic negative selection process, as the depletion Ab cocktail includes Abs to glycophorin. Purity of isolated eosinophils was greater than 99% of nucleated cells with no more than 3% contaminating RBCs and viability greater than 99%, as determined by microscopic analysis and trypan blue exclusion, respectively. Human T cells were isolated from the PBMC layer by negative selection using the human CD4+ T-cell enrichment kit (StemSep; StemCell Technologies), per manufacturer's instructions. For overnight incubations, cells were resuspended at 106 cells/mL in RPMI supplemented with 10% FBS (Life Technologies, Grand Island, NY) and 1% penicillin/streptomycin (Life Technologies) with or without GM-CSF (R&D Systems, Minneapolis, MN) at the indicated concentrations and in the presence or absence of inhibitors or dimethylsulfoxide (DMSO) vehicle control, and cultured at 37°C.
Reagents and Abs
Anti-J1 Ab (clone 188331; R&D Systems) was used for immunoprecipitation and Western blotting of J1. Abs specific for extracellular (ab10580; Abcam, Cambridge, MA) and intracellular (clone H-114; Santa Cruz Biotechnology, Santa Cruz, CA) domains of J1 were used for flow cytometry and microscopy. Abs against J2 (clone H-143), Notch 1 (clone H-131), and Notch 2 (clone 25-255), used for flow cytometry, were from Santa Cruz Biotechnology. Anti-Notch 1 (clone A-6; Thermo Scientific, Fremont, CA) was used for immunoprecipitation and Western blotting of Notch 1. Anti-Notch 1 (ab8925; Abcam) was used for detection of the cleaved intracellular domain of Notch 1. Abs against Aph-1 used for flow cytometry and immuno–electron microscopy (EM) were from Abcam. Secondary Abs for immuno-EM were affinity-purified Fab conjugated to 1.4 nm gold (1:100; Nanogold; Nanoprobes, Stony Brook, NY). γ-Secretase inhibitors and their predetermined optimal concentrations included the following: γ-secretase inhibitor II (GSI; Calbiochem, San Diego, CA; 10 μM); N-[N-(3,5-difluorophenacetyl)-L-alanyl)-S-phenylglycine t-butyl ester (DAPT; Calbiochem; 0.2 μM); and L-685,458 (Sigma-Aldrich, St Louis, MO; 0.05 μM). For neutralization, Abs against Notch 1 (clone A-6; Thermo Scientific) were included at a final concentration of 10 μg/mL.
Lysate preparation and Western blotting
Pelleted cells were lysed in lysis buffer (1% Triton X-100, 50 mM Tris-HCl, 300 mM NaCl, 5 mM EDTA (ethylenediaminetetraacetic acid), 0.02% sodium azide, and 1/100 diluted protease inhibitor cocktail (Sigma-Aldrich). Lysates were preincubated with protein A beads conjugated to isotype control Abs. Unbound supernatants were immunoprecipitated with Abs against J1 or Notch 1. Recovered beads were washed well and resuspended in 30 μL reducing buffer. Bound protein was eluted by boiling for 5 minutes. Samples were run on 4% to 12% bis-Tris gels (Invitrogen, Carlsbad, CA) under denaturing conditions, transferred to polyvinylidene difluoride (PVDF) membranes, and blocked overnight with 5% milk before probing with anti-J1 or anti–Notch 1 Abs (2 μg/mL), followed by anti–mouse horseradish peroxidase (HRP)–conjugated secondary Abs (Jackson ImmunoResearch Laboratories, West Grove, PA). Membranes were developed (WestPico chemiluminescence kit; Pierce, Rockford, IL) per the manufacturer's instructions.
Immuno-EM
Pre-embedding immunonanogold EM was performed in frozen 10-μm sections, as previously described.21 Pre-embedding immuno-EM optimizes antigen preservation and is more sensitive to detect small molecules than postembedding labeling done after conventional EM processing. Moreover, to reach antigens at membrane microdomains such as small vesicles, we used labeling with very small (1.4 nm) gold particles (Nanogold). For controls, primary Ab was replaced by an irrelevant isotype control Ab, and primary Ab was omitted. Specimens were examined on a transmission electron microscope (CM 10; Philips, Eindhoven, The Netherlands) at 60 KV.
Flow cytometry and immunofluorescence
For extracellular detection, nonpermeabilized eosinophils were stained with relevant primary Abs or appropriate isotype controls for 30 minutes at 4°C, followed by incubation with Alexa 488 (Invitrogen)– or fluorescein isothiocyanate (FITC; Jackson ImmunoResearch Laboratories)–conjugated secondary Abs (diluted 1/100), for 15 minutes at 4°C in the dark. Cells were washed well and stored in fluorescence-activated cell sorter (FACS) buffer (HBSS−/− + 0.5% BSA) supplemented with 0.5% paraformaldehyde (PFO) at 4°C until analysis. For staining with intracellularly targeted Abs, eosinophils were fixed for 5 minutes at room temperature in 2% PFO, followed by permeabilization for 5 minutes at 4°C in 0.1% saponin-containing FACS buffer supplemented with 2.5% normal goat serum and 2.5% normal human serum. Permeabilized cells were incubated with primary relevant Abs or appropriate isotype controls for 30 minutes at 4°C, followed by incubation with FITC-conjugated secondary Ab (diluted 1/100; Jackson ImmunoResearch Laboratories) in the continued presence of 0.1% saponin. Cells were washed well, resuspended in FACS buffer, and stored at 4°C until analysis.
Data were acquired using a FACScan with CellQuest acquisition and analysis software (BD Biosciences, Franklin Lakes, NJ). Data are presented as percentage of increase in mean fluorescence intensity (MFI), calculated by the following equation: [(geometric MFI of relevant Ab − geometric mean MFI of isotype control Ab) / (geometric MFI of isotype control Ab)] × 100.
Detection of apoptotic cells
Cells were incubated with FITC-conjugated annexin V (1:100; Biovision, Mountain View, CA) and propidium iodide (PI; 1 μg/mL final concentration; Invitrogen) for 30 minutes on ice before analysis by flow cytometry.
RNA preparation and real-time reverse transcription–polymerase chain reaction
Cell pellets were resuspended in 500 μL RNALater (Ambion, Austin, TX) and incubated at 4°C for at least 15 minutes before extraction of total RNA. RNA was extracted from cell pellets using the RNeasy system (QIAGEN, Valencia, CA), per the manufacturer's instructions. cDNA was synthesized using iScript cDNA Synthesis kit (Bio-Rad, Hercules, CA). Real-time reverse transcription–polymerase chain reaction (RT-PCR) was performed with the 7300 thermocycler (Applied Biosystems, Austin, TX) using the iTaq system with carboxy-X-rhodamine (ROX; Bio-Rad). Primers and TaqMan-specific probes for all Notch ligands and Notch receptors, and for 18S rRNA, were from Applied Biosystems. All reactions were performed in triplicate wells, and 18S rRNA levels were used as an internal endogenous control. Results are presented as log 10 (RQ), in which RQ is the relative quantification (or fold-change), as determined by the equation RQ = 2−ΔΔCt, where ΔCt and ΔΔCt (Ct indicates threshold cycle) are defined by the following equations:
Measurement of eosinophil shape change using flow cytometry
Cell pellets were fixed for 5 minutes in 4% PFO at room temperature, washed, and resuspended in FACS buffer until analysis of forward scatter (FSC) parameter by flow cytometry. Fold increase in FSC percentage was calculated as FSC % = (FSC from stimulated eosinophils/FSC from unstimulated eosinophils) × 100. Mean changes in FSC were calculated from triplicate wells per condition.
F-actin staining of eosinophils
Cells were fixed for 5 minutes in 4% PFO at room temperature, permeabilized on ice for 30 minutes in FACS buffer supplemented with 0.5% Triton X-100 and 5% normal goat serum, and incubated for 1 hour with Alexa 488–conjugated phalloidin (1:70; Molecular Probes, Eugene, OR) and Hoescht stain (1:10 000; Molecular Probes) diluted in permeabilization buffer.
Chemokinesis assay
Eosinophils (106/mL) were primed for 5 hours at 37°C with 1 pM GM-CSF or medium alone in the presence or absence of γ-secretase inhibitors or neutralizing Abs. After priming, cells were washed in RPMI supplemented with 0.1% ovalbumin (OVA), resuspended in RPMI plus OVA containing fresh inhibitors, and added (2 × 105 cells in 100 μL) in triplicate to the upper chambers of 24-well transwell plates (Corning Glass, Corning, MA), separated from medium-containing lower wells by polycarbonate inserts (5 μM pore size). Plates were incubated for an additional hour at 37°C before recovering, and migrated cells from the lower compartment were counted. Cells were quantitated from triplicate wells by averaging total events counted in 15 seconds using a flow cytometer.
Statistical analysis
Levels of significance between individual groups or dose-response curves were analyzed by unpaired Student t tests or 2-way ANOVA with Bonferroni posttests, respectively. P values less than .05 were considered statistically significant.
Results
Mature human blood eosinophils express Notch receptors
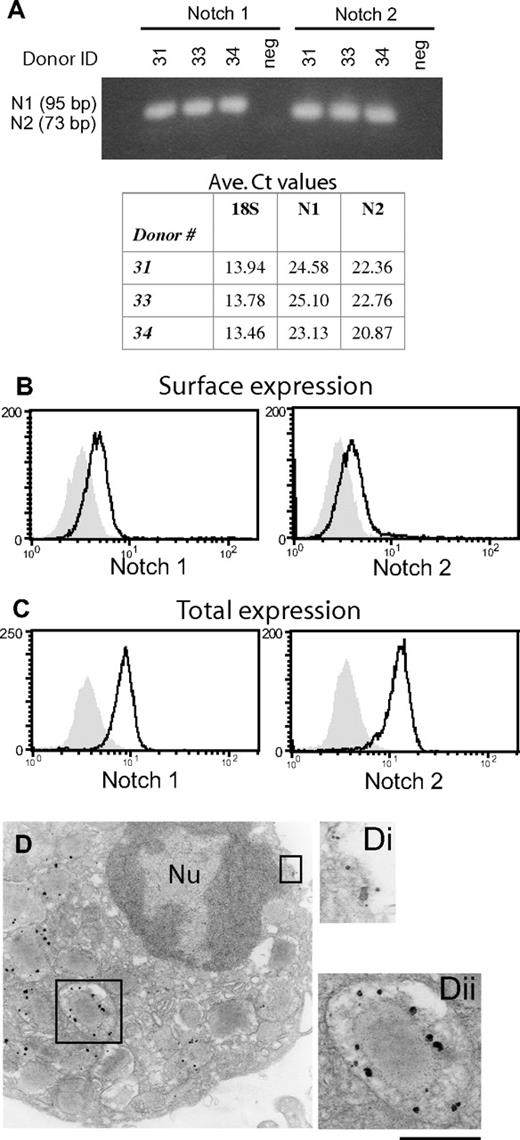
Notch signaling influences eosinophilopoeisis, which may suggest Notch receptor expression by immature eosinophils.19 To determine whether circulating eosinophils might express Notch receptors, RNA was extracted from human blood eosinophils and probed for Notch receptors 1, 2, 3, and 4 mRNA. mRNAs encoding Notch receptors 1 and 2 (Figure 1A), but not 3 or 4 (data not shown), were readily detected in freshly isolated eosinophils.
Human blood eosinophils express Notch receptors. (A) Total RNA extracted from freshly isolated human eosinophils from 3 separate donors was reverse transcribed, and cDNA was probed by real-time PCR for Notch receptor 1 and 2 mRNA expression. neg, no RNA-negative control. Table includes average Ct values for N1 and N2 for comparison with 18S housekeeping controls. Intact (B) or saponin-permeabilized (C) eosinophils were analyzed by flow cytometry for expression of surface or intracellular Notch receptors, respectively. Shaded histogram represents isotype control. (D) Freshly isolated eosinophils were processed for immuno-EM with Abs against Notch receptor 1. Notch 1 expression was observed at the plasma membrane (Di) and within granules (Dii). Bars represent 1 μm (D) and 480 nm (Di,ii). Nu indicates nucleus.
Human blood eosinophils express Notch receptors. (A) Total RNA extracted from freshly isolated human eosinophils from 3 separate donors was reverse transcribed, and cDNA was probed by real-time PCR for Notch receptor 1 and 2 mRNA expression. neg, no RNA-negative control. Table includes average Ct values for N1 and N2 for comparison with 18S housekeeping controls. Intact (B) or saponin-permeabilized (C) eosinophils were analyzed by flow cytometry for expression of surface or intracellular Notch receptors, respectively. Shaded histogram represents isotype control. (D) Freshly isolated eosinophils were processed for immuno-EM with Abs against Notch receptor 1. Notch 1 expression was observed at the plasma membrane (Di) and within granules (Dii). Bars represent 1 μm (D) and 480 nm (Di,ii). Nu indicates nucleus.
Notch 1 protein expression was confirmed by immunoprecipitation from eosinophil whole-cell lysates with anti-N1 Abs (Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). To further investigate localization of eosinophil-expressed Notch receptor proteins, freshly isolated blood eosinophils were incubated, before and after plasma membrane permeabilization, with Abs against extracellular domains of Notch receptors 1 and 2. Notch 1 was detected on the cell surface of eosinophils from 4 of 5 donors (Figure 1B), and within permeabilized cells from all 5 donors (Figure 1C). (See Figure S1B for comparison of relative Notch 1 expression between eosinophils and CD4+ T cells.) Likewise, cell-surface Notch 2 expression was detected on eosinophils from only 2 of 5 donors (Figure 1B), whereas eosinophils from all 5 donors exhibited intracellular Notch 2 receptor expression (Figure 1C).
In full corroboration, immuno-EM localization revealed Notch 1 expression at the cell periphery (Figure 1Di), with a greater density of staining observed intracellularly (Figure 1D). Although cytoplasmic vesicular compartments were labeled, the most robust signal localized to secretory granules (Figure 1Dii).
Eosinophil expression of Aph-1
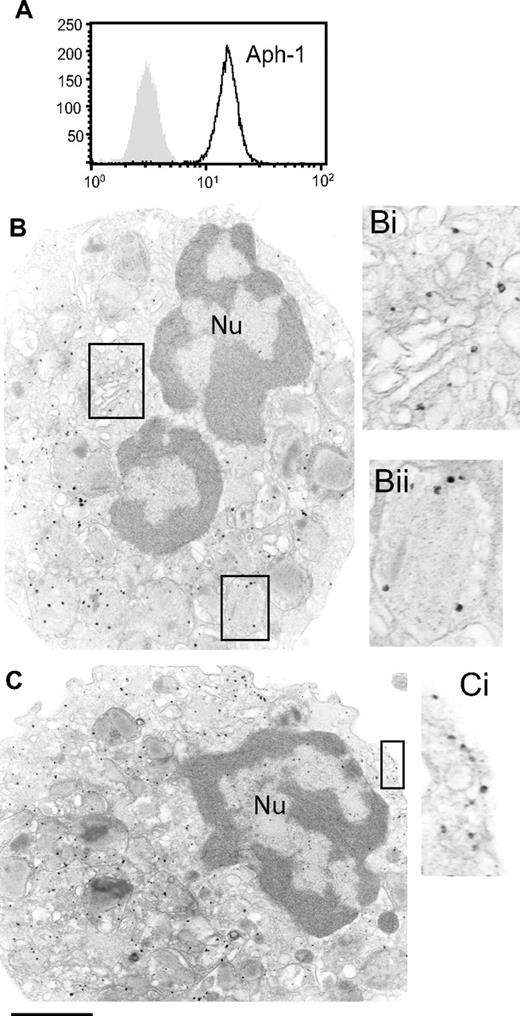
Notch receptor activation requires sequential cleavage events mediated through α- and γ-secretase activities. Functional γ-secretase complexes comprising 4 proteins: presenilin, nicastrin, Pen-2, and Aph-1.22,23 To confirm the potential for classical Notch receptor activation in human eosinophils, we analyzed the expression and cellular localization of γ-secretase complexes as represented by Aph-1 expression. Aph-1, a key component of the functional γ-secretase complex, is thought to be involved in the early stages of γ-secretase complex formation within the endoplasmic reticulum (ER),24-26 and is present as a component of active γ-secretase within the Golgi.27 Freshly isolated human eosinophils stained positively with Abs against Aph-1 (Figure 2A). As anticipated, Aph-1 was expressed within eosinophil Golgi (Figure 2B,Bi). Importantly, Aph-1 was also strongly expressed in association with the plasma membrane (Figure 2C,Ci) and granules (Figure 2B,Bii), reminiscent of the pattern of Notch receptor 1 expression.
Human eosinophils express Aph-1. Freshly isolated human blood eosinophils were analyzed for intracellular Aph-1 expression by flow cytometry of saponin-permeabilized cells (A) and immuno-EM (B,C). Aph-1 expression was observed within the Golgi (Bi), within granules (Bii), and at the cell periphery (Ci). Bars, 870 nm (B,C); 440 nm (Bi,ii and Ci). Nu indicates nucleus. Scale bar equals 1.2 cm.
Human eosinophils express Aph-1. Freshly isolated human blood eosinophils were analyzed for intracellular Aph-1 expression by flow cytometry of saponin-permeabilized cells (A) and immuno-EM (B,C). Aph-1 expression was observed within the Golgi (Bi), within granules (Bii), and at the cell periphery (Ci). Bars, 870 nm (B,C); 440 nm (Bi,ii and Ci). Nu indicates nucleus. Scale bar equals 1.2 cm.
Mature human blood eosinophils express Jagged and Delta family Notch ligands
To determine whether circulating eosinophils might express Notch ligands in addition to Notch receptors, freshly isolated blood eosinophils were evaluated by real-time RT-PCR for expression of the 5 known Notch ligands: J1, J2, DL17, DL3, and DL4. J1 mRNA was identified in eosinophils from 21 of 22 donors analyzed (95%), whereas J2, DL1, and DL4 mRNAs were detected in cells from 6 of 13 (46%), 3 of 7 (43%), and 5 of 7 (71%) donors, respectively. DL3 mRNA was not detected in any donor tested. Gel analyses of recovered PCR products revealed amplicons of the expected sizes for J1, J2, DL1, and DL4, further confirming specificity of results (data not shown). Table 1 shows results from 5 donors probed simultaneously for mRNA expression of all Notch ligands, and results from all donors analyzed are included in Table S1.
Human blood eosinophils constitutively express Jagged 1 and differentially express Jagged 2, Delta 1, and Delta 4
| Donor no. . | Sex . | Eos count*, ×106 . | Jagged 1 . | Jagged 2 . | Delta 1 . | Delta 3 . | Delta 4 . |
|---|---|---|---|---|---|---|---|
| 4 | F | 27.9 | + | + | + | − | + |
| 11 | M | 39.5 | + | − | − | − | − |
| 16 | F | 8.3 | + | − | − | − | + |
| 18 | F | 21.2 | + | + | − | − | + |
| 24 | F | 15.5 | + | − | − | − | − |
| Donor no. . | Sex . | Eos count*, ×106 . | Jagged 1 . | Jagged 2 . | Delta 1 . | Delta 3 . | Delta 4 . |
|---|---|---|---|---|---|---|---|
| 4 | F | 27.9 | + | + | + | − | + |
| 11 | M | 39.5 | + | − | − | − | − |
| 16 | F | 8.3 | + | − | − | − | + |
| 18 | F | 21.2 | + | + | − | − | + |
| 24 | F | 15.5 | + | − | − | − | − |
Eosinophil (Eos) count is total number of eosinophils isolated from 320 mL whole blood.
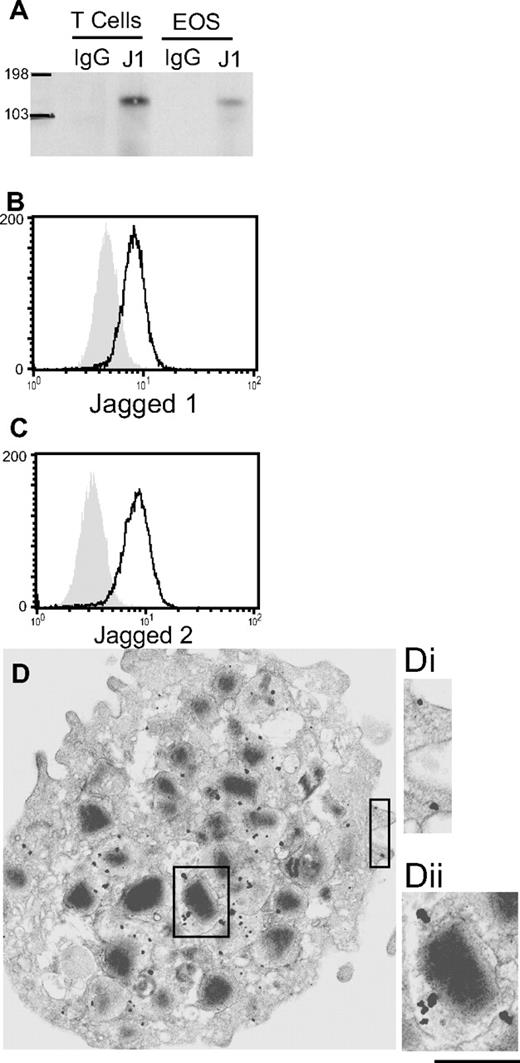
In light of constitutive J1 mRNA expression by blood eosinophils, we investigated protein expression of Jagged family molecules. Western blotting of anti-J1-Ab–immunoprecipitated eosinophil lysates revealed J1 protein expression (Figure 3A). (See Figure S1C for comparison of eosinophil and T cell J1 expression levels.) Using a different anti-J1 Ab to confirm these results by flow cytometry, J1 protein was detected in eosinophils from 11 of 12 donors evaluated (Figures 3B and 4A), extending the findings of constitutive J1 mRNA expression in eosinophils (Table 1) to document eosinophil expression of J1 protein. Total J2 protein expression by eosinophils was detected by flow cytometry in 7 of 12 donors (Figures 3C and 4A), approximating the percentage of donors found to express detectable J2 mRNA.
Human blood eosinophils express Notch ligands J1 and J2 protein. (A) Freshly isolated blood eosinophils or CD4+ T cells (positive control) were lysed and immunoprecipitated with anti-J1 or isotype control Abs. Immunoprecipitated proteins were immunoblotted with anti-J1 Abs. n = 3. Saponin-permeabilized eosinophils were analyzed by flow cytometry for expression of J1 (B) or J2 (C). Shaded histograms = isotype control. (D) Freshly isolated eosinophils were processed for immuno-EM and stained with Abs against J1. J1 expression was observed at the plasma membrane (Di) and within eosinophil-specific granules (Dii). Bars represent 1.1 μm (D) and 650 nm (Di,ii).
Human blood eosinophils express Notch ligands J1 and J2 protein. (A) Freshly isolated blood eosinophils or CD4+ T cells (positive control) were lysed and immunoprecipitated with anti-J1 or isotype control Abs. Immunoprecipitated proteins were immunoblotted with anti-J1 Abs. n = 3. Saponin-permeabilized eosinophils were analyzed by flow cytometry for expression of J1 (B) or J2 (C). Shaded histograms = isotype control. (D) Freshly isolated eosinophils were processed for immuno-EM and stained with Abs against J1. J1 expression was observed at the plasma membrane (Di) and within eosinophil-specific granules (Dii). Bars represent 1.1 μm (D) and 650 nm (Di,ii).
Notably, detection of cell-surface J1, using an extracellularly targeted anti-J1 Ab by flow cytometry or immunofluorescence microscopy, revealed J1 surface expression in only 2 of 6 experiments (Figure S1D), suggesting J1, like Notch receptors, might be sequestered within circulating eosinophils. In full support, immuno-EM of freshly isolated eosinophils revealed detectable J1 (Figure 3D,Di) and J2 (data not shown) expression at the cell periphery, with more dense expression of J1 and J2 protein localized to specific granules (Figure 3D,Dii, and data not shown).
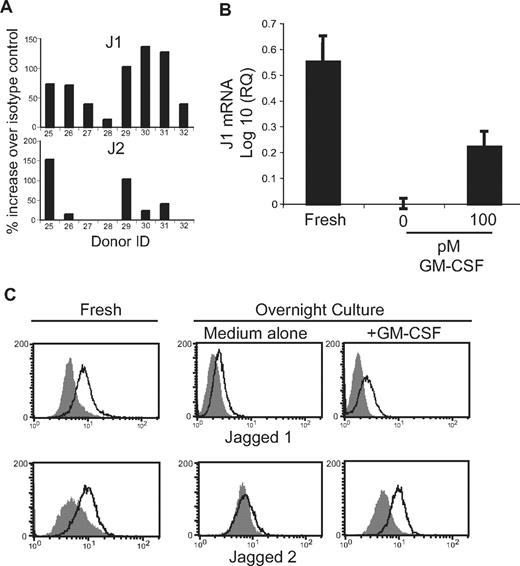
J1 and J2 protein expression is influenced by GM-CSF
Observed levels of J1 and J2 proteins between donors ranged from 6 to 150% of isotype control fluorescence intensities (Figure 4A). Expressions of J1 and J2 were not statistically correlated (Pearson coefficient = 0.34, R2 = 0.1125); however, J2 was not detected in cells with relatively low (< 50-fold increase over isotype control) levels of J1 protein. It is unclear whether the varying levels of J1 and J2 expression by eosinophils singularly represent in vivo expression levels and/or reflect ex vivo down-regulation of their expression. That J1 and J2 expression may be transitory is supported by the finding that J1 mRNA expression decreased upon culture in the absence of exogenous stimuli and remained higher in the presence of GM-CSF (Figure 4B). Likewise, detection of both J1 and J2 proteins is decreased, often to undetectable levels, after overnight culture in the absence of exogenous stimuli (Figure 4C). Protein levels of both J1 and J2 were at least partially maintained upon inclusion of 100 pM of the eosinophil-activating cytokine GM-CSF in the culture medium (Figure 4C), suggesting environmental signals may be integral to maintaining Notch ligand expression by eosinophils in vivo. Despite GM-CSF–induced enhancement of total Notch ligand expression by eosinophils, we failed to detect an increase in surface expression by flow cytometry of J1, Notch 1, or Notch 2 after overnight incubation with 100 pM GM-CSF, despite evidence by electron microscopy of significant eosinophil activation and granule mobilization at this dose (data not shown; n = 3).
J1 and J2 protein expression is influenced by GM-CSF. (A) Saponin-permeabilized eosinophils were analyzed by flow cytometry for expression of J1 and J2. Data are presented as percentage of increase in MFI over isotype control. (B) RNA extracted from eosinophils immediately after isolation (Fresh) or after overnight culture in the presence or absence of 100 pM GM-CSF was evaluated by real-time RT-PCR for J1 mRNA expression. Data are presented as the log 10 of the relative quantification, using overnight culture in medium alone as the baseline value. Data are from 1 experiment representative of 3 separate experiments. (C) Total J1 (top panels) or J2 (bottom panels) protein was detected in saponin-permeabilized eosinophils immediately after isolation (Fresh) or after overnight culture in the presence or absence of 100 pM GM-CSF. Shaded histograms = isotype control.
J1 and J2 protein expression is influenced by GM-CSF. (A) Saponin-permeabilized eosinophils were analyzed by flow cytometry for expression of J1 and J2. Data are presented as percentage of increase in MFI over isotype control. (B) RNA extracted from eosinophils immediately after isolation (Fresh) or after overnight culture in the presence or absence of 100 pM GM-CSF was evaluated by real-time RT-PCR for J1 mRNA expression. Data are presented as the log 10 of the relative quantification, using overnight culture in medium alone as the baseline value. Data are from 1 experiment representative of 3 separate experiments. (C) Total J1 (top panels) or J2 (bottom panels) protein was detected in saponin-permeabilized eosinophils immediately after isolation (Fresh) or after overnight culture in the presence or absence of 100 pM GM-CSF. Shaded histograms = isotype control.
Eosinophil-expressed Notch ligands are functional
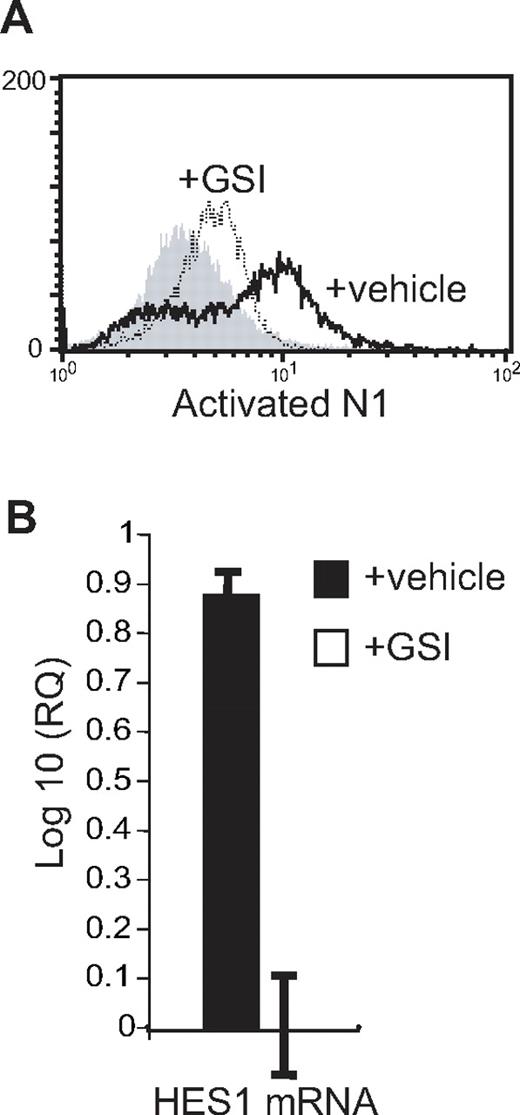
Dual expression by mature eosinophils of Notch ligands and receptors enabled us to investigate the functional potential of eosinophil-expressed Notch ligands through autocrine assays. Upon Notch receptor activation, sequential specific cleavage events result in the release of a portion of the Notch intracellular domain (NICD), which subsequently translocates into the nucleus, where it binds CBF1, suppressor of hairless, Lag-1 (CSL), converting this complex into a transcriptional activator, turning on specific gene transcription.28 To monitor activation of Notch receptors, purified eosinophils were stimulated with GM-CSF in the presence of either the Notch signaling inhibitor GSI or vehicle control, and assessed by flow cytometry at 30, 60, and 120 minutes for the presence of NICD using an Ab recognizing the cleaved, but not the intact, intracellular domain of Notch receptor 1. Although precise kinetics of Notch receptor activation between individual donors varied from 30 to 120 minutes of stimulation, detection of activated NICD in GM-CSF–stimulated eosinophils was diminished in the presence of GSI in 5 of 7 experiments (Figure 5A).
Eosinophil-expressed Notch ligands are functional. (A) Eosinophils were stimulated for 60 minutes with 100 pM GM-CSF in the presence of 10 μM GSI (dotted line) or vehicle alone (heavy line) before permeabilization and detection by flow cytometry of the cleaved, intracellular domain of activated Notch receptor 1. Shaded histogram represents isotype control. (B) RNA was extracted from eosinophils after overnight stimulation with 100 pM GM-CSF in the presence of 10 μM GSI or vehicle control, and analyzed by real-time RT-PCR for the presence of HES1 mRNA. Data are presented as the log 10 of the relative quantification using the GSI-treated sample as the baseline value. Data are from a representative of 3 separate experiments.
Eosinophil-expressed Notch ligands are functional. (A) Eosinophils were stimulated for 60 minutes with 100 pM GM-CSF in the presence of 10 μM GSI (dotted line) or vehicle alone (heavy line) before permeabilization and detection by flow cytometry of the cleaved, intracellular domain of activated Notch receptor 1. Shaded histogram represents isotype control. (B) RNA was extracted from eosinophils after overnight stimulation with 100 pM GM-CSF in the presence of 10 μM GSI or vehicle control, and analyzed by real-time RT-PCR for the presence of HES1 mRNA. Data are presented as the log 10 of the relative quantification using the GSI-treated sample as the baseline value. Data are from a representative of 3 separate experiments.
The basic helix-loop-helix transcription factor, hairy/enhancer of split homologue-1 (HES1), is an early downstream target of Notch signaling in many cell types.9 To confirm the functional potential of eosinophil-expressed Notch ligands using a downstream target, HES1 mRNA expression was compared in GM-CSF–stimulated eosinophil cultures in the presence or absence of GSI. As demonstrated in Figure 5B, HES1 mRNA levels were effectively suppressed in the absence of Notch signaling. These data demonstrate that Notch ligands expressed by human eosinophils are functional, that the GSI is successfully inhibiting Notch receptor signaling, and that autoregulatory Notch signaling occurs in human eosinophils.
Treatment with GSI promotes eosinophil viability
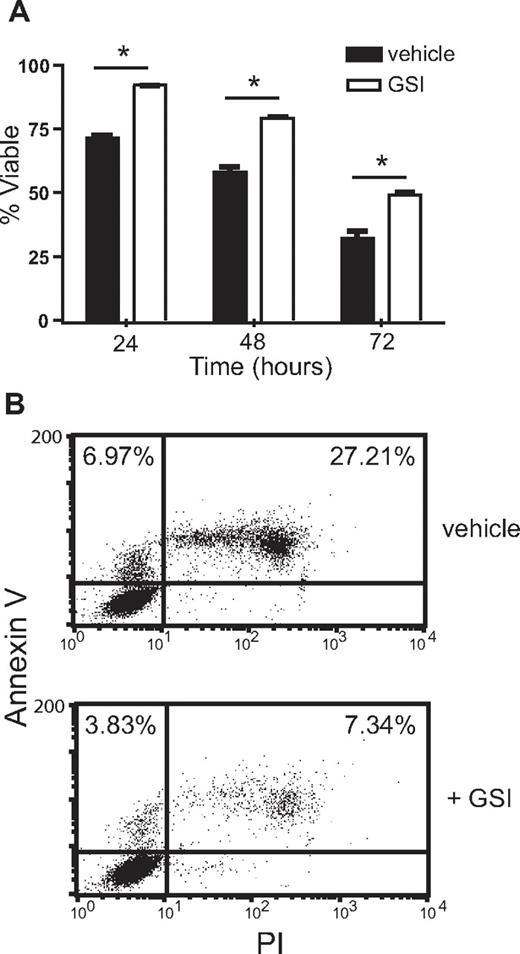
To determine whether Notch signaling might regulate eosinophil activities, eosinophil longevity was evaluated in the presence or absence of GSI. In the absence of exogenously added growth factors, 70% to 75% of eosinophils remained viable after overnight incubation in serum-containing medium. However, when GSI was included in the culture medium, eosinophil viability remained at 85%-90% after 24-hour culture. Inhibition of Notch signaling improved cell viability at 24, 48, or 72 hours by approximately 20% (Figure 6A; see Figure S2 for GSI dose response). Similar results were obtained using either uptake of PI or trypan blue as indicators of cell death (data not shown).
Treatment with γ-secretase inhibitor promotes eosinophil viability. (A) Eosinophils incubated for 24, 48, or 72 hours in the presence of 10 μM GSI or vehicle alone were recovered and incubated on ice with PI (1 μg/mL) for 30 minutes before analysis of PI incorporation by flow cytometry. Data shown are means (± SD) from a representative of 3 independent experiments. (B) Eosinophils recovered after 24 hours of incubation in the presence of 10 μM GSI or vehicle alone were analyzed for exposed annexin V and uptake of PI to differentiate dead from apoptotic cells. Percentages are calculated from total events and represent apoptotic (annexin V+, PI−) and dead (annexin V+, PI+) populations. Data shown are from 1 representative of 3 independent experiments.
Treatment with γ-secretase inhibitor promotes eosinophil viability. (A) Eosinophils incubated for 24, 48, or 72 hours in the presence of 10 μM GSI or vehicle alone were recovered and incubated on ice with PI (1 μg/mL) for 30 minutes before analysis of PI incorporation by flow cytometry. Data shown are means (± SD) from a representative of 3 independent experiments. (B) Eosinophils recovered after 24 hours of incubation in the presence of 10 μM GSI or vehicle alone were analyzed for exposed annexin V and uptake of PI to differentiate dead from apoptotic cells. Percentages are calculated from total events and represent apoptotic (annexin V+, PI−) and dead (annexin V+, PI+) populations. Data shown are from 1 representative of 3 independent experiments.
To confirm that increased cell viability in the presence of GSI reflected a diminished rate of apoptosis, eosinophils recovered after 24 hours of culture were simultaneously stained with PI and annexin V to detect dead and apoptotic cells, respectively. The presence of increased numbers of apoptotic cells (annexin V+, PI−) in the control, vehicle-treated wells indicates that GSI treatment increases eosinophil viability through inhibition of apoptosis (Figure 6B).
Treatment with GSI diminishes activation-induced actin rearrangement in human blood eosinophils
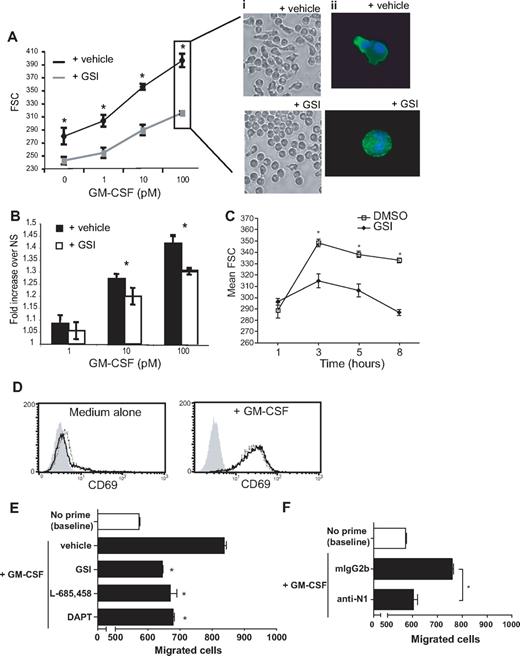
To explore additional potential Notch signaling–mediated autoregulatory effects on eosinophils, activation-induced shape changes were monitored in eosinophils after stimulation with GM-CSF in the presence or absence of GSI. As demonstrated in Figure 7A, Ai and Aii, eosinophils stimulated with GM-CSF undergo a dose-dependent shape change defined by a rearrangement of the actin cytoskeleton that can be objectively quantified through comparison of forward side scatter (FSC) properties using flow cytometry, as previously described.29 Briefly, the FSC parameter of flow cytometry is a measurement of the light-scattering profile of individual cells. Thus, a round cell will elicit a decreased FSC value accordant with its minimal light-scattering potential in comparison with an elongated cell. (Total cellular events are included as histograms in Figure S3A.) Notch signaling did not appear to influence rapid changes in shape occurring within 1 hour of GM-CSF stimulation. However, at time points greater than 3 hours, a dose-dependent impairment in GM-CSF–induced eosinophil shape change was observed in the presence of increasing concentrations of GSI (Figure 7C; see Figure S3B for GSI dose response), further defined by a lack of polarized actin rearrangement (Figure 7Aii). Eosinophil viability after 24-hour incubations in the presence of GM-CSF was greater than 98% in both vehicle- and GSI-treated groups, as determined by trypan blue exclusion and uptake of PI (data not shown), suggesting cell death does not contribute to GSI-inhibitable shape changes. Cultures of eosinophils alone, without exogenous stimuli, revealed a basal level of Notch-mediated shape change (compare 0-pM data points in the presence or absence of GSI; Figure 7A). Thus, to determine whether the apparent inhibition of GM-CSF–induced shape change was solely a reflection of decreased basal eosinophil activation or indicative of synergistic effects between Notch and GM-CSF signaling, FSC values were normalized to the nonstimulated (NS) values of the corresponding condition. By this reanalysis, GM-CSF–induced shape change (fold increase in FSC above nonstimulated baseline) remains significantly impaired in the presence of GSI (Figure 7B), suggestive of synergy between GM-CSF and Notch signaling pathways. Notably, impairment of GM-CSF–induced eosinophil polarization in the presence of GSI did not reflect an overall defect in eosinophil activation, as GM-CSF–induced surface expression of CD69 was not impaired on eosinophils in the presence of GSI after 24 hours of stimulation (Figure 7D).
Notch signaling is required for GM-CSF–induced eosinophil shape change and chemokinesis. (A) Eosinophils were cultured overnight with increasing doses of GM-CSF in the presence of 10 μM GSI or vehicle alone, fixed with 4% PFO, and analyzed using the FSC parameter of flow cytometry to assess cell shape changes. Data represent average FSC geometric means (± SD) for 1 experiment representative of 8 independent experiments. (i) Live cells were imaged using phase microscopy with an inverted microscope before fixation (imaged with a Nikon TE-300 inverted microscope coupled to a Retiga Exi cooled digital camera; images acquired using QImaging software). (ii) Fixed and permeabilized cells were stained with Alexa 488–conjugated phalloidin to detect F-actin filaments and Hoescht for nuclear staining, as described in “Methods” (imaged with a BX62 Olympus upright microscope, 60× PlanApo objective with numerical aperture of 1.42, coupled to a Hamamatsu Orca AG fire-wire cooled digital camera; images acquired using IPLab). (B) Data from panel A were normalized to the relevant nonstimulated values and replotted. (A,B) Statistical significances between curves were confirmed using a 2-way ANOVA test, followed by Bonferroni posttests to determine significances at individual doses. (C) Eosinophils were stimulated with 100 pM GM-CSF in the presence of 10 μM GSI or vehicle alone for 1, 3, 5, or 8 hours. (D) Eosinophils were cultured overnight in medium alone (top panel) or with 100 pM GM-CSF (bottom panel) in the presence of 10 μM GSI (dotted line) or vehicle alone (solid line) before staining for CD69 surface expression. Data shown are from 1 experiment representative of 3 independent experiments. Shaded histogram represents isotype control. (E,F) Eosinophils were primed for 5 hours with 1 pM GM-CSF in the presence or absence of γ-secretase inhibitors (E) or neutralizing Abs against Notch 1 (F). After pretreatment, cells were added to upper wells of chemotaxis chambers, and chemokinesis within 1 hour was determined by quantifying cells migrating to the lower wells. *P < .05 versus vehicle alone or isotype control–treated samples.
Notch signaling is required for GM-CSF–induced eosinophil shape change and chemokinesis. (A) Eosinophils were cultured overnight with increasing doses of GM-CSF in the presence of 10 μM GSI or vehicle alone, fixed with 4% PFO, and analyzed using the FSC parameter of flow cytometry to assess cell shape changes. Data represent average FSC geometric means (± SD) for 1 experiment representative of 8 independent experiments. (i) Live cells were imaged using phase microscopy with an inverted microscope before fixation (imaged with a Nikon TE-300 inverted microscope coupled to a Retiga Exi cooled digital camera; images acquired using QImaging software). (ii) Fixed and permeabilized cells were stained with Alexa 488–conjugated phalloidin to detect F-actin filaments and Hoescht for nuclear staining, as described in “Methods” (imaged with a BX62 Olympus upright microscope, 60× PlanApo objective with numerical aperture of 1.42, coupled to a Hamamatsu Orca AG fire-wire cooled digital camera; images acquired using IPLab). (B) Data from panel A were normalized to the relevant nonstimulated values and replotted. (A,B) Statistical significances between curves were confirmed using a 2-way ANOVA test, followed by Bonferroni posttests to determine significances at individual doses. (C) Eosinophils were stimulated with 100 pM GM-CSF in the presence of 10 μM GSI or vehicle alone for 1, 3, 5, or 8 hours. (D) Eosinophils were cultured overnight in medium alone (top panel) or with 100 pM GM-CSF (bottom panel) in the presence of 10 μM GSI (dotted line) or vehicle alone (solid line) before staining for CD69 surface expression. Data shown are from 1 experiment representative of 3 independent experiments. Shaded histogram represents isotype control. (E,F) Eosinophils were primed for 5 hours with 1 pM GM-CSF in the presence or absence of γ-secretase inhibitors (E) or neutralizing Abs against Notch 1 (F). After pretreatment, cells were added to upper wells of chemotaxis chambers, and chemokinesis within 1 hour was determined by quantifying cells migrating to the lower wells. *P < .05 versus vehicle alone or isotype control–treated samples.
GM-CSF–primed chemokinesis is mediated through Notch receptor 1
Actin-mediated cell shape change is considered a prerequisite for cell migration and chemotaxis. GM-CSF priming induces random, chemokinetic migration of eosinophils30 and enhances eosinophil chemotactic responses to subsequent stimulation (ie, with IL-8 or leukotriene B4 [LTB4]) in vitro.31 To determine whether Notch signaling might also be required for optimal GM-CSF–induced chemokinetic responses, eosinophils were primed with GM-CSF for 5 hours in the presence or absence of GSI before subsequent assessment of cell migration in a chemokinesis assay. As shown in Figure 7E, inclusion of GSI was sufficient to counteract the GM-CSF–priming effect on cell migration. To validate our findings, 2 additional γ-secretase inhibitors with distinct compositions (DAPT and L-685,458) were used in parallel and revealed a similar requirement for intact Notch signaling for GM-CSF–primed chemokinesis (Figure 7E).
Although the γ-secretase inhibitors used in this study effectively depress Notch activation in human eosinophils (as confirmed in Figure 5), and are used at concentrations designed to favor selective Notch signaling inhibition, it is plausible that Notch-independent side effects of the inhibitors may be responsible for the observed effects. To directly link the Notch pathway to GM-CSF–primed effects on eosinophils, chemokinesis assays were performed in the presence or absence of neutralizing Abs against Notch receptor 1. Compared with cells treated with irrelevant isotype control Abs, inclusion of anti-Notch 1 Abs during priming effectively inhibited GM-CSF–induced eosinophil chemokinesis (Figure 7F).
Discussion
Enhanced eosinophil chemotaxis, activation, and survival within inflamed tissues are key components in the development of tissue eosinophilia and subsequent eosinophil effector responses. Delineating the mechanistic bases for these critical functions is requisite for the development of more specifically targeted therapeutic approaches. Our study is the first to demonstrate that mature human blood eosinophils express functional Notch receptors and Notch ligands and are capable of Notch-mediated autocrine signaling. Inhibition of Notch signaling resulted in enhanced viability, decreased actin polarization, and diminished chemokinesis of eosinophils, identifying autoregulatory roles for Notch signaling in mature human eosinophils, which may provide novel therapeutic targets for intervention in eosinophil-associated diseases.
Previous work by Kang et al demonstrated a role for Notch signaling in eosinophil maturation.19 In this study, we have demonstrated that mature human eosinophils within the blood circulation maintain protein expression of Notch receptors and ligands. Parallel intra- and extracellular staining revealed Notch ligands and receptors to be more robustly expressed within eosinophils as opposed to on the cell surface, indicating Notch receptors and ligands may be sequestered within the cell. In full support, immuno-EM demonstrated strong association of Notch receptors and ligands with eosinophil-specific granules. Intracellular granules may serve as a sequestered repository for Notch receptors and ligands, preventing unwarranted cell-autonomous Notch activation while maintaining a rapidly mobilizable cache of signaling components. Alternatively, localization of Notch receptors, ligands, and γ-secretase complexes all to eosinophil granules might hint at an as yet undefined intracellular signaling mechanism focally centered at the granules. Functional responses presented in this work, however, are most likely resultant from canonical, cell-surface expression, as evidenced by the ability of exogenously added anti-Notch 1–neutralizing Abs to inhibit GM-CSF–primed chemokinesis (Figure 7F).
Constitutive J1 expression and apparent donor-dependent variability in expression of J2, DL1, and DL4 indicate plasticity in the potential eosinophil Notch ligand repertoire, and suggest that eosinophils may maintain the capacity to initiate varied effects dependent on specific ligand expression. That Notch ligand expression might be influenced by environmental cues is evidenced by the ability of GM-CSF stimulation to enhance J1 and J2 protein expression (Figure 4B,C). Thus, it is reasonable to speculate that activating signals within inflamed tissues promote Notch activation in eosinophils through regulation of specific Notch ligand expression, and may provide a mechanism for inflammation-induced eosinophil autoregulation.
Eosinophil-expressed Notch receptors and ligands are functional, as evidenced by autocrine, GSI-inhibitable induction of both the cleaved fragment of activated Notch receptor 1 and downstream transcription of the Notch-responsive gene HES1 (Figure 5). In addition to demonstrating the functional competence of eosinophil-expressed Notch ligands, these data establish that Notch-mediated autoregulatory signaling can occur in human blood eosinophils. Our data specifically identify downstream effects of autoregulatory Notch signaling on eosinophil: (1) viability; (2) GM-CSF–induced shape changes; and (3) GM-CSF–primed chemokinesis.
Inclusion of GSI in eosinophil cultures improved cell viability (Figure 6). Although the precise mechanism(s) is yet to be determined, cell death in the presence of intact Notch signaling occurred by apoptosis, as determined by staining with annexin V and PI. These findings may be particularly relevant in light of a recent clinical trial that revealed a mean increase of 48% in blood eosinophils in patients receiving a γ-secretase inhibitor compared with those patients receiving placebo treatment (P < .001 between treatment and placebo groups).18 Whether this increase in eosinophil numbers reflects γ-secretase inhibitor–mediated enhancement of eosinophil maturation,19 reduction in eosinophil apoptosis (as demonstrated in this study; Figure 6), or a combination of both mechanisms is yet to be determined. Moreover, more data are needed to evaluate potential tissue eosinophilia and to determine the clinical significance(s) of the rise in eosinophil counts.
Treatment with GSI also effectively inhibited eosinophil shape changes, as determined by diminished eosinophil FSC and loss of actin polarization (Figure 7A,Aii). Although the inhibitory effects of GSI were observed in the absence of exogenous stimulation, it is unclear whether basal-level Notch-mediated shape change occurred independently of cytokine signals, as eosinophils are known to actively secrete numerous cytokines with the potential for autostimulation, including IL-5, GM-CSF, eotaxin, and RANTES (regulated on activation normal T cell expressed and secreted).32 Effects of Notch signaling on eosinophil shape change were more pronounced upon inclusion of GM-CSF, indicative of synergy between GM-CSF and Notch signaling pathways. Notably, our finding that GM-CSF–induced CD69 expression remained intact in the presence of GSI suggests that inhibition of Notch signaling does not block eosinophil activation in general.
In our studies, synergy between Notch and GM-CSF signaling was not observed in relatively short (1-hour) eosinophil cultures, regardless of whether or not cells were pretreated with GSI in advance of GM-CSF stimulation (Figure 7C and data not shown). Several plausible explanations may account for the delayed action of Notch-mediated effects, including the following: (1) induction of specific Notch ligand(s) or critical expression thresholds may be necessary before induction of significant Notch signaling. (2) The contributions of Notch signaling to enhancement of GM-CSF–induced effects may be dependent upon downstream proteins requiring longer than 1 hour to be transcribed and translated. (3) A shared signaling pathway component may be available for GM-CSF signaling at basal levels in circulating cells, whereas Notch signaling contributes to its replenishment. Mitogen-activated protein (MAP) kinase and nuclear factor (NF)–κB pathways are attractive candidates, as both are required for GM-CSF–induced eosinophil shape change,33 and Notch signaling has been implicated in cross-talk with both of these pathways.34
Downstream of polarized cytoskeletal rearrangements, inhibition of Notch signaling decreased GM-CSF–primed eosinophil chemokinesis (Figure 7E,F). Importantly, similar results were obtained using 3 distinct γ-secretase inhibitors to block the Notch signaling pathway, or neutralizing Abs against Notch receptor 1, confirming the γ-secretase dependence of the observed inhibition, and directly linking the Notch pathway to GM-CSF–primed eosinophil chemokinesis, respectively.
In this study, we demonstrate expression of Notch receptors and ligands by mature human blood eosinophils and identify Notch signaling–dependent autoregulatory functions. Taken together, this work suggests autocrine Notch signaling, enhanced in response to tissue- or inflammatory-derived signals, influences eosinophil migration and longevity, and as such may participate in the development of tissue eosinophilia and exacerbation or remediation of eosinophil effector functions. The significance of these studies to eosinophil-associated pathologies lies in the identification of Notch signaling pathway components as potential targets for therapeutic intervention to control eosinophil activation and viability. Moreover, as γ-secretase inhibitors continue to be formulated as potential therapies for Alzheimer disease and cancers, these findings may provide insights into potential unanticipated adverse effects on eosinophils.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Stephen Blacklow (Brigham and Women's Hospital, Boston, MA) for helpful discussions throughout the course of this study.
This work was supported in part by National Institutes of Health (Bethesda, MD) grants AI020241 and AI051645 to P.F.W., and an Interest Section Award and the Women in Allergy Junior Faculty Development Award to L.A.S. from the American Academy of Allergy, Asthma & Immunology (AAAAI; Milwaukee, WI).
National Institutes of Health
Authorship
Contribution: L.A.S., A.L.R., L.E.R., and R.C.N.M. performed research; L.A.S., L.E.R., A.L.R., and P.F.W. analyzed and interpreted data; R.C.N.M. and A.M.D. interpreted EM data; and L.A.S. and P.F.W. designed research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lisa A. Spencer, 330 Brookline Ave, E/CLS Room 935, Boston, MA 02215; e-mail: lspencer@bidmc.harvard.edu.

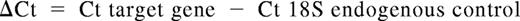
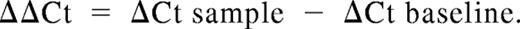
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal