Abstract
Optimization of therapy for childhood acute lymphoblastic leukemia (ALL) requires a greater understanding of the cells that proliferate to maintain this malignancy because a significant number of cases relapse, resulting from failure to eradicate the disease. Putative ALL stem cells may be resistant to therapy and subsequent relapses may arise from these cells. We investigated expression of CD133, CD19, and CD38 in pediatric B-ALL. Cytogenetic and molecular analyses demonstrated that karyotypically aberrant cells were present in both CD133+/CD19+ and CD133+/CD19− subfractions, as were most of the antigen receptor gene rearrangements. However, ALL cells capable of long-term proliferation in vitro and in vivo were derived from the CD133+/CD19− subfraction. Moreover, these CD133+/CD19− cells could self-renew to engraft serial nonobese diabetic–severe combined immunodeficient recipients and differentiate in vivo to produce leukemias with similar immunophenotypes and karyotypes to the diagnostic samples. Furthermore, these CD133+/CD19− ALL cells were more resistant to treatment with dexamethasone and vincristine, key components in childhood ALL therapy, than the bulk leukemia population. Similar results were obtained using cells sorted for CD133 and CD38, with only the CD133+/CD38− subfraction demonstrating xenograft repopulating capacity. These data suggest that leukemia-initiating cells in childhood B-ALL have a primitive CD133+/CD19− and CD38− phenotype.
Introduction
Although the outcome for children with pediatric acute lymphoblastic leukemia (ALL) has improved considerably in recent years, a significant proportion of cases relapse, usually with disease that is resistant to treatment. Relapsed disease is the second most common cause of death among children in the United Kingdom. Intensification of treatment to prevent or treat relapsed patients may not be a feasible approach because of an increased risk of significant adverse effects. The complexity of the leukemogenic process, together with our limited understanding of the biology of this disease, presents a challenge to developing novel therapeutic approaches. Fundamental questions are the stage at which the leukemia originates within the hematopoietic hierarchy and the identity and characteristics of the cells that are responsible for maintaining this disease. High-speed multiparameter flow cytometry has permitted the characterization of normal and leukemia progenitor cells by enabling purification of cell populations for subsequent functional analyses. Such functional and immunophenotypic studies have indicated that leukemias could be considered stem cell disorders, in that the continued growth and dissemination of the malignant cells may be dependent on a small population with self-renewal ability. The demonstration of leukemia stem cells in acute myeloid leukemia (AML),1-5 chronic myeloid leukemia (CML),6 and more recently in pediatric ALL7-11 has added considerable support to this theory. Together these findings suggest that most leukemias are organized in a hierarchical manner, similar to their normal hematopoietic counterparts, with leukemia stem cells generating progeny that have an abnormal differentiation program
Cancer stem cells are not restricted to hematologic malignancies; they have recently been identified in tumors of the central nervous system,12 breast,13 and colon cancer.14,15 Significantly, these cancer stem cells have been shown to express CD133, a primitive cell antigen,16-18 that has been shown to be a more specific marker of hematopoietic stem cells than CD34.19-21 In glioblastoma, CD133+ cells have been shown to initiate and maintain tumor progression in vivo12 and are resistant to chemotherapeutic agents22 and ionizing radiation.23 The implications of these findings are that current treatment strategies may eventually fail because the CD133+ tumor-initiating cells are resistant to therapy. Such resistance of stem cells to therapy may be pertinent for childhood ALL. Whereas more differentiated cells may be killed by current therapeutic approaches, the leukemia stem cells may survive and subsequently contribute to disease relapse.
There have been conflicting reports on the expression of CD133 in ALL. Whereas some found high levels of CD133 expression in particular cases,24,25 others detected only low levels17 or none at all.18 However, the conclusions of these studies were based on flow cytometric analyses of patients' cells at diagnosis and were not verified by functional studies. We and others have recently shown that leukemia cells capable of reconstituting Ph+ and B-cell precursor (BCP) ALL in vivo have a primitive CD34+/CD10−/CD19−/CD38− phenotype, unlike the majority of leukemia cells in these subtypes.7-10 This suggests that the immunophenotype of the bulk ALL population is not representative of the leukemia-initiating cells that have in vivo reconstitution and self-renewal capacities. Given the critical role leukemia stem cells may play in disease progression and the increasing evidence for CD133 expression on cancer stem cells, we have investigated the expression of CD133 on childhood BCP ALL using functional in vitro and in vivo assays, drug sensitivity, and clonality analyses to further characterize the leukemia-initiating cells in this malignancy.
Methods
Patient cells
Bone marrow (BM) cells from children (median age, 6 years 9 months; range, 2-16 years) with BCP ALL at presentation or relapse were collected with approval of the Research Ethics Committee of the United Bristol Healthcare National Health Service (NHS) Trust, and informed consent was obtained in accordance with the Declaration of Helsinki. Detailed characteristics of the patients included in this study are shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Patient samples were selected on the basis of availability of material for study only, and none has been used in previous investigations.8,11 Normal bone marrow (NBM) was obtained from healthy donors with appropriate consent. Cells were separated using Ficoll-Hypaque (Sigma-Aldrich, Poole, United Kingdom) to obtain a mononuclear cell (MNC) population, then frozen in Iscove modified Dulbecco medium (IMDM; Invitrogen, Paisley, United Kingdom) with 50% fetal calf serum (Invitrogen) and 10% DMSO (Manor Park Pharmaceuticals, Bristol, United Kingdom) and stored in liquid nitrogen.
Cell sorting
Thawed ALL and NBM cells were suspended in Hanks balanced salt solution (Sigma-Aldrich) plus 2% human albumin solution (HAS; BPL Commercial, Elstree, United Kingdom) at 107 cells/mL. Cells were stained with monoclonal antibodies CD133/2-phycoerythrin (PE) (Miltenyi Biotec, Bisley, United Kingdom) and CD19-fluorescein isothiocyanate (FITC; clone 4G7) or CD38-FITC (clone HB7). Separate aliquots were stained with irrelevant IgG1 antibodies as isotype controls (all BD Biosciences, Oxford, United Kingdom). Cells were washed twice in Hanks balanced salt solution + 2% HAS and maintained on ice before sorting. Cells were sorted using a MoFlo Cell sorter (Beckman Coulter, High Wycombe, United Kingdom), on the basis of fluorescence intensity after gating on cells with low forward and side scatter. Sort gates were set up to exclude more than or equal to 99.9% of the cells in the isotype controls, and gate separations of at least 10 channels were used to discriminate positive from negative fractions to ensure purity (Figure 1A). The purity of sorted subfractions was checked during and after sorting, as previously described.8,11 This was further confirmed in the CD19− populations by checking for the absence of CD19 using antibody conjugated with allophycocyanin (BD Biosciences). The purity of the sorted subfractions was always more than 97%. CD19-allophycocyanin was also used in combination with CD133-PE and CD38-FITC for 3-color sorting of ALL and NBM samples. Cells were sorted into IMDM plus 50% fetal calf serum, washed, resuspended at known cell concentrations, and then used in functional assays and cytogenetic and genotypic analyses as described in the following sections.
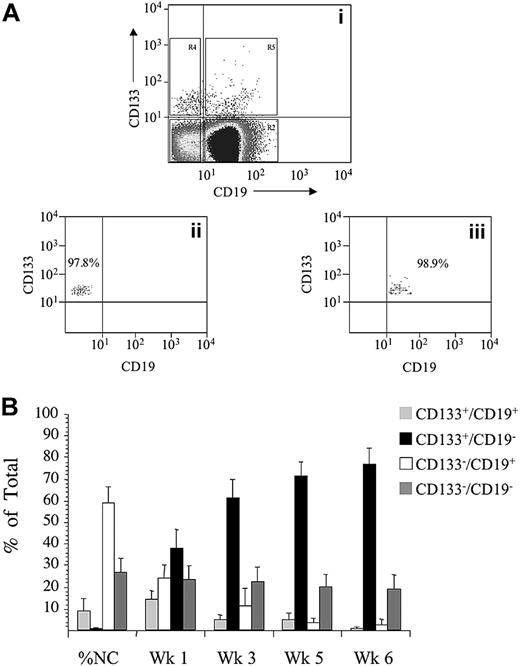
Purification strategy and proliferation of sorted ALL cells in vitro. ALL blast cells were gated on the basis of low forward and side scatter and subsequently gated for expression of CD133-PE and CD19-FITC, R2-R5 (A). Sort gates were separated by at least 10 channels, as shown. Quadrants in dot plot 1 were set up to exclude more than or equal to 99.9% of cells in isotype controls. Reanalysis, after sorting, of CD133+/CD19− cells gated in R4 and CD133+/CD19+ cells gated in R5, is shown in dot plots 2 and 3, respectively. ALL cells from patients 1 to 12 were sorted for expression of CD133 and CD19, and the proliferative capacity of the each of the sorted subfractions and unsorted controls was evaluated in suspension culture (SC) (B). Cultures were maintained with weekly half-media changes. Absolute cell counts, derived from each sorted population and unsorted controls, were determined by flow cytometry at weeks 1, 3, 5, and 6. These values were then used to calculate the proportion of the total viable cells, represented by each sorted population. The proportion of the total cells derived from the each sorted subfraction is presented as the mean plus or minus SE.
Purification strategy and proliferation of sorted ALL cells in vitro. ALL blast cells were gated on the basis of low forward and side scatter and subsequently gated for expression of CD133-PE and CD19-FITC, R2-R5 (A). Sort gates were separated by at least 10 channels, as shown. Quadrants in dot plot 1 were set up to exclude more than or equal to 99.9% of cells in isotype controls. Reanalysis, after sorting, of CD133+/CD19− cells gated in R4 and CD133+/CD19+ cells gated in R5, is shown in dot plots 2 and 3, respectively. ALL cells from patients 1 to 12 were sorted for expression of CD133 and CD19, and the proliferative capacity of the each of the sorted subfractions and unsorted controls was evaluated in suspension culture (SC) (B). Cultures were maintained with weekly half-media changes. Absolute cell counts, derived from each sorted population and unsorted controls, were determined by flow cytometry at weeks 1, 3, 5, and 6. These values were then used to calculate the proportion of the total viable cells, represented by each sorted population. The proportion of the total cells derived from the each sorted subfraction is presented as the mean plus or minus SE.
Suspension culture assay
Suspension cultures were initiated with unsorted ALL cells or with sorted subfractions at up to 5 × 105 cells/mL, supplemented with interleukin (IL)-3, IL-7, and stem cell factor, and maintained for up to 6 weeks with regular measurements of cell viability and absolute cell counts, by flow cytometry and cytogenetic or morphologic analyses, as described previously.8,11
Identification of clonal receptor gene rearrangements
Diagnostic samples and sorted ALL subfractions were screened for the presence of immunoglobulin (Ig) and T-cell receptor (TCR) rearrangements (IGH, IGK-Kde, TCRG, TCRD, and TCRB). DNA was extracted using QIAamp DNA Micro kit (QIAGEN, Crawley, United Kingdom). In these cases, the CD133+/CD19+ and CD133+/CD19− subfractions composed only 1.2 × 103 to 3 × 103 and 1.5 × 103 to 9 × 103 cells, respectively. Consequently, DNA extracted from these subfractions required amplification using a GenomiPhi DNA Amplification kit (GE Healthcare, Little Chalfont, United Kingdom) according to the manufacturer's instructions. The polymerase chain reaction (PCR) primers and conditions used were as described previously.26,27 A positive control gene PCR was set up in parallel to confirm successful DNA amplification. PCR products were subjected to heteroduplex analysis,28 cloned, and the monoclonal products were sequenced. Each rearrangement was characterized by comparison of sequences with germline sequences contained in published databases available online.29,30
Transplantation of leukemia cells into NOD/SCID mice
Nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice were bred and maintained at the University of Bristol. One hour before transplantation, 6- to 8-week-old NOD/SCID mice were irradiated with 2.5 Gy acute X-rays from a linear accelerator at a rate of 7.5 cGy/second. Unsorted ALL cells and sorted subfractions were resuspended in 0.3 mL IMDM plus 5% HAS and injected into the lateral tail vein. Animals were maintained for 8 to 10 weeks and killed electively or until they began to exhibit clinical symptoms of disease. On death, the gross anatomy of each mouse was inspected and femoral BM samples were removed for flow cytometric and cytogenetic/histologic analyses. Animal studies were performed under protocols licensed by the United Kingdom Home Office.
MNCs harvested from some NOD/SCID BM were used for serial transplantation experiments to evaluate self-renewal. For comparison with primary transplants, equal numbers of CD45+ cells were inoculated into the serial recipients. These cells were not enriched for any particular phenotype before evaluation in sequential xenografts.
Evaluation of transplanted cells
The immunophenotype of each xenograft was examined using antibodies against CD45 and appropriate lineage markers, as described.8,11 In every case, cells were stained with CD45 along with one or 2 of the following lineage markers: CD3, CD10, CD19, CD33, CD34 (all BD Biosciences), or CD133 to compare the immunophenotype of the engrafted cells to that of the patient at diagnosis/relapse. We defined human cell engraftment as expression of more than or equal to 0.1% CD45+ cells in a sample. Aliquots of cells from NOD/SCID BM were plated onto slides for cytogenetic or morphologic analysis.
In vitro drug sensitivity
Unsorted ALL cells and sorted subfractions were cocultured with and without increasing concentrations of the glucocorticoid dexamethasone or the Vinca alkaloid vincristine (both Sigma-Aldrich) for 48 and 72 hours, respectively. Unsorted and hematopoietic stem cell-enriched (CD133+/CD38−/CD19−) normal BM samples were also cocultured with drugs to compare the effects on normal and leukemia populations. Apoptosis and viability were assessed by flow cytometry using a human annexin V-FITC and propidium iodide kit (arcus Biologicals, Modena, Italy), according to the manufacturer's instructions. The functional capacity of drug-treated cells was investigated in suspension culture, as described in “Suspension culture assay.”
Cytogenetic and morphologic analysis
Cytogenetic analysis by fluorescence in situ hybridization (FISH) was performed on samples at diagnosis, on cells immediately after sorting, cells harvested from cultures at weeks 2, 4, and 6, cells harvested from NOD/SCID marrow, and on drug-treated cells by the Regional Cytogenetics Unit, Southmead Hospital. At least 50 nuclei per sample were scored on preparations from cultured cells and NOD/SCIDs. If cytogenetic analysis was not possible, cytospins of cultured cells and NOD/SCID BM were prepared, then fixed and stained with Hema-Gurr (BDH, Poole, United Kingdom).
Statistical analysis
Statistical analyses were performed using matched paired t tests or analysis of variance with the Tukey post hoc analysis of means.
Results
Phenotypic characterization of ALL cells with long-term proliferative ability in vitro
Cells from 12 children with ALL were stained with antibodies against CD133 and CD19, the 4 subfractions were sorted and set up in suspension culture to evaluate long-term proliferative ability (Figure 1B). At sorting the majority of cells were CD133−/CD19+ (60 ± 7.9%), whereas 29% plus or minus 6.8% were CD133−/CD19− and the CD133+/CD19+ and CD133+/CD19− subfractions represented only 9.7% plus or minus 6.3% and 0.8% plus or minus 0.4%, respectively. After 1 week in suspension culture, similar numbers of cells were detected in each subfraction. However, by week 3, the majority of cells were derived from the CD133+/CD19− subfraction (62% ± 8%). Cells derived from the CD133+/CD19− subfraction also accounted for the majority of cells at week 5 (72% ± 6%) and week 6 (77% ± 7%). The proportion of cells derived from this subfraction was significantly higher than from any other evaluated (F = 51, P < .001, Fcritical = 2.8). In cultures of CD133+/CD19− cells, there was a 10- to more than 18 000-fold expansion in absolute cell numbers from more than or equal to 150 cells at initiation up to 7.9 × 106 at week 6. This expansion was significantly higher than the other subfractions and the unsorted cells (F = 9.6, P < .01, Fcritical = 4.1). Results for individual patients are shown in Table 1. Cytogenetic analyses were performed on the sorted subfractions immediately after sorting in 6 cases and confirmed that cells with an aberrant karyotype were present in both CD133+/CD19+ (50%-82% FISH+) and CD133+/CD19− subfractions (76%-92% FISH+). In 10 patients, FISH analyses on cultured cells at weeks 4 and 6 confirmed both the CD133+/CD19+ (50%-100%) and CD133+/CD19− (60%-100%) populations contained cytogenetically aberrant cells, confirming maintenance of leukemia in vitro. In the other 2 patients, morphologic analyses demonstrated that the majority of cultured cells derived from these subfractions had blast morphology (CD133+/CD19+, 80%-100%; CD133+/CD19−, 90%-100%).
Growth of childhood ALL cell population in suspension culture
| Patient no. . | Karyotype . | Subfraction . | Percentage NC . | Absolute cell counts × 104 . | Percentage FISH+ (week 4/week 6) . | |
|---|---|---|---|---|---|---|
| Initiation . | Week 6 . | |||||
| 1 | t(12;21) | CD133+/CD19+ | 0.96 | 0.54 | 0 | — |
| CD133+/CD19− | 0.48 | 0.72 | 23.8 | 100 | ||
| 2 | Hyperdiploid | CD133+/CD19+ | 72.26 | 100 | 0.1 | 75 |
| CD133+/CD19− | 3.92 | 80.6 | 792.8 | 92 | ||
| 3 | del 9p, 17p | CD133+/CD19+ | 33.54 | 50 | 0.64 | 50 |
| CD133+/CD19− | 0.21 | 1.7 | 140.8 | 62 | ||
| 4 | t(12;21) | CD133+/CD19+ | 0.55 | 1.8 | 0.96 | 50 |
| CD133+/CD19− | 0.03 | 0.06 | 83.2 | 79 | ||
| 5 | t(12;21) | CD133+/CD19+ | 1.78 | 2.31 | 0.64 | * |
| CD133+/CD19− | 0.09 | 0.4 | 454.4 | 76 | ||
| 6 | t(12;21) | CD133+/CD19+ | 0.42 | 1 | 0.13 | — |
| CD133+/CD19− | 0.28 | 1 | 140.8 | 85 | ||
| 7 | t(12;17) | CD133+/CD19+ | 0.49 | 2.7 | 0 | — |
| CD133+/CD19− | 0.06 | 0.05 | 131.2 | — | ||
| 8 | t(12;21) | CD133+/CD19+ | 3.81 | 3 | 6.4 | 100 |
| CD133+/CD19− | 0.25 | 0.015 | 278.4 | 100 | ||
| 9 | Complex | CD133+/CD19+ | 1.92 | 9.32 | 0 | — |
| CD133+/CD19− | 3.15 | 9.16 | 185.6 | 93 | ||
| 10 | +6 | CD133+/CD19+ | 0.33 | 0.22 | 1.14 | * |
| CD133+/CD19− | 0.09 | 0.05 | 163.2 | 60 | ||
| 11 | 46XX | CD133+/CD19+ | 0.03 | 0.11 | 0 | — |
| CD133+/CD19− | 0.20 | 0.37 | 768.3 | — | ||
| 12 | t(12;21) | CD133+/CD19+ | 0.17 | 0.22 | 0 | — |
| CD133+/CD19− | 0.35 | 0.14 | 288 | 82 | ||
| Median, CD133+/CD19+ | 0.76 | 2.06 | 0.12 | 62.5 | ||
| Median, CD133+/CD19− | 0.23 | 0.39 | 174.4 | 83.5 | ||
| Patient no. . | Karyotype . | Subfraction . | Percentage NC . | Absolute cell counts × 104 . | Percentage FISH+ (week 4/week 6) . | |
|---|---|---|---|---|---|---|
| Initiation . | Week 6 . | |||||
| 1 | t(12;21) | CD133+/CD19+ | 0.96 | 0.54 | 0 | — |
| CD133+/CD19− | 0.48 | 0.72 | 23.8 | 100 | ||
| 2 | Hyperdiploid | CD133+/CD19+ | 72.26 | 100 | 0.1 | 75 |
| CD133+/CD19− | 3.92 | 80.6 | 792.8 | 92 | ||
| 3 | del 9p, 17p | CD133+/CD19+ | 33.54 | 50 | 0.64 | 50 |
| CD133+/CD19− | 0.21 | 1.7 | 140.8 | 62 | ||
| 4 | t(12;21) | CD133+/CD19+ | 0.55 | 1.8 | 0.96 | 50 |
| CD133+/CD19− | 0.03 | 0.06 | 83.2 | 79 | ||
| 5 | t(12;21) | CD133+/CD19+ | 1.78 | 2.31 | 0.64 | * |
| CD133+/CD19− | 0.09 | 0.4 | 454.4 | 76 | ||
| 6 | t(12;21) | CD133+/CD19+ | 0.42 | 1 | 0.13 | — |
| CD133+/CD19− | 0.28 | 1 | 140.8 | 85 | ||
| 7 | t(12;17) | CD133+/CD19+ | 0.49 | 2.7 | 0 | — |
| CD133+/CD19− | 0.06 | 0.05 | 131.2 | — | ||
| 8 | t(12;21) | CD133+/CD19+ | 3.81 | 3 | 6.4 | 100 |
| CD133+/CD19− | 0.25 | 0.015 | 278.4 | 100 | ||
| 9 | Complex | CD133+/CD19+ | 1.92 | 9.32 | 0 | — |
| CD133+/CD19− | 3.15 | 9.16 | 185.6 | 93 | ||
| 10 | +6 | CD133+/CD19+ | 0.33 | 0.22 | 1.14 | * |
| CD133+/CD19− | 0.09 | 0.05 | 163.2 | 60 | ||
| 11 | 46XX | CD133+/CD19+ | 0.03 | 0.11 | 0 | — |
| CD133+/CD19− | 0.20 | 0.37 | 768.3 | — | ||
| 12 | t(12;21) | CD133+/CD19+ | 0.17 | 0.22 | 0 | — |
| CD133+/CD19− | 0.35 | 0.14 | 288 | 82 | ||
| Median, CD133+/CD19+ | 0.76 | 2.06 | 0.12 | 62.5 | ||
| Median, CD133+/CD19− | 0.23 | 0.39 | 174.4 | 83.5 | ||
NC indicates nucleated cells; and —, not applicable.
FISH probe hybridization unsuccessful.
Analysis of clonal IgH/TCR gene rearrangements
Material was available from 5 patients to investigate antigen receptor gene rearrangements in the sorted subfractions and compared with those detected at diagnosis (Table 2). Each patient had at least one clonal rearrangement used for routine monitoring of residual disease throughout the course of treatment. In 2 cases (patients 7 and 14), the unique rearrangements detected at diagnosis were also present in each sorted subfraction. However, in the other 3 patients, differences in the rearrangements were observed. The VH2 rearrangement identified at diagnosis in patient 9 was detected in each subfraction except CD133−/CD19−. In patient 10, 2 different VK3 rearrangements were detected in the CD133+/CD19+ subfraction. Neither of these rearrangements was observed in the diagnostic sample or in any other sorted cell population. In addition, a change in the VK1 rearrangement was observed in CD133−/CD19+ cells, which was not detected in any other population from this patient. Similarly, in patient 18, VH3 rearrangements were observed in the CD133−/CD19+ subfraction that were not identified in the other subfractions or at diagnosis.
Sequence analysis of antigen receptor rearrangements in sorted cell populations
| Patient no. and sample source . | Germline regions . | Junctional region . | |
|---|---|---|---|
| 7 | |||
| Diagnostic, unsorted | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| CD133+CD19+ | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| CD133+CD19− | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| CD133−CD19+ | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| CD133−CD19− | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| 9 | |||
| Diagnostic, unsorted | VH2-5/DH2-2/JH6*02 | −1/8/−20 | −6/12/9 |
| CD133+CD19+ | VH2-5/DH2-2/JH6*02 | −1/8/−20 | −6/12/9 |
| CD133+CD19− | VH2-5/DH2-2/JH6*02 | −1/8/−20 | −6/12/9 |
| CD133−CD19+ | VH2-5/DH2-2/JH6*02 | −1/8/−20 | −6/12/9 |
| CD133−CD19− | † | ||
| 10 | |||
| Diagnostic, unsorted | VK3-20*01/Kde | 0/7/−6 | |
| CD133+CD19+ | VK3-20*01/Kde | 0/17/−23 | |
| VK3-20*01/Kde | 0/5/−11 | ||
| CD133+CD19− | VK3-20*01/Kde | 0/7/−6 | |
| CD133−CD19+ | VK3-20*01/Kde | 0/7/−6 | |
| CD133−CD19− | VK3-20*01/Kde | 0/7/−6 | |
| Diagnostic, unsorted | VK1-17*01/Kde | −1/0/−3 | |
| CD133+CD19+ | VK1-17*01/Kde | −1/0/−3 | |
| CD133+CD19− | VK1-17*01/Kde | −1/0/−3 | |
| CD133−CD19+ | VK1-17*01/Kde | 0/0/−24 | |
| CD133−CD19− | VK1-17*01/Kde | −1/0/−3 | |
| 14 | |||
| Diagnostic, unsorted | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| CD133+CD19+ | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| CD133+CD19− | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| CD133−CD19+ | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| CD133−CD19− | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| Diagnostic, unsorted | TCR D V2*03/DD3 | 0/8/−23 | |
| CD133+CD19+ | TCR D V2*03/DD3 | 0/8/−23 | |
| CD133+CD19− | TCR D V2*03/DD3 | 0/8/−23 | |
| CD133−CD19+ | TCR D V2*03/DD3 | 0/8/−23 | |
| CD133−CD19− | TCR D V2*03/DD3 | 0/8/−23 | |
| 18 | |||
| Diagnostic, unsorted | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| CD133+CD19+ | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| CD133+CD19− | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| CD133−CD19+ | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| CD133−CD19− | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| Diagnostic, unsorted | VH3-13*01/JH6*03 | 0/0/−6 | |
| CD133+CD19+ | VH3-13*01/JH6*03 | 0/0/−6 | |
| CD133+CD19− | VH3-13*01/JH6*03 | 0/0/−6 | |
| CD133−CD19+ | VH3-13*01/J4*02 | −1/18/−6 | |
| VH3-23*01/DH3-16*01/J6*03 | −2/12/−7 | −13/12/3 | |
| CD133−CD19− | VH3-13*01/JH6*03 | 0/0/−6 | |
| Patient no. and sample source . | Germline regions . | Junctional region . | |
|---|---|---|---|
| 7 | |||
| Diagnostic, unsorted | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| CD133+CD19+ | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| CD133+CD19− | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| CD133−CD19+ | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| CD133−CD19− | VH3-9*01/DH3-3/JH6*02 | −1/6/−14 | −9/3/−14 |
| 9 | |||
| Diagnostic, unsorted | VH2-5/DH2-2/JH6*02 | −1/8/−20 | −6/12/9 |
| CD133+CD19+ | VH2-5/DH2-2/JH6*02 | −1/8/−20 | −6/12/9 |
| CD133+CD19− | VH2-5/DH2-2/JH6*02 | −1/8/−20 | −6/12/9 |
| CD133−CD19+ | VH2-5/DH2-2/JH6*02 | −1/8/−20 | −6/12/9 |
| CD133−CD19− | † | ||
| 10 | |||
| Diagnostic, unsorted | VK3-20*01/Kde | 0/7/−6 | |
| CD133+CD19+ | VK3-20*01/Kde | 0/17/−23 | |
| VK3-20*01/Kde | 0/5/−11 | ||
| CD133+CD19− | VK3-20*01/Kde | 0/7/−6 | |
| CD133−CD19+ | VK3-20*01/Kde | 0/7/−6 | |
| CD133−CD19− | VK3-20*01/Kde | 0/7/−6 | |
| Diagnostic, unsorted | VK1-17*01/Kde | −1/0/−3 | |
| CD133+CD19+ | VK1-17*01/Kde | −1/0/−3 | |
| CD133+CD19− | VK1-17*01/Kde | −1/0/−3 | |
| CD133−CD19+ | VK1-17*01/Kde | 0/0/−24 | |
| CD133−CD19− | VK1-17*01/Kde | −1/0/−3 | |
| 14 | |||
| Diagnostic, unsorted | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| CD133+CD19+ | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| CD133+CD19− | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| CD133−CD19+ | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| CD133−CD19− | TCR G V9*01/TCR G J2*01 | −6/1/−1 | |
| Diagnostic, unsorted | TCR D V2*03/DD3 | 0/8/−23 | |
| CD133+CD19+ | TCR D V2*03/DD3 | 0/8/−23 | |
| CD133+CD19− | TCR D V2*03/DD3 | 0/8/−23 | |
| CD133−CD19+ | TCR D V2*03/DD3 | 0/8/−23 | |
| CD133−CD19− | TCR D V2*03/DD3 | 0/8/−23 | |
| 18 | |||
| Diagnostic, unsorted | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| CD133+CD19+ | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| CD133+CD19− | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| CD133−CD19+ | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| CD133−CD19− | VH1-3/DH2-15/JH5*02 | −13/12/−13 | −4/11/−4 |
| Diagnostic, unsorted | VH3-13*01/JH6*03 | 0/0/−6 | |
| CD133+CD19+ | VH3-13*01/JH6*03 | 0/0/−6 | |
| CD133+CD19− | VH3-13*01/JH6*03 | 0/0/−6 | |
| CD133−CD19+ | VH3-13*01/J4*02 | −1/18/−6 | |
| VH3-23*01/DH3-16*01/J6*03 | −2/12/−7 | −13/12/3 | |
| CD133−CD19− | VH3-13*01/JH6*03 | 0/0/−6 | |
Clonal rearrangement not present.
Engraftment of ALL subfractions in NOD/SCID mice
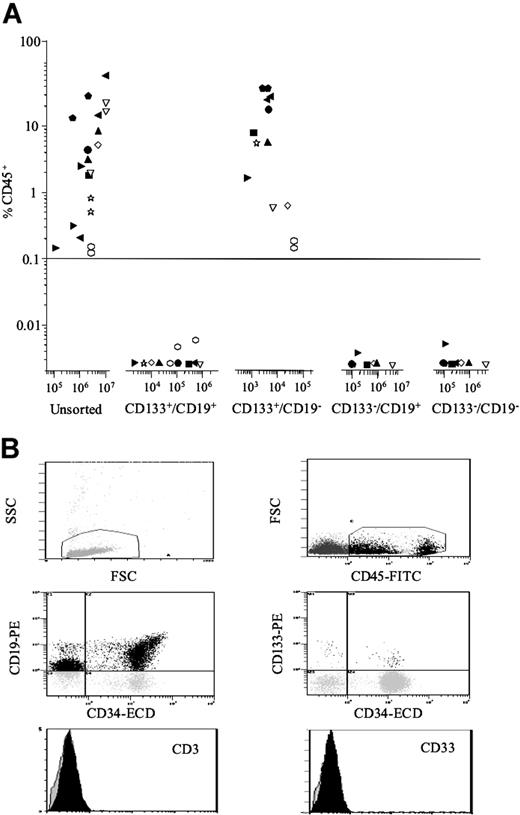
Cells from 10 patients were sorted for CD133/CD19, and the ability of each of the 4 subfractions to engraft NOD/SCID mice was evaluated (Figure 2A). Inoculation of 105 to 107 unsorted cells resulted in engraftment levels varying from 0.12% to 56% CD45+. In recipients of sorted populations, engraftment was only observed using CD133+/CD19− cells (range, 0.14%-35% CD45+ using 743-50 000 cells). To achieve equivalent levels of engraftment, using unsorted cells, 40- to more than 1000-fold more cells had to be inoculated (P < .001). There was no detectable engraftment with the CD133+/CD19+ subfraction or with either of the 2 CD133− populations, despite injecting more than 400-fold more cells in the latter 2 cases.
In vivo repopulating capacity and phenotypic analysis of engrafted NOD/SCID BM. ALL cells from 10 patients (1, 2, 5-11, 14) were sorted for expression of CD133 and CD19. Both unsorted cells and the sorted subfractions were evaluated for their ability to engraft irradiated NOD/SCID recipients (A). Each patient is represented by a specific symbol, and each symbol depicts the engraftment as measured by CD45+ cells present in the BM of the NOD/SCID recipients. BM cells removed from NOD/SCID mice that had been engrafted with CD133+/CD19− cells were analyzed in more detail using additional lineage markers (B). Cells were initially gated on the basis of low forward and side scatter; then CD45+ cells were gated and the expression of the lymphoid and myeloid antigens on these gated cells was investigated. The peaks representing CD3 and CD33 antibodies are shown in black, and isotype controls are represented as translucent peaks on the overlay histograms. The figure shows the immunophenotype of cells removed from an NOD/SCID mouse inoculated with CD133+/CD19− cells from patient 14.
In vivo repopulating capacity and phenotypic analysis of engrafted NOD/SCID BM. ALL cells from 10 patients (1, 2, 5-11, 14) were sorted for expression of CD133 and CD19. Both unsorted cells and the sorted subfractions were evaluated for their ability to engraft irradiated NOD/SCID recipients (A). Each patient is represented by a specific symbol, and each symbol depicts the engraftment as measured by CD45+ cells present in the BM of the NOD/SCID recipients. BM cells removed from NOD/SCID mice that had been engrafted with CD133+/CD19− cells were analyzed in more detail using additional lineage markers (B). Cells were initially gated on the basis of low forward and side scatter; then CD45+ cells were gated and the expression of the lymphoid and myeloid antigens on these gated cells was investigated. The peaks representing CD3 and CD33 antibodies are shown in black, and isotype controls are represented as translucent peaks on the overlay histograms. The figure shows the immunophenotype of cells removed from an NOD/SCID mouse inoculated with CD133+/CD19− cells from patient 14.
Immunophenotypic analyses of engrafted CD45+ cells revealed that they had the same characteristic immunophenotype as the patients at diagnosis: high expression of CD10 (> 42%) and CD19 (> 64%) and lack of expression of CD3 and CD33 (Figure 2B). Details of extended immunophenotypes are provided (Table S2). Expression of CD34 was also high in the engrafted cells (> 56%), with the exception of NOD/SCIDs inoculated with cells from patients 2, 10, and 11 (2%-16% CD34+). However, this low CD34 expression was consistent with the immunophenotypes of these patients at diagnosis. FISH analyses revealed that 50% to 100% of the engrafted cells had an abnormal karyotype, consistent with the number of abnormal cells detected at diagnosis and confirmed engraftment of leukemia cells.
BM cells harvested from mice inoculated with CD133+/CD19− cells from 5 of the 10 patients were transplanted into secondary recipients to evaluate self-renewal ability (Table 3). In each case, similar levels of engraftment were observed between the primary (5%-35% CD45+) and secondary recipients (7%-30% CD45+). The immunophenotypes of the cells harvested from the secondary recipients were consistent with those observed in the corresponding primary animals. FISH analyses confirmed that leukemia cells were being transplanted in the serial xenografts.
Immunophenotypic and karyotypic analyses of BM cells from serial xenografts
| Patient no. . | Xenograft . | No. of cells injected . | % CD45+ in BM . | % FISH+ . | Immunophenotype* . |
|---|---|---|---|---|---|
| 1 | Primary | 6.5 × 103 | 20 ± 4 | 100 | CD3−/CD10+/CD19+/CD33−/ CD34+ |
| Secondary | 6.5 × 103 | 25 ± 3 | 100 | CD3−/CD10+/CD19+/CD33−/CD34+ | |
| 5 | Primary | 1.8 × 103 | 5 | 82 | CD3−/CD10+/CD19+/CD34+ |
| Secondary | 1.8 × 103 | 7 ± 3 | 76 | CD3−/CD10+/CD19+/CD34+ | |
| 6 | Primary | 5.4 × 103 | 18 ± 6 | 85 | CD3−/CD10+/CD19+/CD33−/CD34+ |
| Secondary | 5.4 × 103 | 15 ± 5 | 72 | CD3−/CD10+/CD19+/CD33−/CD34+ | |
| 8 | Primary | 7.9 × 103 | 8 ± 2 | 90 | CD3−/CD10+/CD19+/CD33−/CD34+ |
| Secondary | 7.9 × 103 | 11 ± 3 | 85 | CD3−/CD10+/CD19+/CD33−/CD34+ | |
| 14 | Primary | 5.4 × 103 | 35 ± 6 | — | CD3−/CD10+/CD19+/CD33−/CD34+ |
| Secondary | 5.4 × 103 | 30 ± 7 | — | CD3−/CD10+/CD19+/CD33−/CD34+ |
| Patient no. . | Xenograft . | No. of cells injected . | % CD45+ in BM . | % FISH+ . | Immunophenotype* . |
|---|---|---|---|---|---|
| 1 | Primary | 6.5 × 103 | 20 ± 4 | 100 | CD3−/CD10+/CD19+/CD33−/ CD34+ |
| Secondary | 6.5 × 103 | 25 ± 3 | 100 | CD3−/CD10+/CD19+/CD33−/CD34+ | |
| 5 | Primary | 1.8 × 103 | 5 | 82 | CD3−/CD10+/CD19+/CD34+ |
| Secondary | 1.8 × 103 | 7 ± 3 | 76 | CD3−/CD10+/CD19+/CD34+ | |
| 6 | Primary | 5.4 × 103 | 18 ± 6 | 85 | CD3−/CD10+/CD19+/CD33−/CD34+ |
| Secondary | 5.4 × 103 | 15 ± 5 | 72 | CD3−/CD10+/CD19+/CD33−/CD34+ | |
| 8 | Primary | 7.9 × 103 | 8 ± 2 | 90 | CD3−/CD10+/CD19+/CD33−/CD34+ |
| Secondary | 7.9 × 103 | 11 ± 3 | 85 | CD3−/CD10+/CD19+/CD33−/CD34+ | |
| 14 | Primary | 5.4 × 103 | 35 ± 6 | — | CD3−/CD10+/CD19+/CD33−/CD34+ |
| Secondary | 5.4 × 103 | 30 ± 7 | — | CD3−/CD10+/CD19+/CD33−/CD34+ |
Primary indicates primary NOD/SCID recipient; Secondary, secondary NOD/SCID recipient; and —, not applicable.
The cutoff point for positivity was set at 20% of cells staining more intensely than the isotype control, in accordance with standard diagnostic immunophenotyping procedures defined by the British Committee for Standards in Haematology.31
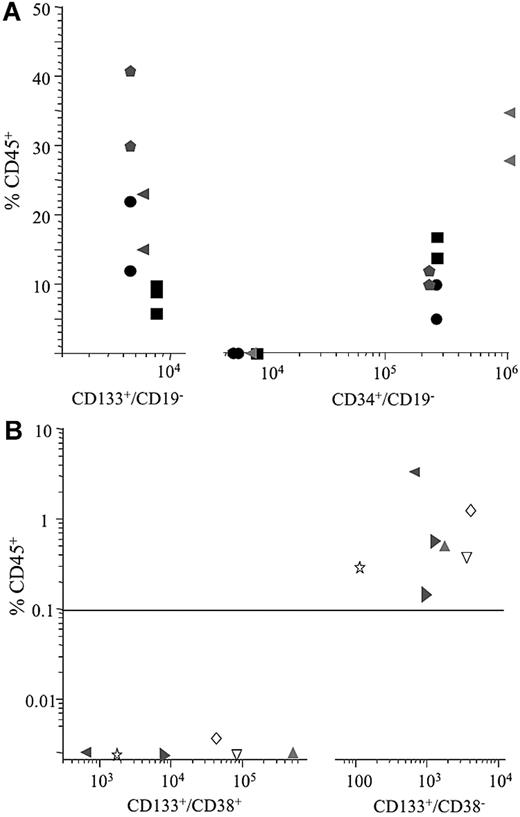
Where sample size permitted, the CD34+/CD19− subfraction was also enriched to compare the leukemia-initiating cell content of this subfraction with the CD133+/CD19− population. When similar numbers of CD133+/CD19− and CD34+/CD19− cells were inoculated, engraftment was only observed in recipients of CD133+/CD19− cells. In these 4 cases, engraftment could not be achieved using inocula of 5.4 × 103 to 8 × 103 CD34+/CD19− cells; at least 2.2 × 105 CD34+/CD19− cells were required for engraftment of leukemia. In contrast, a 30- to 100-fold enrichment of NOD/SCID repopulating cells was achieved by sorting for the CD133+/CD19− subfraction where engraftment was achieved using 5.4 × 103 to 6.5 × 103 cells (Figure 3A).
In vivo repopulating capacity of ALL subpopulations. ALL cells from 4 patients (1, 6, 8, and 14) were sorted for both CD133/19 and CD34/19 (A), and 6 patients (1, 5, 7, and 9-11) were sorted for expression of CD133 and CD38 (B). The closed symbols in panel A represent inoculation of CD133+/CD19− cells; open symbols, CD34+/CD19− cells from the same patient. Numbers on the x-axes represent the number of cells inoculated into recipient animals.
In vivo repopulating capacity of ALL subpopulations. ALL cells from 4 patients (1, 6, 8, and 14) were sorted for both CD133/19 and CD34/19 (A), and 6 patients (1, 5, 7, and 9-11) were sorted for expression of CD133 and CD38 (B). The closed symbols in panel A represent inoculation of CD133+/CD19− cells; open symbols, CD34+/CD19− cells from the same patient. Numbers on the x-axes represent the number of cells inoculated into recipient animals.
Subsequently, cells from 6 of these patients were sorted for expression of CD133 and CD38 and evaluated in the NOD/SCID assay (Figure 3B). Because no engraftment had been observed with the CD133− populations from these patients, in the initial studies, only CD133+/CD38+ and CD133+/CD38− subfractions were sorted to maximize cell numbers for evaluation. In this subgroup, the proportions of CD133+/CD38+ and CD133+/CD38− cells ranged from 0.06% to 1.3% and 0.02% to 0.3%, respectively. No engraftment was observed in recipients of between 670 and 5 × 105 CD133+/CD38+ cells. In contrast, engraftment (0.13%-3.1% CD45+) was observed in recipients of between 650 and 4 × 103 CD133+/CD38− cells. Cells removed from the primary recipient of CD133+/CD38− cells from patient 1 were capable of engrafting secondary recipients (2.6%-3% CD45+). Cytogenetic analyses confirmed the engrafted cells were of leukemia origin.
Multicolor enrichment of ALL cells
In an attempt to further enrich these ALL cells, 3-color sorting was performed on samples from 5 patients. The CD133+/CD19−/CD38− subfraction represented 0.027% plus or minus 0.014% of cells (range, 0.005%-0.079%). The number of cells recovered from sorting this population ranged from 610 to 2500 (mean ± SD, 1612 ± 310). Because cell numbers were so limiting, functional evaluations were performed in suspension culture only. In cultures of CD133+/CD19−/CD38− cells, there was a 7- to more than 7800-fold expansion (median, 682-fold) in absolute cell numbers. This expansion was significantly higher than the unsorted cells (median, 1.8-fold, P = .013). Expansion in cultures of CD133+/CD19−/CD38− cells was also greater than that observed with the CD133+/CD19− population (range, 1.6- to 141-fold; median, 20-fold) in these patients. FISH analyses on the cultured CD133+/CD19−/CD38− populations confirmed the presence of cytogenetically aberrant cells (62%-88% FISH+).
In vitro drug sensitivity
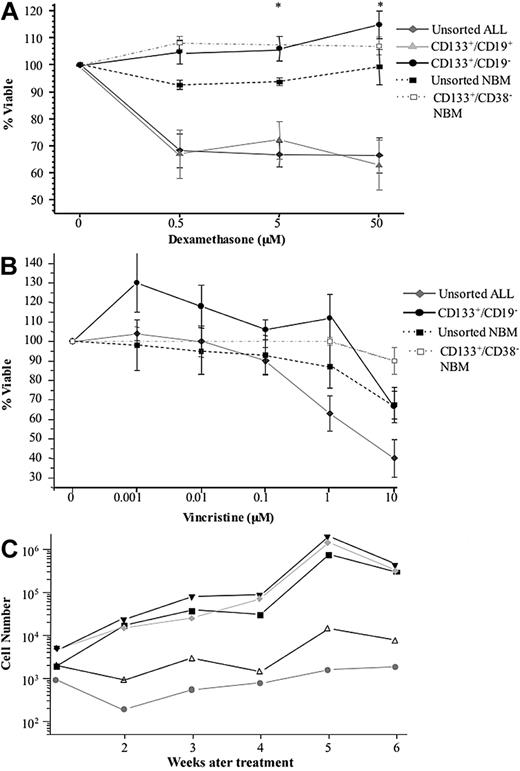
To assess the relative efficacy of dexamethasone on CD133+ ALL subpopulations and on the bulk leukemia, cells from 10 patients were cocultured with the glucocorticoid. Figure 4A shows the percentage viable relative to untreated controls after 48-hour exposure. The viability of the bulk leukemia and CD133+/CD19+ cells was reduced after treatment with dexamethasone. In contrast, no toxicity was observed in the CD133+/CD19− subfractions with higher cell viability observed in the treated samples than in the untreated controls. The viability of CD133+/CD19− cells was significantly higher than the unsorted bulk leukemia at 5 μM and 50 μM (F = 5.8, P < .01, Fcritical = 5.6). FISH and morphologic analyses confirmed that leukemia cells survived drug treatment (> 68% FISH+). Normal unsorted BM and samples enriched for CD133+/CD38− normal hematopoietic stem cells were largely unaffected by dexamethasone. There were no significant differences between the effects of dexamethasone on the stem cell-enriched fractions from ALL or from NBM samples (P > .2). A similar trend was observed when the sensitivity of the CD133+/CD19− population to the Vinca alkaloid vincristine was examined (Figure 4B). A dose of 1 μM vincristine was sufficient to kill 50% of the unsorted ALL cells. In contrast, a 10-fold higher dose was required to induce a 35% kill in the CD133+/CD19− population, a concentration that was also lethal to 33% unsorted NBM cells. Whereas CD133+/CD38− NBM cells were more resistant to 10 μM vincristine, the proportion of cells surviving this dose was not significantly different from those of unsorted NBM cells (P > .08) or to CD133+/CD19− ALL cells (P > .09). The functional capacity of drug-treated cells from 5 patients was assessed in suspension culture. In 4 cases, the treated cells were capable of expanding and cell numbers were maintained in the 5th case. The levels of expansion ranged from 7.2- to 400-fold (Figure 4C). Growth of treated CD133+/CD19− ALL cells in suspension culture was significantly greater than that of treated bulk leukemia cells (P < .03). FISH analyses confirmed that the majority of cultured cells (88%-100%) had an abnormal karyotype.
In vitro sensitivity of ALL cells to dexamethasone and vincristine. Sorted populations from 10 patients (2, 3, 5, 7, 9, 11, 14, and 19-21) were cultured with dexamethasone for 48 hours (A). Cell viability was assessed by annexin V–propidium iodide staining. Data are mean plus or minus SE. * CD133+/CD19− cells were significantly less sensitive to treatment with dexamethasone than the CD133+/CD19+ and the unsorted bulk leukemia population (P < .01). Cells from patients 5, 11, 14, and 20 to 22 were cultured with vincristine for 72 hours and the viability assessed in the same way (B). The percentage viable is represented relative to untreated controls. Subsequently, growth of drug-treated CD133+/CD19− cells from patients 5, 14, 20, 21, and 22 was evaluated in suspension culture (C). The cultures were maintained with weekly half-media changes, and the absolute number of viable cells present at each week was determined using flow cytometry. The results for the 5 patients are shown, each represented by a different symbol.
In vitro sensitivity of ALL cells to dexamethasone and vincristine. Sorted populations from 10 patients (2, 3, 5, 7, 9, 11, 14, and 19-21) were cultured with dexamethasone for 48 hours (A). Cell viability was assessed by annexin V–propidium iodide staining. Data are mean plus or minus SE. * CD133+/CD19− cells were significantly less sensitive to treatment with dexamethasone than the CD133+/CD19+ and the unsorted bulk leukemia population (P < .01). Cells from patients 5, 11, 14, and 20 to 22 were cultured with vincristine for 72 hours and the viability assessed in the same way (B). The percentage viable is represented relative to untreated controls. Subsequently, growth of drug-treated CD133+/CD19− cells from patients 5, 14, 20, 21, and 22 was evaluated in suspension culture (C). The cultures were maintained with weekly half-media changes, and the absolute number of viable cells present at each week was determined using flow cytometry. The results for the 5 patients are shown, each represented by a different symbol.
Discussion
Purification of leukemia stem cells is important both for our understanding of the biology of leukemias and for the development of novel treatment strategies. Several lines of evidence indicate a central role for leukemia stem cells in the pathogenesis of these malignancies and exemplify the need to develop strategies that target these cells. Current chemotherapeutic regimens were developed against differentiated malignant cells, not putative stem cells. Consequently, leukemia stem cells may survive, even though more differentiated cells are killed and may in time contribute to disease relapse. Clearly, there is a need to characterize and specifically target these leukemia-initiating cells. However, the complexity of the leukemogenic process, together with our limited understanding of the pathogenic and regulatory mechanisms of leukemia stem cells, presents a challenge to translating biologic findings into novel therapeutic approaches. To further define the properties of leukemia-initiating cells in childhood BCP ALL, we have investigated the expression of the primitive stem cell marker, CD133, using preclinical in vivo and in vitro model systems.
Although CD133 is coexpressed on CD34+ cells in normal hematopoietic tissue, functional studies have revealed that a very rare population of CD133+/CD34−/CD38−/Lin− cells have long-term in vitro and in vivo repopulating ability.19 Our previous studies in childhood BCP ALL and in T-ALL have demonstrated that the immunophenotype of the bulk leukemia is not indicative of the cells that can proliferate to repopulate serial NOD/SCID xenografts.8,11 Instead, these putative ALL stem cells are phenotypically similar to their counterparts in normal hematopoiesis. Therefore, to determine whether this similarity in antigenic profile also applies to expression of CD133, we investigated the proliferative ability of sorted ALL cells in functional assays. Whereas the majority of cells in this cohort of patients were CD133−/CD19+, the cells that were capable of long-term growth in vitro were derived from the CD133+/CD19− subfraction. FISH analyses performed on cells immediately after sorting and on cells removed during the culture period revealed that cytogenetically abnormal cells were detected in both CD133+/CD19+ and CD133+/CD19− populations, confirming growth of leukemia cells in vitro. Aberrant cells have been reported in CD34+/CD19+ and CD34+/CD19− populations in childhood ALL.8,9,32,33 The presence of identical cytogenetic alterations in both CD133+ and CD133− cells has also been reported in AML,34 CML,35 and in glioblastoma cases,12 suggesting that they are clonally derived.
To further investigate this clonal origin, the genotype of the sorted populations was analyzed to determine whether there were any differences between the subfractions and the primary leukemias at diagnosis. In 2 of the 5 patients, the clonal Ig and TCR rearrangements detected in the diagnostic samples were observed in all 4 sorted ALL subfractions. In the other cases, some diagnostic rearrangements were not present and other rearrangements were detected. However, in every patient, diagnostic clonal rearrangements were identified in the CD133+/CD19− subfractions. Subclonal diversity, as indicated by the presence of multiple antigen receptor gene rearrangements, has been previously reported in childhood BCP ALL.36,37 In a study investigating t(12;21) ALL cases, multiple independent antigen receptor rearrangements were identified and initial clonal expansion could occur at various stages of early B-cell differentiation but most often before IgH VDJ recombination.37 The presence of unique TCR Vδ2-Dδ3 rearrangements, characteristic of the patients' leukemia, has also been reported in CD34+/CD38− childhood ALL cells in remission cases.38 Our findings concur with these previous reports that patient-specific gene rearrangements can be detected in relatively immature leukemia cells. Furthermore, the presence of residual leukemia cells in remission samples suggests that primitive CD34+/CD38− ALL cells might be resistant to induction therapy.38 Credible support for this hypothesis was reported recently in the demonstration of minor intrinsically resistant subclones in childhood ALL at diagnosis that could expand to become the major clone at relapse. In addition, the quantity of the relapse clone in diagnostic samples had an impact on clinical outcome in that it was inversely correlated with the time to first relapse.39 In our study, it is possible that enriching these cell populations by sorting permitted detection of subclones not observed during routine diagnostic screening. It will be interesting to explore this premise by analyzing a more extensive repertoire of clonal antigen receptor rearrangements. Similarly, investigating the antigen receptor rearrangements detected after passage in NOD/SCID mice will be informative because we and others have reported the emergence of new markers in NOD/SCID repopulating cells.11,40 Limited amounts of available material precluded such an extensive analysis in this study.
Whereas in vitro phenotypic and genotypic studies can add to the evidence for the presence of leukemogenic mutations in primitive progenitor cells, rigorous functional in vivo analyses are required to provide compelling evidence of stem cell properties. CD133 was expressed at low levels in the majority of patient samples in this investigation; only 4 of 22 cases were CD133+ using a 20% cutoff level.31 In this respect, our findings are similar to previous reports17,24 but lower than those in a study of 119 cases where 47 (39%) were CD133+.25 These differences may be attributable to the use of different CD133 antibody clones. However, despite the low CD133 expression, only the CD133+/CD19− subfraction was capable of NOD/SCID engraftment in each case. Sorting for CD133+/CD19− cells resulted in a 3-log enrichment of NOD/SCID repopulating cells compared with the bulk population and led to further enrichment than sorting for CD34+/CD19− cells. Cytogenetic and flow cytometric analyses confirmed that the engrafted cells had the same karyotype and immunophenotype as those of the patients at diagnosis. The majority of cells recovered from NOD/SCID xenografts expressed the B-lineage markers CD10 and CD19, suggesting that they had undergone differentiation in vivo. Furthermore, these primitive ALL cells could be transplanted into secondary recipients, with no loss of engrafting capacity and preservation of the immunophenotype and karyotype of the transplanted leukemia, providing definitive evidence of self-renewal ability. Similar results were found when cells were sorted for expression of CD133 and CD38 with only the CD133+/CD38− cells demonstrating NOD/SCID engrafting capacity. The use of some anti-CD38 antibodies (clones HIT2 and AT13/5) has recently been reported to have an inhibitory effect on engraftment of AML cells in mouse models.41 Using unsorted cells, we confirmed that there was no reduction of engraftment of cells stained with the CD38 (clone HB7) antibody, used in this study, compared with cells stained with isotype controls (Figure S1). So the lack of engraftment with CD38+ cells in this cohort of patients could not be attributed to antibody-mediated clearance of repopulating cells.
It should be possible to further enrich NOD/SCID leukemia-initiating cells using a 3-color sorting strategy to purify the CD133+/CD19−/CD38− population. Such a strategy was used in 5 patients, but insufficient numbers of CD133+/CD19−/CD38− cells were recovered to conduct functional evaluations in vivo. However, these cells were capable of long-term proliferation and expansion in vitro. Furthermore, expansion in cultures of CD133+/CD19−/CD38− cells was greater than that observed in cultures of CD133+/CD19− cells from the same patients. Consequently, the 3-color sorting permitted further purification of ALL cells with long-term proliferative ability in vitro. To our knowledge, this is the first report of expression of CD133 on childhood BCP ALL cells with long-term proliferative ability in vitro and in vivo. These results also substantiate our previous findings that the immunophenotype of the bulk leukemia is not indicative of the cells that can proliferate to repopulate serial NOD/SCID xenografts because in most patient samples only a very small fraction of the bulk leukemia cells expressed CD133.
Crucially, these CD133+/CD19− ALL cells were more resistant to treatment with dexamethasone and vincristine than the unsorted bulk leukemia population. The proportions of CD133+/CD19− ALL cells that survived treatment with these agents were not significantly different from that of normal hematopoietic stem cells. In addition, CD133+/CD19− cells remained capable of long-term proliferation in vitro after treatment. These chemotherapeutic agents are commonly used in the induction phase of therapy for pediatric ALL.42,43 The CD133+/CD19− cells were derived from patient samples both at diagnosis and in relapse, so the observed resistance could not be attributed to disease status or initial risk stratification. The effects of these drugs on the bulk ALL population were similar to published reports.40,44 These groups also reported in vitro resistance of unsorted leukemia samples to dexamethasone, with IC50 values more than 10 μM in 50% of cases.40,44 Our own findings highlight the importance of evaluating the effects of therapeutic agents on ALL stem cells, which may be less sensitive and consequently may have the potential to cause disease relapse.
These findings further refine the characteristics of leukemia stem cells in childhood B-ALL. ALL stem cells were first reported in the CD34+/CD38− population in Ph+ ALL cases.7 We have previously shown the CD34+/CD10− or CD19− subfractions in BCP ALL contained NOD/SCID repopulating cells.8 However, there have been some conflicting reports on the phenotype of ALL cells with the capacity to engraft xenogeneic recipients.9,10,33,45 One study found that both CD19+ and CD19− cells from 3 patients could engraft NOD/SCID mice to modest levels but could only detect the ETV6–RUNX1 translocation in recipients of CD19+ cells.9 We have sorted for more specific cell populations, resulting in enhanced engraftment levels, and the ETV6–RUNX1 translocations could be detected in recipients of CD133+/CD19− cells in our investigation. Furthermore, serial transplantation studies demonstrated that CD133+/CD19− ALL cells were capable of self-renewal and expansion of the progenitor cell pool. Another study of 4 pediatric ETV6–RUNX1 cases found that engraftment of NOD/SCID mice could be achieved using the CD34+/CD38−/CD19+ subfraction but not the CD34+/CD38+/CD19+ subfraction.10 The CD19− populations were not assessed by Hong et al,10 but our findings that NOD/SCID repopulating cells from various ALL subtypes lacked expression of CD38 concur with their results. In contrast, Kong et al reported that both CD34+/CD38+/CD19+ and CD34+/CD38−/CD19+ populations in 2 infant ALL cases and one undefined subtype could engraft immune-deficient mice.45 Another study reported variable results in 6 high-risk and 2 standard-risk cases, when investigating CD34/CD19 populations, with at least 2 subfractions capable of engrafting NOD/SCIDs.30 Most experiments conducted by le Viseur et al33 involved intrafemoral inoculation of unsorted ALL cells. Once engrafted, leukemia cells were collected and then sorted for expression of CD34/CD19 before inoculation into serial recipients. So the majority of ALL cells used for sorting had already undergone a round of preselection by their ability to repopulate primary NOD/SCID mice. Cells from 3 of the high-risk cases were sorted for CD34+/CD19+, CD34+/CD19−, and CD34−/CD19+ subfractions before evaluation in primary recipients. In one case, all 3 subfractions engrafted; only the CD34+/CD19− subfraction from the second case engrafted primary animals, and engraftment was achieved in 2 animals inoculated with CD34+/CD19+ and CD34−/CD19+ cells, respectively, in the third. However, regardless of the sorting strategy used, fewer CD34+/CD19− cells (≥ 2 × 103) were required for engraftment compared with the more mature populations.33 In our study, we have shown that leukemia-initiating cells can be further enriched using CD133, where less than 750 CD133+/CD19− or CD133+/CD38− cells could engraft NOD/SCIDs. It is possible that there is heterogeneity at the stem cell level between infant and pediatric leukemias and high-risk or standard-risk cases, which may explain some of the discordant results. Nevertheless, the use of different mouse strains, different sorting strategies together with differences in inoculation and sampling techniques, may also be contributing factors.
Further refinement of the phenotype of the ALL stem cell compartment is required to elucidate the nature of these cells and ultimately to improve clinical outcomes. In this study, we have confirmed our previous findings and demonstrated that ALL-initiating cells can be enriched using the most primitive stem cell antibody routinely available for hematopoietic cell purification. Furthermore, we have demonstrated that current therapeutic agents are less toxic to these primitive leukemia cells. The knowledge gained from these studies could be applied to the development of novel agents, directed at leukemia stem cell-specific targets, rather than the bulk differentiated progeny. The model systems used here could subsequently be used to measure the efficacy of such novel therapies and to provide valuable information on their mechanism of action.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mr Paul Archer and Mrs Tracey Perry, University of Bristol, Mr Paul Virgo, Immunology Unit Southmead Hospital, staff of the Regional Cytogenetics Unit, Southmead Hospital and staff of the cell sorting facility, University of Wales College of Medicine for excellent technical assistance, Dr Jeremy Hancock for helpful discussion on the sequencing data, and the patients and their families who gave permission for their cells to be used for research. The authors acknowledge the Sequencing Service (School of Life Sciences, University of Dundee, United Kingdom, http://www.dnaseq.co.uk/) for DNA sequencing.
This work was supported by grants from the Leukemia Research Fund, United Kingdom and the National Blood Authority.
Authorship
Contribution: C.V.C. processed samples, performed the in vitro experiments and gene rearrangement analyses, and helped write the report; P.D. performed drug sensitivity assays and data analyses; R.S.E. helped perform the in vivo experiments and commented on the manuscript; P.R.K. collated the clinical data, contributed to the interpretation of results, and commented on the manuscript; and A.B. conceived and designed the study, supervised its execution, performed the in vivo experiments, and wrote the report. A.B. had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Allison Blair, Bristol Institute for Transfusion Sciences, NHSBT Filton, 500 North Bristol Park, Bristol, BS34 7QH, United Kingdom; e-mail: allison.blair@nbs.nhs.uk.