Abstract
Fibrinogen residue Bβ432Asp is part of hole “b” that interacts with knob “B,” whose sequence starts with Gly-His-Arg-Pro-amide (GHRP). Because previous studies showed BβD432A has normal polymerization, we hypothesized that Bβ432Asp is not critical for knob “B” binding and that new knob-hole interactions would compensate for the loss of this Asp residue. To test this hypothesis, we solved the crystal structure of fragment D from BβD432A. Surprisingly, the structure (rfD-BβD432A+GH) showed the peptide GHRP was not bound to hole “b.” We then re-evaluated the polymerization of this variant by examining clot turbidity, clot structure, and the rate of FXIIIa cross-linking. The turbidity and the rate of γ-γ dimer formation for BβD432A were indistinguishable compared with normal fibrinogen. Scanning electron microscopy showed no significant differences between the clots of BβD432A and normal, but the thrombin-derived clots had thicker fibers than clots obtained from batroxobin, suggesting that cleavage of FpB is more important than “B:b” interactions. We conclude that hole “b” and “B:b” knob-hole binding per se have no influence on fibrin polymerization.
Introduction
The fibrinogen molecule consists of 3 pairs of nonidentical polypeptide chains: Aα, Bβ, and γ. These chains are folded into 3 distinct structural regions: 2 distal D regions and 1 central E region, linked by coiled-coil connectors forming a trinodular structure.1 Each D region contains polymerization holes “a” and “b” located in the C-terminus of the γ- and Bβ-chains, respectively. The central E region contains 2 sets of polymerization knobs “A” and “B” that are cryptic in fibrinogen but become exposed in fibrin after thrombin cleaves fibrinopeptide A (FpA) and fibrinopeptide B (FpB) from the N-terminus of the Aα- and Bβ-chains, respectively.1,2 The exposed knob “A” binds to hole “a” of another fibrin molecule forming “A:a” interactions that lead to a double-stranded protofibril with a half-staggered overlap between molecules in different strands. Cleavage of FpB occurs primarily during protofibril growth, leading to the binding of knob “B” to hole “b” forming “B:b” interactions.1
The location of the binding holes and possible models for knob-hole interactions are known from X-ray crystallographic studies using 2 synthetic peptide analogs of knobs “A” and “B.” When fibrinogen fragment D and double-D are crystallized in the presence of both peptide analogs, the knob “A” peptide mimic, Gly-Pro-Arg-Pro-amide (GPRP), forms H-bond interactions with residues γ364Asp, γ330Asp, γ329Gln, and γ340His found in hole “a.”3-5 In the same manner, the knob “B” peptide mimic, Gly-His-Arg-Pro-amide (GHRP), interacts with residues Bβ397Glu, Bβ398Asp, and Bβ432Asp in hole “b.”3,5,6
The functional role of “A:a” interactions was demonstrated in several studies with reported dysfibrinogenemias,7,8 as well as studies with γ364Asp variant fibrinogens.9 Although the importance of “A:a” interactions have been well documented, the role of “B:b” interactions is still unclear. Previously published data on BβD432A fibrinogen, a variant with mutation in hole “b,” showed normal polymerization.10 Without structural data for BβD432A, it is reasonable to anticipate that BβAsp432 may not be critical in binding knob “B” and that there may be alternative and new sets of interactions in BβD432A that would still allow “B:b” interactions to occur. We also based this hypothesis on recent studies with γD364A variant fibrinogen.11 The 2 aspartate residues, BβAsp432 in hole “b” and γAsp364 in hole “a,” form ionic interactions with the positively charged free amino-terminus of their corresponding knob. Residues BβAsp432 and γAsp364 are located at equivalent positions in the β- and γ-chain polymerization holes, respectively.3,5 Whereas γD364A showed severely impaired function, structural data on this variant surprisingly showed that hole “a” binds knob “A” peptide mimic, GPRP, suggesting that the residue γAsp364 per se is not critical for “A:a” interactions.11 In contrast to γD364A, we now report that BβD432A does not bind knob “B” peptide mimic, GHRP, and that the residue BβAsp432 is critical for “B:b” interactions. This surprising finding led us to further examine the polymerization of BβD432A and re-evaluate the importance of hole “b” and “B:b” interactions. Our biochemical data suggest that BβD432A, indeed, has normal polymerization, despite having impaired hole “b.” In summary, our findings suggest that hole “b” may have very little influence on polymerization and that other events, such as FpB release, are more critical to polymerization, rather than the actual “B:b” knob-hole binding.
Methods
Reagents
All chemicals were of reagent grade and were purchased from Sigma-Aldrich (St Louis, MO), unless specified otherwise. The peptide GHRP was synthesized by the Protein Chemistry Laboratory at the University of North Carolina at Chapel Hill. Cell culture media with normal recombinant fibrinogen were obtained from the National Cell Culture Center (Minneapolis, MN). Monoclonal IF-1 antibody was purchased from Kamiya Biomedical (Seattle, WA).
Expression and purification of recombinant fibrinogen BβD432A
Normal and variant BβD432A fibrinogens were synthesized in CHO cells as described previously.10 Large-scale protein expression was carried out in serum-free medium in roller bottles. Media containing secreted fibrinogen were harvested periodically. After addition of protease inhibitors, the media were stored at −20°C.12
Recombinant fibrinogen was purified as described.13 Briefly, fibrinogen was precipitated from the media with ammonium sulfate in the presence of a cocktail of protease inhibitors. The precipitate was resuspended in buffer containing 10 mM CaCl2 and applied to a Sepharose 4B column coupled with the fibrinogen specific monoclonal antibody, IF-1. Fibrinogen was eluted from the column with buffer containing 5 mM EDTA; dialyzed against HEPES buffered saline (HBS) (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 150 mM NaCl) and 1 mM CaCl2 for one exchange then extensively dialyzed against HBS; and stored at −80°C. The integrity of the polypeptide chains and purity of the recombinant protein were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) under reducing and nonreducing condition following the method of Laemmli.14
Preparation and purification of fragment D
Fragment D was prepared from BβD432A by controlled trypsin digestion, according to the method used for normal recombinant fibrinogen.5 Briefly, CaCl2 (final concentration of 20 mM) was added to 10 mg BβD432A fibrinogen in HBS and digestion was initiated by adding 100 μL immobilized TPCK trypsin (Pierce Biotechnology, Rockford, IL). The reaction was allowed to proceed over a period of days until SDS-PAGE revealed essentially 2 bands that correspond to the molecular weights of fragments D and E. Digestion was stopped by removing the trypsin beads with centrifugation at 1200g (Sorvall RC-3; DuPont Instruments, Wilmington, DE) for 10 minutes.
Fragment D was purified by peptide affinity chromatography based on previously described methods.15 Briefly, the digest was loaded onto a polymeric resin covalently linked to the peptide GPRPAA. Fragment D was eluted using 50 mM sodium acetate (pH 5.3) and 1 M NaBr. Purified fragment D was dialyzed against 50 mM Tris-HCl, pH 7.4, concentrated 100-fold with a centrifugal filter device (50-kDa MW cutoff; Millipore, Billerica, MA), and stored at 4°C.
Crystallization of fragment D from BβD432A
Purified BβD432A fragment D was cocrystallized with GHRP by sitting-drop diffusion at 4°C similar to methods previously described for normal fragment D crystallization.3,5 Crystals were grown from drops containing 5 μL BβD432A fragment D initially at 10 mg/mL in 50 mM Tris-HCl, pH 7.4, and GHRP at 4 mM mixed with an equal volume of well solution containing 50 mM Tris-HCl, pH 8.5, 2 mM NaN3, 12.5 mM CaCl2, and 11% to 14% (wt/vol) PEG 3350. Single crystals appeared in days, after streak seeding with crystals of normal recombinant fragment D.
X-ray diffraction data collection and structure determination
Data collection for crystals of BβD432A fibrinogen fragment D (rfD-BβD432A+GH) was carried out at 100 K using SER-CAT beamline 22-ID at the Advanced Photon Source (Argonne, IL). A single crystal was cryoprotected in loop containing the cystallant solution plus 20% glycerol and 4 mM GHRP, then flash-frozen in liquid nitrogen. Diffraction data were processed with DENZO and ScalePack.16
The structure of rfD-BβD432A+GH was solved by molecular replacement using MolRep17 in CCP4i18 with one molecule of the normal recombinant fragment D structure, rfD (PDB code: 1LT9) as the search model. The solution gave a correlation coefficient and R-factor of 53.0% and 45.8%, respectively. The resulting model was improved by rigid body refinement in Refmac519 reserving 5% of the reflections for the calculation of the free R-factor. After one round of refinement, the model was improved by manual fitting in the program O20 using sigmaA-weighted |2Fo − Fc| and |Fo − Fc| electron density maps.21 Several cycles of restrained and individual temperature factor refinement were done before water molecules were added to complete the model. Furthermore, cis-peptide bonds were added at positions Bβ407 and γ339. Final refinement was done with the TLS protocol in Refmac5.22 After all these refinement steps, the Rcrystal and Rfree converged to the values reported in Table 1.
Crystallographic data and refinement statistics
| Statistics . | . |
|---|---|
| Data statistics | |
| Resolution, Å | 50.0-2.15 |
| Space group | P212121 |
| Cell constants, Å | a = 54.5, b = 146.6, c = 229.0 |
| Molecules/asymmetric unit | 2 |
| Total observations | 877 794 |
| Unique reflections | 82 662 |
| Mean redundancy | 6.7 |
| *Rsym, % | 6.4 (33.1) |
| Completeness, % | 94.8 (83.4) |
| Mean I/s | 29.5 (4.2) |
| Refinement statistics | |
| Resolution, Å | 50.0-2.4 |
| †Rcryst, % | 21.4 |
| ‡Rfree, % | 24.2 |
| Average B factor, Å 2 | 33.6 |
| No. of model atoms | 10 676 |
| No. of solvent sites | 260 |
| RMS deviations from ideals | |
| Bond length, Å | 0.008 |
| Bond angles, deg | 1.08 |
| Statistics . | . |
|---|---|
| Data statistics | |
| Resolution, Å | 50.0-2.15 |
| Space group | P212121 |
| Cell constants, Å | a = 54.5, b = 146.6, c = 229.0 |
| Molecules/asymmetric unit | 2 |
| Total observations | 877 794 |
| Unique reflections | 82 662 |
| Mean redundancy | 6.7 |
| *Rsym, % | 6.4 (33.1) |
| Completeness, % | 94.8 (83.4) |
| Mean I/s | 29.5 (4.2) |
| Refinement statistics | |
| Resolution, Å | 50.0-2.4 |
| †Rcryst, % | 21.4 |
| ‡Rfree, % | 24.2 |
| Average B factor, Å 2 | 33.6 |
| No. of model atoms | 10 676 |
| No. of solvent sites | 260 |
| RMS deviations from ideals | |
| Bond length, Å | 0.008 |
| Bond angles, deg | 1.08 |
The atomic coordinates have been deposited in the Protein Data Bank23 under the access code 3E1I (rfD-BbD432A+GH).
Rsym = Σ|I − 〈I〉|, where I is the observed intensity and 〈I〉 is the average intensity of multiple symmetry-related observations of that reflection.
Rcryst = Σ(|Fobs| − |Fcalc|)/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factors, respectively.
Rfree is the R-factor based on the 5% of the data withheld from structural refinement.
Thrombin-catalyzed polymerization
Thrombin-catalyzed polymerization of BβD432A was monitored as the turbidity changes at 350 nm using SpectraMax PC microtiter plate reader (Molecular Devices, Sunnyvale, CA) essentially as described.24 Briefly, 10 μL of 1 U/mL thrombin was added to reaction wells containing 90 μL of 0.22 mg/mL fibrinogen in HBS with no added calcium and with CaCl2 at final concentrations of 10 μM, 1 mM, and 10 mM. Polymerization was monitored for 60 minutes at room temperature. Three distinct parameters were analyzed: (1) the time it takes for the absorbance to rise above zero (lag time); (2) the portion of the curve with the steepest slope after the absorbance starts to rise (Vmax); and (3) the final maximum absorbance (ODmax).
Scanning electron microscopy
Fibrin clot and specimen preparations were based on a previously described method with some modifications.24 For each polymerization condition, scanning electron microscopy (SEM) was performed on 2 clots each with 2 separate microscopy preparations for normal and BβD432A. Clots were polymerized in microtiter plate wells at 0.5 mg/mL fibrinogen and 0.4 U/mL batroxobin or thrombin in HBS and 1 mM CaCl2 for 2 hours. The clots were then fixed in 2% glutaraldehyde overnight, rinsed 3 times with HBS, then dehydrated with a series of ethanol solutions (20%-100%). The samples were then critical-point dried in a Balzers CPD020 (Oerlikon Balzers, Balzers, Liechtenstein), mounted, sputter-coated with approximately 20 nm gold-palladium, and viewed on a Zeiss Supra 25 FESEM (Carl Zeiss MicroImaging, Thornwood, NY). All images were taken at 50 200× magnification, with a 3.0-mm working distance and 4.99-kV accelerating voltage. Fiber diameters were calculated using the measurement tool in Photoshop (Adobe Systems, San Jose, CA).
Factor XIIIa–catalyzed cross-linking of fibrin
Factor XIII (Enzyme Research Laboratories, South Bend, IN) was activated with human α-thrombin for 60 minutes at 37°C in HBS with 5 mM CaCl2. To examine the cross-linking of fibrin, fibrinogen (5 μg), in HBS and 5 mM CaCl2, was incubated at 37°C with a mixture of FXIIIa (3.3 U/mL) and human α-thrombin (0.07 U/mL). To examine the cross-linking of batroxobin-catalyzed fibrin, hirudin (10 U/mL) was added to the thrombin-activated FXIIIa before incubation with fibrinogen and batroxobin (0.08 U/mL). The reactions were stopped by the addition of SDS-PAGE sample buffer with 2-mercaptoethanol followed by heating at 100°C. Sample components were separated by 8% SDS-PAGE and stained with Coomassie brilliant blue R-250. Densitometric analyses of stained gels were performed using the gel analysis tools in ImageJ (National Institutes of Health [NIH], Bethesda, MD) and γ-γ/Bβ ratios were calculated.
Statistical analysis
The statistical significance of differences between normal control and variant fibrinogen was determined using unpaired t tests. A difference was considered significant when the P value was less than .05.
Results
Structure of rfD-BβD432A+GH
To understand the molecular basis for the normal turbidity of BβD432A, we determined the crystal structure of fragment D from this variant, crystallized in the presence of GHRP. We obtained high-quality diffraction data and were able to solve the structure to 2.4 Å. A total of 643 amino acid residues in one fragment D molecule (α134-190, β161-458, and γ105-392) can be elucidated clearly from the electron density. The substitution of the Asp to Ala at position Bβ432 was evident. Alignment of the Cα atoms in normal and BβD432A fragment D showed excellent agreement with RMSD of 0.75 Å, demonstrating that the global structure of rfD-BβD432A+GH was not changed by the alanine substitution.
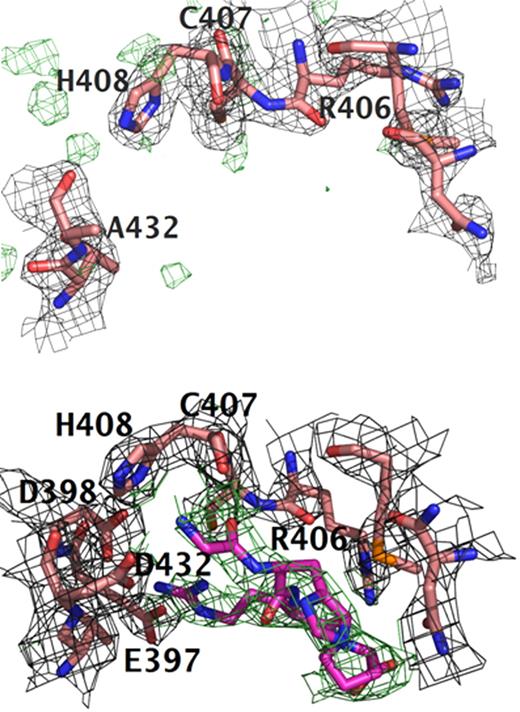
Surprisingly, the structure of rfD-BβD432A+GH showed that GHRP is not bound to hole “b.” As shown in Figure 1A, there was no electron density in |2Fo − Fc| and |Fo − Fc| maps for the peptide in hole “b.” Moreover, the structure of hole “b” in rfD-BβD432A+GH is reminiscent of the structure of normal recombinant fragment D in the absence of peptides (rfD). We did see GHRP bound in hole “a” (Figure 2). Thus, rfD-BβD432A+GH structure resembled that of rfD-BβD398A+GH, the structure of another variant with mutation in hole “b,” BβD398A, crystallized in the presence of GHRP. The rfD-BβD398A+GH structure showed that GHRP was not bound to hole “b” but was bound to hole “a” in the γ-chain forming noncanonical “B:a” interactions.10 As with the rfD-BβD398A+GH structure, comparison of the 2 molecules in the asymmetric unit of rfD-BβD432A+GH showed 2 different structures for the 2 “B:a” interactions with calcium present in the γ2 site in only one of the molecules (Figure 2). The γ2 calcium is located in the γ294-301 loop, which is in close proximity to hole “a.” This calcium binding site has also been observed in structures of normal fragment D and its cross-linked form,25 as well as in recombinant rfD-BβD398A+GH,10 when the proteins were crystallized in the presence of GHRP alone. Studies with recombinant fibrinogen γD298,301A showed modest impairment in polymerization, suggesting that γ2 does not significantly modulate polymerization.26
Comparison of polymerization holes “b.” Structures of (A) rfD-BβD432A+GH and (B) normal with both GPRP and GHRP (rfD+BOTH). For both structures, the difference electron density |Fo − Fc| is contoured at 3.0σ and is shown in green. The |2Fo − Fc| electron density maps are contoured at 1.0σ shown in gray. Clear positive difference electron density for GHRP is evident in hole “b” of normal rfD+BOTH and is absent in the rfD-BβD432A+GH structure. This suggests that BβD432A has an impaired hole “b” and does not bind the peptide. Note the positions of BβGlu397 and BβAsp398. In normal rfD+BOTH, both residues flip toward hole “b” to interact with GHRP. In rfD-BβD432A+GH, these residues are nowhere near hole “b.”
Comparison of polymerization holes “b.” Structures of (A) rfD-BβD432A+GH and (B) normal with both GPRP and GHRP (rfD+BOTH). For both structures, the difference electron density |Fo − Fc| is contoured at 3.0σ and is shown in green. The |2Fo − Fc| electron density maps are contoured at 1.0σ shown in gray. Clear positive difference electron density for GHRP is evident in hole “b” of normal rfD+BOTH and is absent in the rfD-BβD432A+GH structure. This suggests that BβD432A has an impaired hole “b” and does not bind the peptide. Note the positions of BβGlu397 and BβAsp398. In normal rfD+BOTH, both residues flip toward hole “b” to interact with GHRP. In rfD-BβD432A+GH, these residues are nowhere near hole “b.”
Electron density corresponding to GHRP in hole “a” of the 2 molecules in the asymmetric unit of rfD-BβD432A+GH. The |Fo − Fc| electron density around GHRP (shown as green sticks) is contoured at 3.0σ. The 2 calcium binding sites γ1 and γ2 are shown as orange spheres. Note that the calcium in γ2 site is absent in the other molecule of the asymmetric unit.
Electron density corresponding to GHRP in hole “a” of the 2 molecules in the asymmetric unit of rfD-BβD432A+GH. The |Fo − Fc| electron density around GHRP (shown as green sticks) is contoured at 3.0σ. The 2 calcium binding sites γ1 and γ2 are shown as orange spheres. Note that the calcium in γ2 site is absent in the other molecule of the asymmetric unit.
Calcium binding in crystal structure
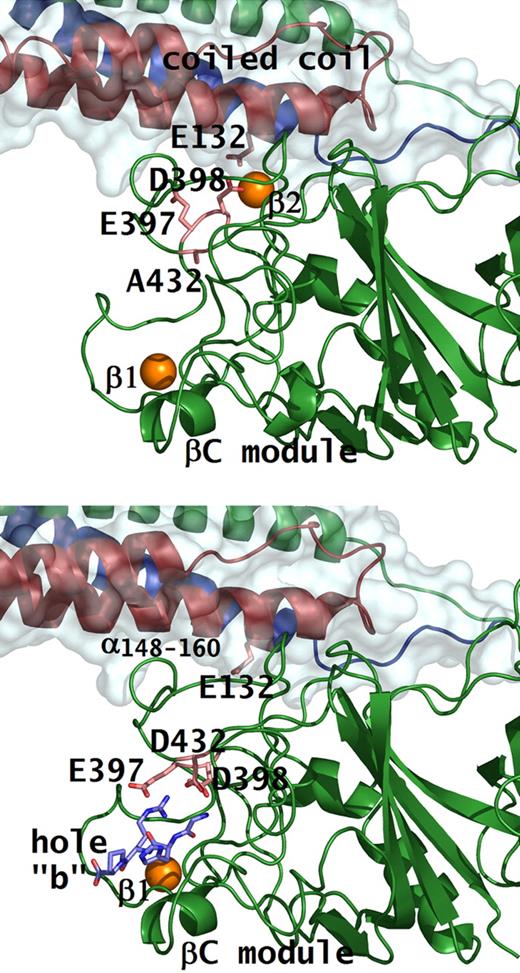
It is well known from crystal structures that fragment D has a high-affinity calcium binding site in the γ-chain, designated as γ1. Another calcium binding site that is analogous to γ1, is located at an equivalent position in the β-chain adjacent to hole “b,” designated as β1. Both the γ1 and β1 calcium binding sites are present in the rfD-BβD432A+GH structure (Figure 3A). An additional calcium binding site, designated as β2, which is located near the junction of the βC-module and the coiled-coil region, is also seen in rfD-BβD432A+GH. This calcium ion anchors the βC-module to the coiled-coil region and is coordinated by the side chains of BβAsp398, BβAsp261, γGlu132, and the backbone carbonyl oxygen of BβGly263.3,5 Binding of peptide to hole “b,” as observed in the structures of normal fragment D, crystallized in the presence of peptides, rfD+BOTH (Figure 3B), causes BβGlu397 and BβAsp398 to flip toward BβAsp432 to interact with the peptide.5,25 Because BβAsp398 is part of the β2 calcium coordination, movement of BβGlu397 and BβAsp398 causes the disruption of the β2 site upon peptide binding. In rfD-BβD432A+GH, peptide binding to hole “b” is lost, hence, the calcium is present in the β2 site.
Conformational change with peptide bound in hole “b.” Structures of rfD-BβD432A+GH (A) and rfD+BOTH (normal fibrinogen with both GPRP and GHRP; B) are shown. Residues involved in β2 calcium and peptide binding are shown as pink sticks. Note the conformational change in residues Bβ397Glu and Bβ398Asp when peptide (shown as blue sticks) is bound to hole “b,” as in the case of normal. In the rfD-BβD432A+GH structure, hole “b” does not bind the peptide so that Bβ398Asp, together with γ132Glu, coordinates the β2 calcium (shown as orange sphere) that tethers the βC-module to the coiled coil.
Conformational change with peptide bound in hole “b.” Structures of rfD-BβD432A+GH (A) and rfD+BOTH (normal fibrinogen with both GPRP and GHRP; B) are shown. Residues involved in β2 calcium and peptide binding are shown as pink sticks. Note the conformational change in residues Bβ397Glu and Bβ398Asp when peptide (shown as blue sticks) is bound to hole “b,” as in the case of normal. In the rfD-BβD432A+GH structure, hole “b” does not bind the peptide so that Bβ398Asp, together with γ132Glu, coordinates the β2 calcium (shown as orange sphere) that tethers the βC-module to the coiled coil.
Thrombin-catalyzed polymerization of BβD432A
Because the loss of “B:b” interactions appeared inconsistent with our prior findings, we re-evaluated the polymerization of this variant. Thrombin-catalyzed polymerization experiments were performed as described before10 and carried out at 3 calcium concentrations: 10 μM CaCl2, 1 mM CaCl2, and 10 mM CaCl2. Consistent with previous data, the variant BβD432A was indistinguishable from normal fibrinogen for essentially all parameters examined (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). All polymerization parameters for BβD432A (lag time, Vmax, and ODmax) were not significantly different compared with normal, with the exception of the lag time at 10 mM CaCl2 in which BβD432A had a longer lag time (P = .01).
To confirm that the final turbidity was indicative of fiber thickness and to quantitate the differences in fiber thickness, we examined thrombin- and batroxobin-catalyzed fibrin clots using scanning electron microscopy. Consistent with our turbidity data, clots formed from BβD432A have similar fiber diameters as those formed from normal fibrinogen (Figure S1). For the clots formed using thrombin, we found the average fiber diameter for BβD432A was 84 plus or minus 18 nm, which was essentially similar to normal with an average fiber diameter of 83 plus or minus 18 nm (P = .8). The batroxobin-derived clots for BβD432A had fiber diameter of 68 plus or minus 13 nm, which was comparable with those of normal with average fiber diameter of 66 plus or minus 15 nm (P = .7). However, batroxobin-derived clots for either BβD432A or normal had significantly thinner fiber diameter compared with the thrombin-derived clots (P = .03 for BβD432A and P = .04 for normal), suggesting that FpB cleavage modulates fiber diameter.
FXIIIa-catalyzed cross-linking
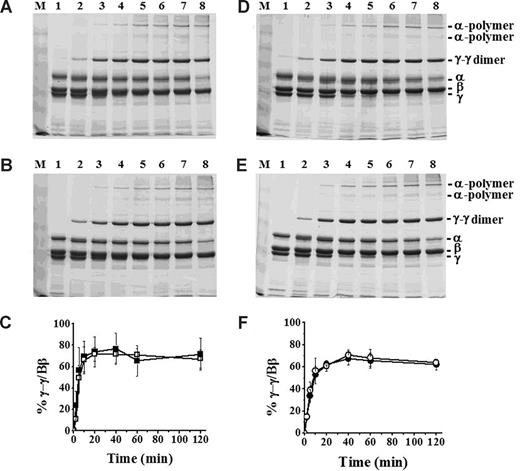
In recent studies, we concluded that “B:b” interactions promote protofibril assembly.9,27,28 FXIIIa-catalyzed γ-γ dimers form early in polymerization, likely as soon as protofibrils are present.29 We therefore analyzed the kinetics of γ-γ dimer formation because this could reflect subtle differences in protofibril formation between normal and BβD432A. Analysis of thrombin-catalyzed FXIIIa cross-linking showed γ-γ dimer formation after 2 minutes in both normal and BβD432A (Figure 4A,B). These γ-γ dimer bands increased in intensity with longer incubation times (Figure 4C). We also performed experiments in the presence of FXIIIa and batroxobin. Similar to thrombin-catalyzed polymerization and cross-linking, the γ-γ dimer bands were weakly evident in both normal and BβD432A after 2 minutes (Figure 4D,E). With longer incubation, these γ-γ dimer bands increased in intensity (Figure 4F). These data indicate that protofibril formation was not delayed for BβD432A, suggesting that protofibril formation in BβD432A is not markedly different from normal.
FXIIIa cross-linking of BβD432A and normal recombinant fibrinogen. Cross-linking was examined by 8% SDS-PAGE under reducing conditions. Fibrinogen (5 μg) was incubated with either FXIIIa and thrombin at final concentrations of 3.3 U/mL and 0.07 U/mL, respectively (A: normal, B: BβD432A) or FXIIIa and batroxobin at final concentrations of 3.3 U/mL and 0.08 U/mL, respectively (D: normal, E: BβD432A). The samples were incubated for 0 minutes (lane 1), 2 minutes (lane 2), 5 minutes (lane 3), 10 minutes (lane 4), 20 minutes (lane 5), 40 minutes (lane 6), 1 hour (lane 7), and 2 hours (lane 8). Data were analyzed by determining the ratio of γ-γ dimers to Bβ-chain by densitometry and are plotted against time (thrombin-polymerized; C: ■ indicates normal; □, BβD432A; and batroxobin-polymerized: F: ● indicates normal; ○, BβD432A). The error bars (C,F) represent SDs of 3 experiments.
FXIIIa cross-linking of BβD432A and normal recombinant fibrinogen. Cross-linking was examined by 8% SDS-PAGE under reducing conditions. Fibrinogen (5 μg) was incubated with either FXIIIa and thrombin at final concentrations of 3.3 U/mL and 0.07 U/mL, respectively (A: normal, B: BβD432A) or FXIIIa and batroxobin at final concentrations of 3.3 U/mL and 0.08 U/mL, respectively (D: normal, E: BβD432A). The samples were incubated for 0 minutes (lane 1), 2 minutes (lane 2), 5 minutes (lane 3), 10 minutes (lane 4), 20 minutes (lane 5), 40 minutes (lane 6), 1 hour (lane 7), and 2 hours (lane 8). Data were analyzed by determining the ratio of γ-γ dimers to Bβ-chain by densitometry and are plotted against time (thrombin-polymerized; C: ■ indicates normal; □, BβD432A; and batroxobin-polymerized: F: ● indicates normal; ○, BβD432A). The error bars (C,F) represent SDs of 3 experiments.
Discussion
Our structural data showed GHRP does not bind to hole “b” in BβD432A. Hence, the variant lacks “B:b” interactions. Our biochemical data, which corroborate and extend previous studies, showed polymerization, fiber diameter, and FXIIIa cross-linking of BβD432A were indistinguishable from normal. These findings are remarkable because they show that formation of cross-linked clots is normal even in the absence of “B:b” interactions. This then raises the question: Do “B:b” interactions actually occur in fibrin; and if they do, are they critical for fibrin clot formation?
Thirty years ago, Blombäck et al showed that fibers obtained from reptilase, which cleaves predominantly FpA, have lower mass-to-length ratio than those derived from thrombin.30 They then proposed a 2-step sequence of events in fibrin polymerization where protofibril formation mediated by “A:a” interactions is followed by lateral aggregation of the protofibrils through “B:b” interactions. Subsequent studies by Weisel31 and Hantgan et al32 showed the extent of lateral aggregation was greater for fibers with cleavage of both fibrinopeptides; the addition of thrombin after activation with reptilase, at a time when protofibrils have formed, caused rapid fiber formation. These studies demonstrated that the release of FpB, exposing knob “B,” has a marked impact, supporting the idea that “B:b” interactions are important in lateral aggregation.
Nevertheless, several observations, including these studies, suggest otherwise. Even though the snake venom enzymes cleave only FpA, they still produce normal-looking fibers, albeit thinner in diameter. Thus, protofibrils are formed and these protofibrils are capable of lateral aggregation even in the absence of “B:b” interactions.31 The knob “A” peptide mimic, GPRP, has been shown to inhibit polymerization,33 whereas knob “B” peptide mimic GHRP does not.34 In fact, GHRP and other knob “B” peptide analogs enhance turbidity during thrombin-catalyzed polymerization.35 Thus, “B:b” interactions are not essential for lateral aggregation.
More recent studies proposed a role for “B:b” interactions in protofibril formation. These studies examined polymerization of single-substituted recombinant fibrinogens, γD318A and γD320A, which have changes in the high-affinity calcium binding site,27 and γD364V, γD364H, and γD364A, which have changes in hole “a.”9 These variants did not polymerize with batroxobin, indicating complete loss of “A:a” interactions. They did, however, polymerize with thrombin, although with markedly delayed kinetics. This thrombin-induced polymer formation was inhibited by GHRP, suggesting that “B:b” interactions support protofibril formation, at least in the absence of normal “A:a” interactions. With normal “A:a” interactions, as seen with reptilase, the subsequent addition of thrombin might stabilize protofibrils, and consequently enhance fiber formation.
In contrast to the previous reports, our results with BβD432A suggest “B:b” interactions are not critical for normal polymerization. To reconcile these differences, we propose that the loss of FpB, rather than the gain of “B:b” interactions, influences polymerization. Because FpB is electronegative, diminished electrostatic repulsion between associating fibrin molecules can be surmised after cleavage of FpB. Although the removal of the 2 FpAs reduces the negative charge, it is only after FpB release that the central domain actually assumes a net positive charge to favor association with the negatively charged D nodules. Aside from electrostatic factors, FpB also modulates fibrinogen structure through intramolecular interactions with the αC domains.36 After FpB release, the αC domains dissociate so they are free to form intermolecular associations enhancing lateral aggregation.36,37 These findings suggest the role of FpB in modulating polymerization, whereas our data indicate that actual occupancy of hole “b” is not critical to fibrin clot formation.
Although hole “b” may have only very little influence on polymerization, recent data indicate that “B:b” interactions play a more important role in fibrinolysis.35 Previous studies have suggested that a consequence of “B:b” interactions may be a global conformational change where the β2 calcium anchor is disrupted and the βC-module disengages from the coiled coil.5 The resulting conformation may lead to increased accessibility to a tPA (or plasminogen) binding site at α148-160 (Figure 3B) when binding of the tethered knob to hole “b” locks and prevents the βC-module from retreating back toward the coiled-coil region.38 Indeed, addition of free, untethered knob “B” peptide mimics that can compete with the authentic tethered knob resulted in a delay in fibrinolysis. Our data from our experiments using BβD432A and the peptide AHRPY, which exclusively binds to hole “b,” are consistent with this observation (S.R.B., Brian Holliday, and S.T.L., unpublished findings, February 2008).
In summary, we have shown that Bβ432Asp is critical for peptide binding. However, the normal polymerization of BβD432A despite absence of “B:b” interactions suggests that occupancy of hole “b” may have little impact on polymerization. It may be that FpB release and electrostatic factors are more critical than actual “B:b” knob-hole binding.
The online version of this article contains a data supplement.
Presented in abstract form at the 21st Congress of the International Society on Thrombosis and Haemostasis, Geneva, Switzerland, July 8, 2007.39
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to Dr Oleg Gorkun for helpful discussions and to Robert A. Campbell for critical comments on the paper. We thank Dr Laurie Betts for guidance in solving the crystal structure and the Southeast Regional Collaborative Access Team (SER-CAT) at Argonne National Laboratory for synchrotron time.
This work was supported by research funding from NIH grant HL 31048 (S.T.L.) and by an American Heart Association (Dallas, TX) Predoctoral Fellowship, grant 0715292U (S.R.B.).
National Institutes of Health
Authorship
Contribution: S.R.B. designed and performed experiments, analyzed the data, and prepared the paper; S.T.L. supervised the study and contributed to various stages of paper preparation; and both authors approved the final paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Susan T. Lord, Department of Pathology and Laboratory Medicine, University of North Carolina at Chapel Hill, CB# 7525, Brinkhous-Bullitt Bldg, Rm 821A, Chapel Hill, NC 27599-7525; e-mail: stl@med.unc.edu.