Abstract
Pattern recognition receptors represent the first line of defense against invading pathogens. Herpes simplex virus (HSV) encodes multiple ligands detected by these receptors, yet persists in the majority of infected individuals indicating a breakdown in host defense against the virus. Here we identify a novel mechanism through which HSV immediate-early protein ICP0 inhibits TLR-dependent inflammatory response by blocking NF-κB and JNK activation downstream of TLR signal activation. This process depends on ICP0-mediated translocation of USP7 (HAUSP) from the nucleus to cytoplasm. We show that nuclear USP7 migrates to the cytoplasm in response to TLR engagement, a process that contributes to termination of TLR response. Cytoplasmic USP7 binds to and deubiquitinates TRAF6 and IKKγ, thus terminating TLR-mediated NF-κB and JNK activation. These findings suggest that USP7 is part of a negative feedback loop regulating TLR signaling and that ICP0 exploits this physiologic process to attenuate innate response to HSV. ICP0 inhibition of the TLR response serves to uncouple the innate and adaptive immune response, thereby playing a key role in HSV pathogenesis and persistence.
Introduction
Herpes simplex virus (HSV) infection initiates a robust innate response encompassing a wide array of cytokines, chemokines, and interferons.1 HSV activates innate immunity through both Toll-like receptor (TLR)–dependent and TLR-independent mechanisms.2,3 The TLR-dependent response entails TLR2,4,5 TLR3,6 and TLR9,7 whereas the non-TLR response is mediated through 2 cytosolic nucleic acid sensors (RIG-I in humans8 and DAI in mice9 ). HSV possesses several molecular components capable of activating an innate response: (1) HSV DNA: either through TLR9-dependent recognition of unmethylated CpG motifs10 or non-TLR DNA sensors11 ; (2) glycoproteins recognized by TLR24,5 ; and (3) dsRNA generated through self-hybridization of viral genes transcribed from complementary DNA strands.12,13 The multiplicity of pattern recognition receptors (PRRs) detecting HSV necessitates a robust ability of the virus to effectively block multiple innate signaling pathways to survive. Previous studies identified several HSV-encoded mechanisms that interfere with antiviral host immunity including nonspecific degradation of host mRNA by the RNAse VHS,14 inhibition of PKR by US1115 and γ34.5,16 inhibition of MHC-I peptide loading by ICP47,17,18 and suppression of interferon response by ICP0.19,20 However, to date, no HSV-encoded protein has been shown to interfere with TLR responses. Our previous studies comparing 2 versions of the HSV amplicon vector, helper virus–containing HSV amplicon (H+-HSV) and helper virus–free HSV amplicon (HF-HSV), in primary chronic lymphocytic leukemia (CLL) B cells identified an immunosuppressive activity associated with the HSV helper virus, which appeared to inhibit development of antitumor T-cell immunity.21 In contrast helper-free HSV amplicon (HF-HSV) possessed an adjuvant immunostimulatory effect that translated into potent anti-CLL response. An understanding of how specific components of HSV activate and/or inhibit innate immunity would allow for rational design of gene therapy vectors specifically tailored for particular clinical applications. HSV amplicon vectors engineered to enhance innate immunity would be useful as a platform for vaccine development, whereas a similar vector with the capacity to inhibit TLR signaling/response is better suited for clinical applications where an inflammatory response is undesirable
Through systematic comparison of innate response elicited by H+-HSV and HF-HSV amplicons, we identified the HSV immediate early (IE) protein ICP0 as an inhibitor of TLR-mediated NF-κB response. ICP0 inhibitory activity is dependent on its association with the USP7 (HAUSP) and requires an intact nuclear localization signal (nls) motif. USP7 down-regulates TLR-mediated NF-κB–dependent inflammatory cytokine/chemokine response through deubiquitination of TRAF6 and IKKγ. Our results show that USP7 serves a regulatory function in TLR response and this function is usurped by HSV ICP0 to suppress host innate response in HSV-infected cells.
Methods
Cells lines, plasmids, antibodies, and reagents
CLL B cells, obtained from patients with confirmed diagnosis of CLL after informed consent was obtained in accordance with the Declaration of Helsinki and with approval of the University of Miami, were isolated by density gradient centrifugation over endotoxin-free Lymphoprep (Nycomed Pharma, Oslo, Norway). HEK293 cells stably transfected with human TLRs were purchased from InvivoGen (San Diego, CA), cultured in DMEM plus 5% FCS under selection with blasticidin (10 μg/mL). Genomic HSV-1 DNA was extracted from HSV virus using the QIAamp DNA mini kit (QIAGEN, Valencia, CA) and used at final concentration of 0.5 μg/mL. HSV ICP0 and its mutants (M4, FXE), USP7, and USP7.C223S were kindly provided by Dr R. Everett (MRC Virology Unit, Institute of Virology, Glasgow, United Kingdom).22-24 ICP0.NLSmut was generated by site-directed mutagenesis of wt-ICP0 NLS motif 500-VRPRKRR-506 into 500-VRPAARA-506 (bold letters represent the consensus NLS motif residues that were mutated to alanines). USP7.NES was generated by introducing nuclear export signal (NES) (LPPLERLTL) from HIV-I Rev protein in-frame between the Ds-Red tag and USP7 in pDs.Red2-USP7 (Clontech, Palo Alto, CA). Full-length HSV ICP22, ICP27, and ICP47 were cloned into pIRES-hrGFPII (Stratagene, La Jolla, CA). P3X.FLAG-TRAF6 and IKKγ were generated by cloning human TRAF6/IKKγ into p3X.FLAG-CMV-26 (Sigma-Aldrich, St Louis, MO). Cyclohexamide (CHX) and MG-132 were purchased from Sigma-Aldrich.
Antibodies
FLAG-tagged proteins (ICP22, ICP27, ICP47, TRAF6, and IKKγ) were detected using anti-FLAG M2 antibody (Sigma-Aldrich). ICP0 and USP7 were detected using mouse anti-ICP0 (H1A027-100; Virusys, Sykesville, MD) and rabbit anti-USP7 (A300-033A; Bethyl), respectively. Anti-HA antibody (12CA5) was purchased from Roche Applied Science (Indianapolis, IN). Anti-JNK (56G8), phospho-JNK (9251), Iκ-Bα (44D4), phospho-Iκ-Bα (5A5), NF-κB p65 (3034), and α/β tubulin (2148) antibodies were purchased from Cell Signaling (Beverly, MA). Anti-TRAF6 and anti-IKKγ antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
HSV amplicon viruses
Coding sequence for a modified version of Escherichia coli β-galactosidase in which CpG sequences were mutated (InvivoGen) and were cloned into the polylinker region of the pHSVPrPUC plasmid and packaged into either helper virus–containing (H+-HSV)25 or helper virus–free (HF-HSV) amplicon stocks as previously described.26 Amplicon titers were determined using a previously published methodology.27
RNA isolation and real-time PCR
RNA was isolated using RNeasy Mini Kits (QIAGEN), and cDNA was synthesized using a mix of oligo dT, random hexamer (Ambion, Austin, TX), and SuperScript RT-III reverse transcriptase (Invitrogen, Frederick, MD). Quantitative real-time polymerase chain reaction (PCR) was performed on an Applied Biosystems ABI Prism 7900HT using Applied Biosystems Taqman Gene Expression Assays (Foster City, CA), and normalized to the housekeeper PGK-1 as follows: ΔC = CTCytokine − CTPGK1. Relative expression was calculated using the formula: 2−ΔΔCT and expressed as arbitrary units.
Transient transfection and reporter gene analysis
For NF-κB reporter assays, we used pNiFty2-Luc (InvivoGen). A constitutively active Renilla luciferase plasmid, pRLTK (0.02 μg per transfection), was used to normalize for transfection efficiency (Promega, Madison, WI).
RNAi
293-TLR4 cells were transfected with either nontargeting shRNA or shRNA specific for USP7, Cyld, or A20 (Open Biosystems, Huntsville, AL) using LipofectamineLTX (Invitrogen) and 24 hours later selected under puromycin (1 μg/mL). Forty-eight hours later, the cells were transfected with pCI-ICP0 versus empty vector (pCI-Neo) together with pNF-κB-Luc reporter construct and 24 hours later, stimulated by LPS for 4 hours before luciferase activity was assayed by quantitative reverse-transcription (qRT)–PCR for IL8 and TNF-α.
Confocal microscopy
293T or 293-TLR4 cells were seeded onto 20-mm coverslips on a 6-well plate and transfected with the indicated plasmids. Thirty-six hours later, the cells were washed with PBS and stained with ProLong antifade mounting reagent (Molecular Probes, Eugene, OR). Images were acquired using Zeiss LSM 510 META laser scanning confocal microscopy (Carl Zeiss, Heidelberg, Germany) through a 63× water-immersion lens, and subsequently processed using LSM (Carl Zeiss) and Adobe Photoshop software (Adobe Systems, San Jose, CA).
Tandem-affinity purification
The cDNA for human USP7 was cloned into the mammalian expression vector pcDNA3.2/capTEV-CT/V5-DEST using Gateway technology (Invitrogen), and tandem-affinity purification performed according to the manufacturer's instructions. Briefly, pcDNA3.2/capTEV-CT/V5-USP7 was expressed in 293-TLR4 and 24 hours later, the cells were split and one half was stimulated with LPS for 1 hour. Cells were lysed in lysis buffer (100 mM Tris-HCl, pH 8.0, 100 mM KCl, 200 μM EDTA, 1.5 mM MgCl2, 1× [700 ng/mL] pepstatin, complete protease inhibitor; Roche Applied Science) by freeze-thaw ×3. The cleared lysates were incubated with streptavidin-agarose for 4 hours at +4°C before being washed; USP7 was cleaved off column by AcTEV protease overnight at +4°C, concentrated, and immunoprecipitated (IPed) on nickel-affinity column; and the eluate was resolved on SDS–polyacrylamide gel electrophoresis (PAGE) and blotted with anti-USP7, TRAF6, and IKKγ.
IP/nuclear:cytoplasmic fractionation
293T cells transfected with the indicated plasmids for 48 hours were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with the protease inhibitor cocktail (SigmaFAST; Sigma-Aldrich) and immunoprecipitated using anti-FLAG beads (Sigma-Aldrich). The agarose beads were washed 3 times with RIPA buffer supplemented with 1 M urea. Endogenous TRAF6/IKKγs were IPed using specific Abs (Santa Cruz Biotechnology) and protein G agarose (Invitrogen).
Nuclear extracts were prepared as follows: 107 THP-1 or HEK293-TLR4 cells were stimulated with LPS at concentration of 1 μg/mL for 30 or 60 minutes, or left untreated before being washed in PBS and incubated for 10 minutes on ice in 400 μL buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail [1 μg/mL leupeptin, 1 μg/mL aprotinin, 10 μM pepstatin]), before they were lysed in 25 μL 10% NP-40. Cell lysates were vortexed and centrifuged and the supernatant containing the cytoplasmic fraction was collected. The pellet containing the nuclear fraction was resuspended in buffer B (20 mM HEPES, pH 7.9, 0.4 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 5% glycerol, and protease inhibitor cocktail) and incubated for 45 minutes at 4°C. The lysate was centrifuged at 16 000g for 5 minutes at 4°C, and the supernatant stored at −80°C.
Electrophoretic mobility shift assay
Nuclear extracts from THP-1 cells were prepared as described in “IP/nuclear: cytoplasmic fractionation.” Biotin 3′-end-labeled DNA duplex, as well as unlabeled duplex, of the NF-κB putative consensus binding motif 5′-AGT TGA GGG GAC TTT CCC AGG C-3 and 3′-TCA ACT CCC CTG AAA GGG TCC G-5′ were purchased from IDT-DNA. Nuclear extract protein (5 μg) was incubated with 2 nM biotin-labeled probe in binding buffer as previously described.28
Deubiquitination assays
293T cells were transfected with HA-ubiquitin, 3X-FLAG-TRAF6, or IKKγ vectors (pCMV-3X.FLAG-26; Sigma-Aldrich) and the indicated plasmids. Thirty-six hours later, cells were lysed in RIPA buffer supplemented with protease inhibitor cocktail, 20 μM of the ubiquitin hydrolase inhibitor N-ethylmaleimide (NEM; Sigma-Aldrich), and then boiled in 1% SDS for 5 minutes to remove noncovalently bound proteins. The cell lysates were then immunoprecipitated overnight at 4°C using anti-FLAG beads (Sigma-Aldrich). The agarose beads were washed 3 times with RIPA buffer followed by elution with 3X FLAG peptide (Sigma-Aldrich). The eluted HA-ubiquitin–conjugated TRAF6 and IKKγ were blotted using mouse polyclonal anti-HA (clone 12CA5; Roche Applied Science) and detected using antimouse TrueBlot Western blot (WB) kit (eBioscience, San Diego, CA).
Statistical analysis
Data were analyzed using one-way ANOVA and results expressed as mean plus or minus SEM. P values less than .05 were considered statistically significant.
Results
Differential activation of innate immune response by HF-HSV versus H+-HSV
We compared 2 HSV-based amplicon vectors for their ability to evoke an innate immune response in primary CLL B cells and a human macrophage cell line THP-1. The 2 HSV amplicon stocks are a helper virus–free HSV amplicon (HF-HSV) packaged using a bacterial artificial chromosome that encodes HSV proteins, and helper virus–containing HSV amplicons (H+-HSV) packaged using an ICP4-deleted HSV helper virus.25 Both viral stocks comprised identical HSV amplicon particles, however, H+-HSV stock contained ICP4-deleted HSV helper virus at a ratio of approximately 1:8 helper virus–amplicon particles. Gene expression from the HSV helper virus, by virtue of the ICP4 deletion, is limited to the remaining 4 immediate early (IE) proteins, ICP0, ICP22, ICP27, ICP47, which are expressed at a higher levels than otherwise achieved with wild-type HSV virus infection.29,30 Although equally efficient at transducing CLL B cells and THP1, H+-HSV and HF-HSV amplicons differ dramatically in their capacity to stimulate an anti-CLL T-cell response.21,31 We questioned whether such an outcome could be a result of quantitative difference in host innate response to the 2 HSV viruses under study. A comparison of the innate cytokine/chemokine response to transduction by H+-HSV and HF-HSV amplicon vectors in primary CLL B cells at different MOIs (Figure 1A; Table 1) and time points (Figure 1B) and in the human macrophage cell line THP-1 (Figure 1C) showed that at comparable MOI, HF-HSV induced a far more potent innate response than H+-HSV. Since H+-HSV amplicon stocks are composed of 2 HSV virus populations, an HSV amplicon that does not encode any viral proteins and HSV helper virus that encodes 4 regulatory proteins (ICP0, ICP22, ICP27, and ICP47), we compared the innate response against HF-HSV versus H+-HSV versus HSV helper virus alone in THP-1 cell line. As shown in Figure 1D, HF-HSV–transduced THP-1 cells generated a more robust innate cytokine response compared with cells transduced with either H+-HSV amplicon or the HSV helper virus alone. Since TLR-mediated innate cytokine response is dependent on both NF-κB and mitogen-activated protein kinases (MAPKs) JNK and p38,32 we investigated these 2 pathways in HSV-transduced THP1. THP1 cells were transduced with HF-HSV amplicon and HSV helper virus, and immunoblotted with antibodies specific for Iκ-Bα, phospho-Iκ-Bα, and phospho-JNK1/2 at 15, 30, and 60 minutes after HSV transduction (Figure 2A), and NF-κB activity was assessed by electrophoretic mobility shift (EMSA) (Figure 2B). As shown in Figure 2A and B, HF-HSV transduction correlated with significantly more NF-κB activity and JNK phosphorylation, compared with HSV helper virus–transduced THP1 cells. These findings were further confirmed by demonstrating increased RelA/p65 nuclear translocation in THP1 cells transduced with HF-HSV compared with HSV helper virus (Figure 2C). These results demonstrate that HF-HSV generates a more robust innate cytokine response compared with HSV helper virus, at least in part due to helper virus inhibition of TLR-mediated NF-κB and JNK activation.
Comparison of innate immune response initiated by HF-HSV and H+-HSV in CLL B cells and THP1 cells. (A-C) CLL B cells (A,B) and THP1 cells (C) were transduced with HF-HSV or H+-HSV amplicons at the indicated MOI; 8 hours later (A,C) innate response was assayed by qRT-PCR for TNF-α, IL6, IP10, and IFN-β mRNA. AU indicates arbitrary units. Data are representative of 3 experiments. *P < .05. In panel B, CLL B cells were transduced with HF-HSV or H+-HSV amplicons at MOI of 0.5, and TNF-α and IP10 response were assayed by qRT-PCR at multiple time points as indicated on the x-axis. (D) Comparison of innate response to HF-HSV, H+-HSV, and the HSV helper virus component of H+-HSV in THP1 cells at MOI of 0.5, 8 hours after transduction, shows that HF-HSV generates more potent innate response than either H+-HSV or HSV helper virus alone. Data are representative of 2 experiments. Error bars represent SEM.
Comparison of innate immune response initiated by HF-HSV and H+-HSV in CLL B cells and THP1 cells. (A-C) CLL B cells (A,B) and THP1 cells (C) were transduced with HF-HSV or H+-HSV amplicons at the indicated MOI; 8 hours later (A,C) innate response was assayed by qRT-PCR for TNF-α, IL6, IP10, and IFN-β mRNA. AU indicates arbitrary units. Data are representative of 3 experiments. *P < .05. In panel B, CLL B cells were transduced with HF-HSV or H+-HSV amplicons at MOI of 0.5, and TNF-α and IP10 response were assayed by qRT-PCR at multiple time points as indicated on the x-axis. (D) Comparison of innate response to HF-HSV, H+-HSV, and the HSV helper virus component of H+-HSV in THP1 cells at MOI of 0.5, 8 hours after transduction, shows that HF-HSV generates more potent innate response than either H+-HSV or HSV helper virus alone. Data are representative of 2 experiments. Error bars represent SEM.
Cytokine secretion in response to HSV viruses by CLL B cells
| . | TNF-α, pg/mL . | IL6, pg/mL . | IP10, pg/mL . | INF-β, pg/mL . |
|---|---|---|---|---|
| Mock | 0.63 ± 0.06 | 242 ± 12 | ND | ND |
| H+-HSV | 1.19 ± 0.02 | 65 ± 3 | 74 ± 24 | 57.8 ± 2.3 |
| HF-HSV | 3.32 ± 0.2 | 1035 ± 31 | 341 ± 38 | 200.8 ± 12.7 |
| . | TNF-α, pg/mL . | IL6, pg/mL . | IP10, pg/mL . | INF-β, pg/mL . |
|---|---|---|---|---|
| Mock | 0.63 ± 0.06 | 242 ± 12 | ND | ND |
| H+-HSV | 1.19 ± 0.02 | 65 ± 3 | 74 ± 24 | 57.8 ± 2.3 |
| HF-HSV | 3.32 ± 0.2 | 1035 ± 31 | 341 ± 38 | 200.8 ± 12.7 |
ND indicates nondetectable.
HSV helper virus encodes an inhibitor of TLR signaling that suppresses NF-κB and MAPK. (A) THP1 cells were transduced with HF-HSV and HSV helper virus and immunoblotted at 15, 30, and60 minutes with the indicated antibodies; blots are representative of 3 experiments. (B) Nuclear extracts from mock-transduced THP1 cells or THP1 cells transduced with HF-HSV or HSV helper virus were studied by NF-κB EMSA. P indicates probe alone; M, mock; HF, HF-HSV amplicon; and H, HSV helper virus. (C) Nuclear and cytoplasmic fractions of THP1 cells mock transduced (M) or transduced with HF-HSV or HSV helper virus were immunoblotted with anti-RelA/p65. (D) THP1 cells were left untreated or preincubated with 50 μM cyclohexamide (CHX) for 30 minutes before transduction with HF-HSV, H+-HSV amplicon, or HSV helper virus at MOI of 0.5. Six hours later, TNF-α and IP10 mRNA was assayed by qRT-PCR. Inhibition of HSV protein expression restored innate response to both H+-HSV amplicon and HSV helper virus. *P < .05. Data are representative of 2 experiments. (E) Expression of ICP0, ICP22, ICP27, and ICP47 was verified by Western blot using anti-FLAG for ICP22, ICP27, and ICP47 and specific anti-ICP0 for ICP0. (F) HEK293-TLR2/6 were transfected by expression vectors for ICP0, ICP22, ICP27, ICP47, or empty vector (EV) as a control and 24 hours later stimulated with Pgn. TNF-α expression was assayed by qRT-PCR 6 hours later. Error bars represent SEM.
HSV helper virus encodes an inhibitor of TLR signaling that suppresses NF-κB and MAPK. (A) THP1 cells were transduced with HF-HSV and HSV helper virus and immunoblotted at 15, 30, and60 minutes with the indicated antibodies; blots are representative of 3 experiments. (B) Nuclear extracts from mock-transduced THP1 cells or THP1 cells transduced with HF-HSV or HSV helper virus were studied by NF-κB EMSA. P indicates probe alone; M, mock; HF, HF-HSV amplicon; and H, HSV helper virus. (C) Nuclear and cytoplasmic fractions of THP1 cells mock transduced (M) or transduced with HF-HSV or HSV helper virus were immunoblotted with anti-RelA/p65. (D) THP1 cells were left untreated or preincubated with 50 μM cyclohexamide (CHX) for 30 minutes before transduction with HF-HSV, H+-HSV amplicon, or HSV helper virus at MOI of 0.5. Six hours later, TNF-α and IP10 mRNA was assayed by qRT-PCR. Inhibition of HSV protein expression restored innate response to both H+-HSV amplicon and HSV helper virus. *P < .05. Data are representative of 2 experiments. (E) Expression of ICP0, ICP22, ICP27, and ICP47 was verified by Western blot using anti-FLAG for ICP22, ICP27, and ICP47 and specific anti-ICP0 for ICP0. (F) HEK293-TLR2/6 were transfected by expression vectors for ICP0, ICP22, ICP27, ICP47, or empty vector (EV) as a control and 24 hours later stimulated with Pgn. TNF-α expression was assayed by qRT-PCR 6 hours later. Error bars represent SEM.
ICP0 encoded by the helper virus inhibits innate responses to HSV infection
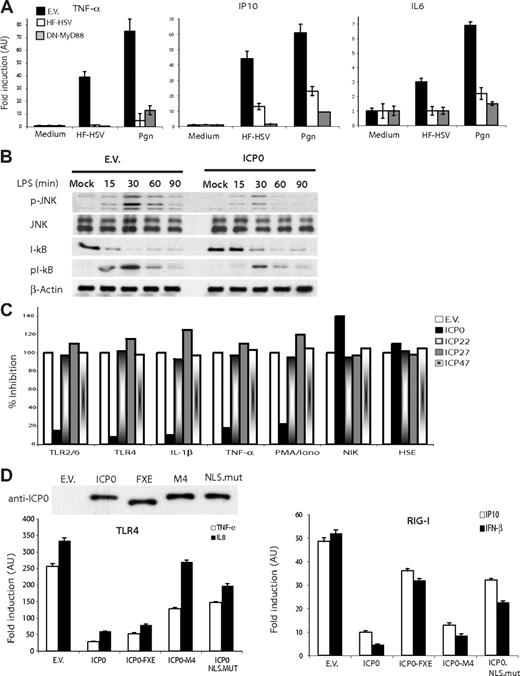
To assess whether HSV helper virus encodes an inhibitor for HSV-triggered innate responses, THP1 cells were transduced with HF-HSV, H+-HSV, or HSV helper virus in the presence or absence of cyclohexamide (CHX) and innate cytokine/chemokine response was assessed by qRT-PCR. As shown in Figure 2D, inhibition of HSV protein expression by CHX unmasked the capacity of both H+-HSV amplicon and HSV helper virus to induce inflammatory cytokine response. We inferred from this observation that one or more of the 4 immediate early (IE) proteins ICP0, ICP22, ICP27, and ICP47 encoded by HSV helper virus are capable of inhibiting innate responses to HSV. A comparison of the 4 IE proteins for their capacity to inhibit TLR2/6 response to peptidoglycan (Pgn) stimulation identified ICP0 as a likely candidate for HSV helper virus–mediated suppression of TLR response (Figure 2F). To establish a role for ICP0 in inhibiting TLR signaling, we compared TLR2 innate response in cells expressing ICP0 or a dominant-negative form of the TLR adaptor MyD88 encoded by its C-terminal TIR domain (DN-MyD88). As shown in Figure 3A, both ICP0 and DN-MyD88 were equally efficient at inhibiting TLR2-mediated innate response to hf-HSV and Pgn. ICP0 also inhibited LPS-induced TLR4-dependent I-κBα phosphorylation, degradation, and JNK1/2 phosphorylation (Figure 3B). The effect of ICP0 on NF-κB and JNK activation downstream of TLR4 signaling closely resembled that seen upon transduction of THP1 cells with HSV helper virus, suggesting that ICP0 is the primary effector of helper virus–mediated TLR inhibition. ICP0 inhibition of the canonical NF-κB pathway upon TLR stimulation was also reproducible in cells stimulated with IL-1β, TNF-α, and PMA/ionomycin. On the other hand, the alternative NF-κB pathway induced by overexpression of the IKKα kinase NIK (NF-κB–inducing kinase) was resistant to ICP0 inhibition (Figure 3C).
ICP0 inhibits TLR response to HSV, and this activity depends on its association with the deubiquitinating enzyme USP7. (A) 293-TLR2/6 cells were transfected with the empty vector (EV), pCI-ICP0, or pcDNA3.1-DN-MyD88 encoding MyD88 TIR domain. Twenty-four hours later, cells were left untreated or stimulated with PGN or HF-HSV for 6 hours. TNF-α, IP10, and IL6 mRNA was assayed by qRT-PCR.(B) 293-TLR4 cells transduced with empty vector (EV) or ICP0 (ICP0) were stimulated with LPS and immunoblotted at the indicated time points with antibodies against I-κBα, phosphorylated-I-κBα, JNK, and phosphorylated JNK. (C) 293T or 293 stably expressing indicated TLRs were transfected with pCI-ICP0, ICP22, ICP27, and ICP47 or empty vector (EV), pcDNA3-NIK, together with NF-κB luciferase reporter. Twenty-four hours later, cells were activated by corresponding TLR ligands, TNF-α (10 ng/mL), IL-1β (10 ng/mL), or PMA/ionomycin (5 ng/mL and 1 μg/mL, respectively) for 6 hours before luciferase activity was measured. To rule out nonspecific interference with mRNA transcription or protein expression, 293T cells were cotransfected with pCI-ICP0, ICP22, ICP27, and ICP47 and heat-shock response element HSE-Luc, and 24 hours later incubated for 30 minutes at 42°C before luciferase activity was measured 6 hours later. None of the ICP proteins interfered with heat-shock response. (D) ICP0 inhibits NF-κB and IRF3 signaling pathways through different mechanisms: HEK293-TLR4 or HEK293-RIG-I cells were transfected with empty vector, ICP0, ICP0-FXE, ICP0-M4, or ICP0.NLS-mut and stimulated with their respective ligands, and innate response (TNF-α and IL8 for HEK293-TLR4 or IFN-β and IP10 for HEK293-RIG-I) was assayed by qRT-PCR. An ICP0 mutant that did not bind USP7 (ICP0-M4) lacked the ability to inhibit NF-κB response to TLR4 activation. Deletion of the E3 ligase RING domain (FXE) attenuated ICP0 inhibition of IFN-β promoter response to RIG-I. ICP0 with mutated NLS motif (ICP0-NLS-mut) failed to inhibit either NF-κB or IRF3 response. Error bars represent SEM.
ICP0 inhibits TLR response to HSV, and this activity depends on its association with the deubiquitinating enzyme USP7. (A) 293-TLR2/6 cells were transfected with the empty vector (EV), pCI-ICP0, or pcDNA3.1-DN-MyD88 encoding MyD88 TIR domain. Twenty-four hours later, cells were left untreated or stimulated with PGN or HF-HSV for 6 hours. TNF-α, IP10, and IL6 mRNA was assayed by qRT-PCR.(B) 293-TLR4 cells transduced with empty vector (EV) or ICP0 (ICP0) were stimulated with LPS and immunoblotted at the indicated time points with antibodies against I-κBα, phosphorylated-I-κBα, JNK, and phosphorylated JNK. (C) 293T or 293 stably expressing indicated TLRs were transfected with pCI-ICP0, ICP22, ICP27, and ICP47 or empty vector (EV), pcDNA3-NIK, together with NF-κB luciferase reporter. Twenty-four hours later, cells were activated by corresponding TLR ligands, TNF-α (10 ng/mL), IL-1β (10 ng/mL), or PMA/ionomycin (5 ng/mL and 1 μg/mL, respectively) for 6 hours before luciferase activity was measured. To rule out nonspecific interference with mRNA transcription or protein expression, 293T cells were cotransfected with pCI-ICP0, ICP22, ICP27, and ICP47 and heat-shock response element HSE-Luc, and 24 hours later incubated for 30 minutes at 42°C before luciferase activity was measured 6 hours later. None of the ICP proteins interfered with heat-shock response. (D) ICP0 inhibits NF-κB and IRF3 signaling pathways through different mechanisms: HEK293-TLR4 or HEK293-RIG-I cells were transfected with empty vector, ICP0, ICP0-FXE, ICP0-M4, or ICP0.NLS-mut and stimulated with their respective ligands, and innate response (TNF-α and IL8 for HEK293-TLR4 or IFN-β and IP10 for HEK293-RIG-I) was assayed by qRT-PCR. An ICP0 mutant that did not bind USP7 (ICP0-M4) lacked the ability to inhibit NF-κB response to TLR4 activation. Deletion of the E3 ligase RING domain (FXE) attenuated ICP0 inhibition of IFN-β promoter response to RIG-I. ICP0 with mutated NLS motif (ICP0-NLS-mut) failed to inhibit either NF-κB or IRF3 response. Error bars represent SEM.
Within the TLR signaling pathway, ICP0 inhibited, in a dose-dependent manner, the NF-κB response to overexpression of various signaling molecules with the exception of constitutively active IKKβ (ΕΕ) (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Subsequent experiments using the macrophage cell line J774 transduced with H+-HSV and HF-HSV amplicon stocks correlated ICP0 mRNA and protein levels to inhibition of HF-HSV–mediated innate response (Figure S2A-C). Taken together, such data would suggest that HF-HSV amplicon's ability to activate host innate immunity can be dissociated from the helper virus ability to suppress such a response, the latter mediated by HSV helper virus–encoded ICP0.
ICP0 inhibition of the TLR-mediated NF-κB response is dependent on its association with the deubiquitinating enzyme USP7
ICP0 is a multidomain protein that mediates proteasomal degradation of several cellular proteins (PML, SP-100, DNA-PK, and CD83) through its N-terminus RING domain E3 ligase function. The latter also inhibits innate response to HSV infection in part through interference with IRF3/IRF7-mediated transcription of ISRE-responsive genes and degradation of IRF3.20,33 ICP0 contains a nuclear localization signal (nls; aa's 500-506)34 that enables it to translocate from cytosol to nucleus and also a USP7 (HAUSP) binding motif (aa's 618-638).22 To identify the underlying mechanism(s) by which ICP0 inhibits the TLR response, we compared a series of ICP0 mutants (details in Table 2) for their ability to inhibit either TLR4-mediated cytokine response to LPS versus RIG-I–mediated IRF3-dependent response to poly I:C (Figure 3D). ICP0 expression vectors with point mutations within the USP7-binding domain (ICP0-M4)22 or nls motif (ICP0-NLS-MUT) lost their capacity to inhibit the NF-κB response (Figure 3D). On the other hand, deletion of the E3 ligase RING domain (ICP0-FXE) compromised ICP0's ability to inhibit IRF3-dependent response, as previously reported,35 but had modest effect on its capacity to inhibit NF-κB–dependent cytokine response. Mutation of ICP0 nls motif (ICP0-NLS-MUT) compromised its capacity to inhibit both NF-κB– and IRF3-dependent pathways. These results suggest that ICP0 inhibition of TLR-induced NF-κB response is mediated through its association with the deubiquitinating (DUB) enzyme USP7 (HAUSP) and requires ICP0 to shuttle to the nucleus where USP7 is expressed. Since USP7 is a nuclear protein, it is conceivable that the inability of ICP0.NLS-MUT to inhibit NF-κB–dependent cytokine response might be secondary to its inability to gain access to nuclear USP7.
ICP0 mutants
| ICP-0 mutant . | Deletion/mutation . | Impact on ICP0 function . |
|---|---|---|
| ICP0-M4 | K620I | Loss of USP7 binding capacity |
| ICP0-FXE | Del of aa's 106-149 | Deletion of E3 ligase RING domain |
| ICP0-NLSmut | 500-VRPRKRR-506 into 500-VRPAARA-506 | Nuclear exclusion |
| ICP-0 mutant . | Deletion/mutation . | Impact on ICP0 function . |
|---|---|---|
| ICP0-M4 | K620I | Loss of USP7 binding capacity |
| ICP0-FXE | Del of aa's 106-149 | Deletion of E3 ligase RING domain |
| ICP0-NLSmut | 500-VRPRKRR-506 into 500-VRPAARA-506 | Nuclear exclusion |
ICP0 alters cellular localization of USP7
TLR signaling initiates at the cell membrane (TLR1, 2, 4, 5, and 6) or within acidic endosomes (TLR3, 7, 8, and 9) and proceeds to liberate NF-κB subunits from I-κB. ICP0 shuttles between the nucleus and cytoplasm, whereas USP7 is a nuclear protein that resides within specialized nuclear structures known as PML or ND10.36 This suggested to us that ICP0 might inhibit TLR signaling by facilitating the export of USP7 from the nucleus to the cytoplasm. To address this question, we studied the subcellular localization of USP7 in the presence and absence of ICP0 and in response to TLR engagement. As shown in Figure 4Ai, wt-ICP0 is detectable within both the nucleus and cytoplasm. Mutations within the NLS sequence (ICP0-NLS-MUT) excluded ICP0 from the nucleus (Figure 4Aii) and precluded its interaction with nuclear USP7 (Figure 4Avi). USP7, which appeared as a nuclear protein in the absence of ICP0 (Figure 4Aiii) translocated to a perinuclear compartment when coexpressed with wt-ICP0 (Figure 4Av). In agreement with previous studies showing that USP7 undergoes proteasomal degradation by ICP0,37 we detected reduced levels of USP7 in cells coexpressing wt-ICP0 (Figure 4Av). ICP0-NLS-MUT failed to alter USP7 localization. Modifying USP7 to encode a nuclear export signal, USP7NES allowed a fraction of USP7 to be detected within the cytoplasm in the absence of ICP0 (Figure 4Aiv). Since ICP0-NLS-MUT failed to inhibit the TLR response, this suggested that translocating USP7 from its nuclear to cytoplasmic location might be a key element in ICP0 inhibition of TLR response.
ICP0 translocates USP7 from a nuclear to cytoplasmic protein. (A) 293T cells were transfected with GFP-tagged wt-ICP0 or ICP0.NLS-mut alone or in combination with DsRed-tagged USP7 at a ratio of at 1:1. Wt-ICP0 was detectable both within the nucleus and cytoplasm (i). On the other hand, wt-USP7 was an entirely nuclear protein (iii). Modification of USP7 by attaching a NES motif allowed it to be expressed both in the nucleus and cytoplasm (iv). In cells coexpressing USP7 and wt-ICP0, USP7 was a predominantly cytoplasmic protein (v) that colocalized with ICP0. As expected, mutation of ICP0 NLS motif excluded it from the nucleus (ii) and abolished its colocalization with USP7 (vi). In these cells, USP7 behaved as in non–ICP0-expressing cells, remaining a nuclear protein. (B) HEK293-TLR4 cells were transfected with DsRed-USP7 and stimulated by LPS. DsRed-tagged USP7 cellular localization was traced over 1 hour after LPS stimulation. USP7 was detectable within the cytoplasm starting 20 minutes after LPS stimulation and lasting for up to an hour. Images were obtained with a Zeiss LSM 510 confocal microscope using 63× water-immersion objective lens.
ICP0 translocates USP7 from a nuclear to cytoplasmic protein. (A) 293T cells were transfected with GFP-tagged wt-ICP0 or ICP0.NLS-mut alone or in combination with DsRed-tagged USP7 at a ratio of at 1:1. Wt-ICP0 was detectable both within the nucleus and cytoplasm (i). On the other hand, wt-USP7 was an entirely nuclear protein (iii). Modification of USP7 by attaching a NES motif allowed it to be expressed both in the nucleus and cytoplasm (iv). In cells coexpressing USP7 and wt-ICP0, USP7 was a predominantly cytoplasmic protein (v) that colocalized with ICP0. As expected, mutation of ICP0 NLS motif excluded it from the nucleus (ii) and abolished its colocalization with USP7 (vi). In these cells, USP7 behaved as in non–ICP0-expressing cells, remaining a nuclear protein. (B) HEK293-TLR4 cells were transfected with DsRed-USP7 and stimulated by LPS. DsRed-tagged USP7 cellular localization was traced over 1 hour after LPS stimulation. USP7 was detectable within the cytoplasm starting 20 minutes after LPS stimulation and lasting for up to an hour. Images were obtained with a Zeiss LSM 510 confocal microscope using 63× water-immersion objective lens.
To assess physiologic relevance of USP7 localization in the context of TLR signaling, we studied whether TLR stimulation induced nuclear export of USP7 independent of HSV. 293-TLR4 cells were LPS stimulated and cellular localization of fluorescent USP7 was traced over 1 hour. As shown in Figure 4B, TLR stimulation induced USP7 translocation to the cytoplasm as early as 20 minutes following LPS stimulation and this effect was maintained for up to 1 hour. These results were confirmed by biochemical fractionation of LPS-stimulated 293-TLR4 (Figure 5A) and THP1 stimulated with either LPS or CpG ODN (Figure 5B). These data represent the first such example linking USP7 to TLR response and would suggest that HSV ICP0 exploits USP7 to suppress host innate response.
Nuclear USP7 migrates to cytoplasm to inhibit TLR signal. Nuclear and cytoplasmic fractions of LPS-stimulated 293-TLR4 (A) and LPS and CpG ODN-stimulated THP1 (B) cells were collected at the indicated time points and immunoblotted with anti-USP7. Nucleolin and α-tubulin were blotted as nuclear and cytoplasmic fraction controls, respectively. (C) 293-TLR4 cells were transfected with nonsilencing shRNA or shRNA against A20, Cyld, or USP7 and knockdown of individual proteins was confirmed. Seventy-two hours later, TLR4 was stimulated with LPS for 6 hours before TNF-α and IL8 mRNA were assayed by qRT-PCR. (D) 293-TLR2/6 cells were transfected with empty vector (EV), full-length USP7 (USP7.FL), USP7.TD (aa's 1-210), USP7-ΔTD (aa's 210-1102), and USP7.C223S, and 24 hours later, TLR2/6 was stimulated with Pgn for 6 hours before TNF-α mRNA was assayed by qRT-PCR. Error bars represent SEM.
Nuclear USP7 migrates to cytoplasm to inhibit TLR signal. Nuclear and cytoplasmic fractions of LPS-stimulated 293-TLR4 (A) and LPS and CpG ODN-stimulated THP1 (B) cells were collected at the indicated time points and immunoblotted with anti-USP7. Nucleolin and α-tubulin were blotted as nuclear and cytoplasmic fraction controls, respectively. (C) 293-TLR4 cells were transfected with nonsilencing shRNA or shRNA against A20, Cyld, or USP7 and knockdown of individual proteins was confirmed. Seventy-two hours later, TLR4 was stimulated with LPS for 6 hours before TNF-α and IL8 mRNA were assayed by qRT-PCR. (D) 293-TLR2/6 cells were transfected with empty vector (EV), full-length USP7 (USP7.FL), USP7.TD (aa's 1-210), USP7-ΔTD (aa's 210-1102), and USP7.C223S, and 24 hours later, TLR2/6 was stimulated with Pgn for 6 hours before TNF-α mRNA was assayed by qRT-PCR. Error bars represent SEM.
USP7 knockdown augments the TLR-mediated NF-κB response
To investigate whether endogenous USP7 played a regulatory role similar to that of A20 and Cyld, 2 other deubiquitinating (DUB) enzymes capable of suppressing NF-κB activity, we compared cytokine/chemokine response of LPS-stimulated HEK293-TLR4 transfected with shRNA targeting USP7, A20, or Cyld. Knockdown of USP7 enhanced innate response in a manner comparable with that seen in A20- and Cyld-depleted cells (Figure 5C). Similar results were obtained using 293-TLR2/6 (Figure S3). On the other hand, overexpression of USP7 inhibited NF-κB reporter response to HEK293-TLR2/6 and TLR9 stimulation and MyD88 overexpression (Figure S4).
USP7 encodes an 1102–amino acid nuclear protein with a TRAF-like domain (TD) at the NH2-terminus (58-196),38 ubiquitin-specific protease domain (USP) catalytic center (208-560),39 and ICP0-binding domain (between aa's 622-801).40 Experiments comparing full-length USP7 (FL-USP7) to USP7-TD (1-210), USP7.ΔTD (210-1102), and USP7.C223S, where the catalytic active site cysteine was mutated to serine, showed that USP7-mediated inhibition of TLR response was dependent on its TD and USP catalytic activity. Deletion or mutation of either diminished USP7 inhibition of TLR response. Overexpression of the TD alone partially suppressed TLR response (Figure 5D).
USP7 binds and deubiquitinates TRAF6 and IKKγ
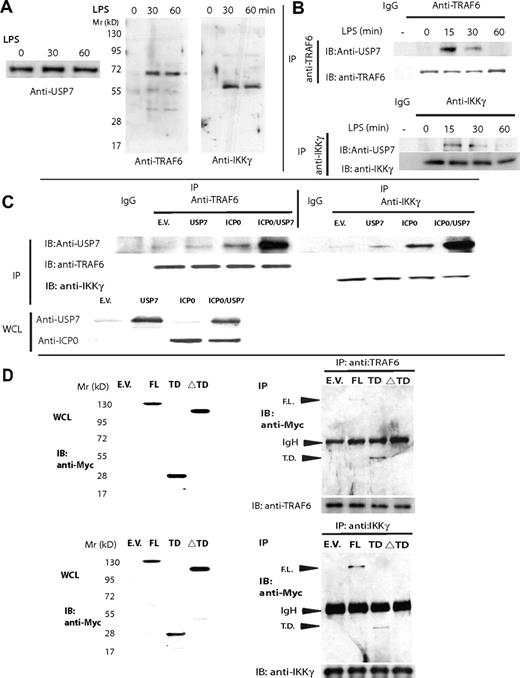
USP7 possesses a TRAF-like domain at the N-terminus of the protein (aa's 58-196) that has been previously shown to interact with TRAF6 in vitro.38 Crystallographic studies of USP7 TRAF-like domain in conjunction with peptides derived from 2 of its substrates, MDM2 and p53, identified the sequence P/AXXS as a consensus binding motif within USP7 substrates41 ; a similar motif, EXXS, was found within the EBNA1 peptide binding USP7.42 We scanned TRAF6 and IKKγ, 2 components of the TLR signaling pathway that undergo stimulus-induced K63 ubiquitination and substrates for other deubiquitinating enzymes, for the P/AXXS sequence. Three such motifs were present within TRAF6 (22AMAS25, 63PLES66, and 276AVHS279) and 2 within IKKγ (374AYLS377, 381ALPS384). In addition TRAF6 also possesses an EXXS motif (126-EILS-129) similar to that of EBNA1. To test whether USP7 interacts with TRAF6/IKKγ upon TLR signaling, we generated 293-TLR4 cells expressing USP7-TAP (tandem affinity purification tag composed of a biotinylation motif and His tag) fusion construct. 293-TLR4-USP7-TAP cells were left untreated or stimulated with LPS for 15 or 30 minutes before they were lysed by freeze-thaw. Protein complexes isolated by TAP were resolved on SDS-PAGE and blotted with anti-TRAF6 and anti-IKKγ (Figure 6A). Upon LPS stimulation, both TRAF6 and IKKγ coimmunoprecipitated (co-IPed) with USP7. To study the kinetics of USP7 interaction with TLR signaling components, endogenous TRAF6 and IKKγ were IPed from LPS-stimulated THP1 cells at 0, 15, 30, and 60 minutes after stimulation, and immunoblotted for USP7. We detected specific binding of USP7 to both TRAF6 and IKKγ at 15 and 30 minutes after LPS stimulation (Figure 6B). Interestingly, even though USP7 was detectable in the cytoplasm for up to 90 minutes after TLR stimulation (Figure 5A,B), minimal binding to either TRAF6 or IKKγ was detectable beyond the initial 30 minutes after LPS stimulation, suggesting that USP7 might preferentially bind the ubiquitinated form of TRAF6/IKKγ. Given the role played by ICP0 to translocate USP7 to the cytoplasm (Figure 4A), we questioned whether ICP0 augmented USP7 binding to TRAF6/IKKγ by imunoprecipitating endogenous TRAF6/IKKγ from cells expressing ICP0, USP7, or both and immunoblotting with anti-USP7. In 293T cells, trace amount of either TRAF6 or IKKγ co-IPed with USP7 in the absence of ICP0. However, coexpression of ICP0, similar to TLR stimulation, significantly augmented USP7 binding to TRAF6/IKKγ (Figure 6C). To investigate which USP7 domain binds to TRAF6 and IKKγ, Myc-tagged full-length USP7 (USP7-FL), USP7-TRAF–like domain (USP7-TD, aa's 1-210), and USP7 deleted for the TRAF domain (USP7.ΔTD, aa's 210-1102) were expressed in HEK293-TLR4 cells, and 30 minutes following LPS stimulation, endogenous TRAF6/IKKγ was immunoprecipitated and blotted for Myc-tagged proteins. Both USP7-FL and USP7-TD bound endogenous TRAF6 and IKKγ but not the USP7.ΔTD (Figure 6D). These results indicate that USP7 binds TRAF6 and IKKγ through its NH2-terminus TRAF-like domain, and this interaction is significantly enhanced upon TLR stimulation and/or coexpression of ICP0.
USP7 interacts with TRAF6 and IKKγ. (A) 293-TLR4 cells expressing pcDNA3.2-capTEV-USP7 were LPS-stimulated at different time points (0, 15, and 30 minutes) before they were lysed by freeze-thaw, TAP-purified, and blotted with anti-USP7, anti-TRAF6, and anti-IKKγ. Data are representative of 2 experiments. (B) THP1 cells were LPS-stimulated at different time points (0, 15, 30, and 60 minutes) before endogenous TRAF6 and IKKγ were immunoprecipitated and blotted with anti-USP7. (C) ICP0 enhances USP7 binding to endogenous TRAF6 and IKKγ: 293T cells were transfected with ICP0 and USP7 either alone or in combination and 24 hours later endogenous TRAF6 and IKKγ were immunoprecipitated and blotted with anti-USP7. Expression of ICP0 significantly enhanced USP7 binding to endogenous TRAF6 and IKKγ. (D) HEK293-TLR4 cells were transfected with Myc-tagged USP7-FL (1-1102), USP7-TD (1-210), or USP7.ΔTD (210-1102), stimulated with LPS for 1 hour, and then lysed and endogenous TRAF6 and IKKγ were immunoprecipitated. Immunoprecipitated TRAF6 and IKKγ bound both USP7-FL (1-1102) and USP7-TD (1-210).
USP7 interacts with TRAF6 and IKKγ. (A) 293-TLR4 cells expressing pcDNA3.2-capTEV-USP7 were LPS-stimulated at different time points (0, 15, and 30 minutes) before they were lysed by freeze-thaw, TAP-purified, and blotted with anti-USP7, anti-TRAF6, and anti-IKKγ. Data are representative of 2 experiments. (B) THP1 cells were LPS-stimulated at different time points (0, 15, 30, and 60 minutes) before endogenous TRAF6 and IKKγ were immunoprecipitated and blotted with anti-USP7. (C) ICP0 enhances USP7 binding to endogenous TRAF6 and IKKγ: 293T cells were transfected with ICP0 and USP7 either alone or in combination and 24 hours later endogenous TRAF6 and IKKγ were immunoprecipitated and blotted with anti-USP7. Expression of ICP0 significantly enhanced USP7 binding to endogenous TRAF6 and IKKγ. (D) HEK293-TLR4 cells were transfected with Myc-tagged USP7-FL (1-1102), USP7-TD (1-210), or USP7.ΔTD (210-1102), stimulated with LPS for 1 hour, and then lysed and endogenous TRAF6 and IKKγ were immunoprecipitated. Immunoprecipitated TRAF6 and IKKγ bound both USP7-FL (1-1102) and USP7-TD (1-210).
TLR-mediated NF-κB response is associated with nondegradative K63-linked polyubiquitination of TRAF6 and IKKγ,43 whereas their deubiquitination by A20 and Cyld terminates NF-κB response. We postulated that deubiquitination of TRAF6 and/or IKKγ by USP7 could explain the latter's capacity to inhibit TLR response. To test this hypothesis, we compared wt ICP0 and its mutants (ICP0-FXE, M4, NLS-MUT) as well as USP7 and a catalytically inactive mutant USP7.C223S for their capacity to deubiquitinate TRAF6 and IKKγ. Both ICP0 and, to a lesser degree, FXE deubiquitinated TRAF6 and IKKγ, although loss of ICP0's ability to associate with USP7 either through mutation of the NLS domain (ICP0-NLS-MUT) or USP7-binding domain (ICP0-M4) ablated its deubiquitinating effect (Figure 7A). wt USP7 (Figure 7B) possessed similar activity, whereas C/S mutation of the catalytic domain compromised it.
Both USP7 and ICPO deubiquitinate TRAF6 and IKKγ. (A,B) 293T cells coexpressing FLAG-tagged TRAF6 or IKKγ, HA-ubiquitin, plus one of the following plasmids: empty vector (EV), wt ICP0, ICP0-FXE, ICP0-NLS-MUT, ICP0-M4 (A) or USP7, USP7-C223S (B). Thirty-six hours later, the cells were lysed and FLAG-tagged proteins were immunoprecipitated and blotted with anti-HA. Whole-cell lysates (WCLs) were immunoblotted for ICP0 and USP7 expression. (C) HEK293-TLR2/6 cells were transfected with nonsilencing shRNA or shRNA targeting USP7 and knockdown was confirmed by WB. Forty-eight hours later, the cells were transfected with ICP0 and NF-κB-Luc reporter and stimulated with Pgn for 6 hours before luciferase activity was assayed. *P < .05. (D) 293T cells were transfected with nontargeting shRNA or shRNA-USP7. Forty-eight hours later, the cells were transfected with FLAG-TRAF6 or IKKγ, HA-ubiquitin, and ICP0 before they were lysed, and FLAG-tagged proteins were immunoprecipitated and blotted with anti-HA. WCLs were immunoblotted with specific antibodies to confirm ICP0 expression and USP7 knockdown. (E) THP1 cells were transduced with HF-HSV amplicon or HSV helper virus or left untransduced. Twenty minutes later the cells were lysed, and endogenous TRAF6 and IKKγ were immunoprecipitated with anti-TRAF6/IKKγ antibodies and blotted with antiubiquitin to assess their ubiquitination status. WCLs were immunoblotted for ICP0 expression. Cell lysates were also fractionated into nuclear and cytoplasmic fractions and blotted for USP7 expression. (F) Overexpression of ICP0 does not deplete endogenous TRAF6 or IKKγ: 293T cells were transfected with increasing concentration of ICP0 together with either FLAG-tagged TRAF6 or IKKγ in the presence or absence of the proteasome inhibitor, MG132 (5 μM added for the last 12 hours), and 24 hours later, cell lysate was blotted using anti-FLAG mAb to assess ICP0 effect on TRAF6 and IKKγ. ICP0 did not deplete either TRAF6 or IKKγ. (G) Comparison of wt-USP7 and USP7.NES ability to deubiquitinate TRAF6 and IKKγ and suppress TLR-induced NF-κB response: 293T cells coexpressing FLAG-tagged TRAF6 or IKKγ, HA-ubiquitin, and USP7 or USP7-NES. Thirty-six hours later, the cells were lysed and FLAG-tagged proteins were immunoprecipitated and blotted with anti-HA. (H) Enhanced deubiquitinating efficacy of USP7-NES against TRAF6/IKKγ compared with wt-USP7 correlated with its ability to suppress TLR-2/6 response to Pgn stimulation, measured as TNF-α mRNA. Error bars represent SEM.
Both USP7 and ICPO deubiquitinate TRAF6 and IKKγ. (A,B) 293T cells coexpressing FLAG-tagged TRAF6 or IKKγ, HA-ubiquitin, plus one of the following plasmids: empty vector (EV), wt ICP0, ICP0-FXE, ICP0-NLS-MUT, ICP0-M4 (A) or USP7, USP7-C223S (B). Thirty-six hours later, the cells were lysed and FLAG-tagged proteins were immunoprecipitated and blotted with anti-HA. Whole-cell lysates (WCLs) were immunoblotted for ICP0 and USP7 expression. (C) HEK293-TLR2/6 cells were transfected with nonsilencing shRNA or shRNA targeting USP7 and knockdown was confirmed by WB. Forty-eight hours later, the cells were transfected with ICP0 and NF-κB-Luc reporter and stimulated with Pgn for 6 hours before luciferase activity was assayed. *P < .05. (D) 293T cells were transfected with nontargeting shRNA or shRNA-USP7. Forty-eight hours later, the cells were transfected with FLAG-TRAF6 or IKKγ, HA-ubiquitin, and ICP0 before they were lysed, and FLAG-tagged proteins were immunoprecipitated and blotted with anti-HA. WCLs were immunoblotted with specific antibodies to confirm ICP0 expression and USP7 knockdown. (E) THP1 cells were transduced with HF-HSV amplicon or HSV helper virus or left untransduced. Twenty minutes later the cells were lysed, and endogenous TRAF6 and IKKγ were immunoprecipitated with anti-TRAF6/IKKγ antibodies and blotted with antiubiquitin to assess their ubiquitination status. WCLs were immunoblotted for ICP0 expression. Cell lysates were also fractionated into nuclear and cytoplasmic fractions and blotted for USP7 expression. (F) Overexpression of ICP0 does not deplete endogenous TRAF6 or IKKγ: 293T cells were transfected with increasing concentration of ICP0 together with either FLAG-tagged TRAF6 or IKKγ in the presence or absence of the proteasome inhibitor, MG132 (5 μM added for the last 12 hours), and 24 hours later, cell lysate was blotted using anti-FLAG mAb to assess ICP0 effect on TRAF6 and IKKγ. ICP0 did not deplete either TRAF6 or IKKγ. (G) Comparison of wt-USP7 and USP7.NES ability to deubiquitinate TRAF6 and IKKγ and suppress TLR-induced NF-κB response: 293T cells coexpressing FLAG-tagged TRAF6 or IKKγ, HA-ubiquitin, and USP7 or USP7-NES. Thirty-six hours later, the cells were lysed and FLAG-tagged proteins were immunoprecipitated and blotted with anti-HA. (H) Enhanced deubiquitinating efficacy of USP7-NES against TRAF6/IKKγ compared with wt-USP7 correlated with its ability to suppress TLR-2/6 response to Pgn stimulation, measured as TNF-α mRNA. Error bars represent SEM.
In agreement with previous results showing that ICP0-M4 mutant that does not bind USP7 was deficient at inhibiting TLR response (Figure 3D), shRNA-mediated knockdown of USP7 compromised ICP0's ability to suppress TLR-induced NF-κB response (Figure 7C) and deubiquitinate TRAF6 and IKKγ (Figure 7D). These results support the hypothesis that ICP0 inhibits TLR response by recruiting USP7 to deubiquitinate TRAF6 and IKKγ. To investigate whether such a model could explain the difference in innate response to HF-HSV amplicon versus HSV helper virus, we compared the ubiquitination status of endogenous TRAF6 and IKKγ in THP1 cells transduced with HF-HSV versus HSV helper virus. As shown in Figure 7E, HF-HSV transduction induced polyubiquitination of endogenous TRAF6 and IKKγ; this effect was significantly attenuated in cells transduced with the helper virus expressing ICP0. A comparison of nuclear and cytoplasmic USP7 fractions from mock-, HF-HSV–, and HSV helper virus–transduced THP1 cells showed that in the presence of HSV helper virus–encoded ICP0, a significant fraction of USP7 translocated to the cytoplasm compared with mock and HF-HSV arms. In HF-HSV–transduced THP1 cells, cytoplasmic fraction of USP7 was intermediate to that seen in mock- and HSV helper–transduced cells, likely on the basis of HF-HSV–mediated TLR stimulation.
Previous studies showed that the ubiquitin-editing enzyme A20 terminates TNF-α receptor signaling by cleaving K63-linked polyubiquitin chains off RIP-1, mediated by its N-terminus OTU domain, followed by K48 polyubiquitination and proteasomal degradation of RIP1 mediated by its C-terminus E3 ligase zinc finger.44 Given the capacity of ICP0 to polyubiquitinate various substrates for proteasomal degradation through its N-terminus RING E3 ligase function, we questioned whether ICP0/USP7 could function in tandem to mimic the effect of A20, with USP7 deubiquitinating TRAF6/IKKγ followed by ICP0-mediated K48 polyubiquitination and degradation. To address this issue, TRAF6 and IKKγ were individually coexpressed with different concentrations of ICP0 in the presence or absence of the proteasome inhibitor MG-132 and protein levels were assessed by WB. As Figure 7F illustrates, there was no evidence for ICP0-mediated depletion of either TRAF6 or IKKγ. This would indicate that the role of ICP0 in TLR suppression is limited to facilitating USP7 nuclear export. In support of this, USP7 modified to express a nuclear export signal, USP7.NES (Figure 4Aiv), showed enhanced ability to suppress TLR2/6 cytokine response, and was more effective at deubiquitinating IKKγ than native USP7 (Figure 7G,H).
Discussion
HSV establishes life-long latency with repeat cycles of reactivation in the majority of nonimmunocompromised hosts. However, this does not translate into effective anti-HSV immunity to eradicate HSV. In this report, we identify HSV-1 ICP0 as a novel inhibitor of NF-κB/MAPK response to TLR signaling. The presence of such an inhibitor was inferred from the marked difference in innate response generated by HF-HSV and H+-HSV. Although several viruses have been reported to encode inhibitors of TLR signaling, there have been no previous reports of an HSV-encoded inhibitor of TLR response. ICP0 plays a central role in HSV reactivation from latency and propagation of lytic infection, conditions where HSV-encoded TLR ligands are abundantly generated. HSV infection of human dendritic cells (DCs) suppresses their antigen-presenting cell (APC) function; the latter is dependent on TLR signaling.45
HSV mutants lacking ICP020 (d109,19 in1312,46 and KM110,47 n212,35 713435,48 ) confirmed its role in suppressing interferon response. Subsequent work linked ICP0 to inhibition of IRF3/IRF733,35 as a mechanism for suppression of interferon response. However, inhibition of IRF3/IRF7 is not sufficient to explain HSV's ability to inhibit TLR2/TLR9-induced inflammatory cytokine response; the latter is mediated by MyD88-dependent NF-κB and MAPK activation, and is independent of IRF3/7. On the other hand, several studies have linked HSV to induction of NF-κB activity in infected cells; the latter appears to be necessary for efficient viral replication.49-51 This raises the question of how HSV could selectively promote sufficient NF-κB activity to promote its propagation yet avoid the robust innate cytokine response driven by the same NF-κB activity? This apparent contradiction is reconciled by the fact that HSV activates NF-κB through multiple signaling pathways. Some of which, such as HSV glycoprotein D activation of the TNF superfamily receptor HVEM (Hve-A),52 drive both the canonical and alternative NF-κB pathway; the latter is resistant to ICP0 suppression. TLR-mediated cytokine response, on the other hand, requires NF-κB and MAPK activity and both are dependent on K63-ubiquitin modification of signaling adaptors including TRAF6 and IKKγ.53-56 This differential requirement for K63-ubiquitination of adaptor proteins by nonoverlapping signaling pathways allows HSV to fine-tune NF-κB activity in infected cells, generating sufficient NF-κB response for its own replication yet suppressing host innate response.
Posttranslational protein modification by ubiquitination regulates a broad range of cellular processes including DNA repair, endocytosis, protein trafficking, signal transduction, and protein degradation.57 Several these processes impact pathogen life cycle, hence manipulation of the ubiquitin-proteasome system by pathogens has become a well-recognized survival strategy by viruses and bacteria.58,59 Stimulus-induced K63-ubiquitination of signaling adaptors following engagement of PRR, antigen receptors, and cytokine receptors has been linked to NF-κB response,60 whereas its reversal by Cyld and A20 inhibits both NF-κB and MAPK response.61,62 Macrophages and DCs from mice with germ line mutation expressing a ubiquitination-defective form of IKKγ (K392R) had blunted TLR cytokine response and were resistant to LPS-induced endotoxic shock.56 Here, we present evidence that ICP0:USP7 complex protects HSV against host innate immunity by directing USP7 to disassemble K63 polyubiquitin chains off TRAF6 and IKKγ. Within the TLR signaling pathway, the autoubiquitination site for TRAF6 has been mapped to lysine 124,55 whereas IKKγ is ubiquitinated at lysine 285 and 399 (392 in mice).63 Both TRAF6 Lys124 and IKKγ Lys399 are within close proximity to at least one of the USP7 consensus binding motifs, potentially enabling USP7 to dock in close proximity to polyubiquitinated lysine residues. Further supporting this hypothesis, the 2 P/AXXS USP7-binding motifs within IKKγ are located 4 residues from the binding site (aa's 388-393)64 of CYLD.
In summary, we show that HSV ICP0 is a potent inhibitor of TLR-mediated NF-κB response, an activity mediated through its partnership with the cellular DUB, USP7. ICP0 accomplishes this by translocating a nuclear protein, USP7, to the cytoplasm where it interacts with 2 adaptors within the TLR signaling pathway, TRAF6 and IKKγ. By translocating USP7 to the cytoplasm, ICP0 exploits USP7 normal response to TLR stimulation to the advantage of the HSV virus, effectively switching off TLR response to viral PAMPs. In support of this, a modified USP7 that constitutively resides in the cytosol was more effective than wt-USP7 at dampening TLR signal. The work shows how a seemingly contradictory set of functions residing on different domains of ICP0 (E3 ligase within the N-terminus RING domain and deubiquitinating activity of USP7 associated with the C-terminus of ICP0) could cooperate to provide comprehensive protection for HSV against the 2 main signaling pathways of innate immunity, namely IRF3/IRF7-mediated interferon response and NF-κB/MAPK–mediated inflammatory cytokine response.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dr Joseph D. Rosenblatt for critical discussion of the paper and Dr Roger Everett for providing ICP0, ICP0-M4, ICP0-FXE, USP7, and USP7.C223S. We thank Ann Casey, Clark Burris, and Louis Lotta for helper virus–free amplicon packaging and Wade Narrow for helper-based amplicon packaging. We acknowledge the Sylvester Comprehensive Cancer Center for providing support for this work.
This study was supported by National Institutes of Health (NIH, Bethesda, MD) grant NCI RO1 CA08798-05.
National Institutes of Health
Authorship
Contribution: S.D., D.S., Y.T., and H.L. performed research; H.J.F. and W.J.B. provided HSV virus; and K.T. designed experiments, performed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Khaled Tolba, Division of Hematology-Oncology, Department of Medicine, Sylvester Comprehensive Cancer Center, Batchelor Research Building, University of Miami, 1580 NW 10th Ave, Miami, FL 33136; e-mail: ktolba@mac.com.
References
Author notes
*S.D. and D.S. contributed equally to this work.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal