Abstract
To evaluate the incidence and clinical impact of WT1 gene mutations in younger adult patients with cytogenetically normal acute myeloid leukemia (CN-AML), sequencing of the complete coding region was performed in diagnostic samples from 617 patients who were treated on 3 German-Austrian AML Study Group protocols. WT1 mutations were identified in 78 (12.6%) of the 617 patients; mutations clustered in exon 7 (54 of 78) and exon 9 (13 of 78), but also occurred in exons 1, 2, 3, and 8. WT1 mutations were significantly associated with younger age, higher serum lactate dehydrogenase levels, higher blood blast counts, and the additional presence of FLT3-ITD (P < .001) and CEBPA mutations (P = .004). There was no difference in relapse-free survival and overall survival between patients with (WT1mut) or without WT1 mutations. Subset analysis showed that patients with the genotype WT1mut/FLT3-ITDpos had a lower complete remission rate (P = .003) and an inferior relapse-free survival (P = .006) and overall survival (P < .001) compared with those with the genotype WT1mut/FLT3-ITDneg. In conclusion, in our large cohort of younger adults with CN-AML, WT1 mutation as a single molecular marker did not impact on outcome. However, our data suggest a negative impact of the genotype WT1mut/FLT3-ITDpos.
Introduction
In the last decade, somatically acquired mutations have been identified in several genes in cytogenetically normal acute myeloid leukemia (CN-AML): the nucleophosmin 1 (NPM1) gene, the fms-related tyrosine kinase 3 (FLT3) gene, the CCAAT/enhancer binding protein alpha (CEBPA) gene, the myeloid-lymphoid or mixed-lineage leukemia (MLL) gene, the neuroblastoma RAS viral oncogene homolog (NRAS) gene, and the runt-related transcription factor 1 (RUNX1) gene.1,2 Besides providing insights into leukemia biology, some of these markers have proved to be important for disease classification and outcome prediction.3,4 Thus, the discovery of novel genetic changes probably will contribute to a refined molecular classification of the disease. Recently, Summers et al5 revived the initial findings on WT1 mutations in AML that were first reported by King-Underwood et al in 1996.6
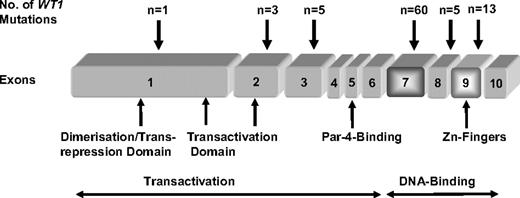
The Wilms' tumor 1 gene (WT1), cloned in 1990, is located at chromosomal band 11p13.7 Its corresponding protein consists of 4 C-terminal Zinc (Zn) fingers (exons 7-10) that are characteristic for transcription factors, whereas the N-terminal domain (exons 1-6), which contains proline-glutamine–rich sequences has been shown to be involved in RNA and protein interactions.8,9 WT1 functions as a potent transcriptional regulator for genes involved in cellular growth and metabolism, such as extracellular matrix components, growth factors, and other transcription factors.10
Genomic deletions affecting the WT1 gene were first identified in patients with WAGR syndrome characterized by Wilms tumor (WT), aniridia, genitourinary malformations, and mental retardation. These patients were found to have constitutional deletions of 11p13.11 Twelve years later, a WT1 mutation was described in a sporadic, unilateral WT as a heterozygous 25-bp deletion that results in loss of one of the 4 Zn finger consensus domains.12 Subsequent studies revealed WT1 mutations as the causative gene alteration in sporadic WT occurring with an incidence of 10% to 15%. Interestingly, in addition to the WAGR syndrome, WT1 germline mutations have also been found in other distinct constitutional syndromes, such as Denys-Drash syndrome and Frasier syndrome, which have a predisposition for the development of WT.6,13,14
The initial finding of a WT1 mutation in a secondary AML of a WAGR patient6 was confirmed and extended by 2 consecutive studies in which 67 samples of different types of leukemia (AML, acute lymphoblastic leukemia, biphenotypic leukemia, undifferentiated leukemia) were analyzed.15,16 The overall incidence of WT1 mutations was 12%, with 4 of 33 AML patients exhibiting WT1 mutations.6,16 None of the patients with WT1 mutation achieved complete remission (CR), and disease-free survival (DFS) and overall survival (OS) appeared to be worse. In a more recent study performed by Summers et al, WT1 mutations were identified in 7 of 70 CN-AML patients.5 Of note, 6 of the 7 patients also had FLT3 internal tandem duplications (ITDs), and all 6 patients were refractory to induction therapy.
Although its role in hematopoiesis is still not clarified, disruption of WT1 function is currently considered to promote stem cell proliferation and to hamper cell differentiation, the hallmarks of the 2 complementation groups of gene mutations that have been postulated for the development of AML.8,10,17,18 In addition, WT1 expression is normally restricted to the primary cells of the developing genitourinary system and to the hematopoietic progenitor cells. However, in AML, WT1 expression can be found in 75% to 100% of AML patients.
The aim of this study was to evaluate the incidence and clinical impact of WT1 mutations in the context of other known gene mutations in a large cohort of younger adult patients with CN-AML who were treated within 3 consecutive treatment trials of the German-Austrian AML Study Group (AMLSG).
Methods
Patients and treatment
Diagnostic bone marrow (BM) or peripheral blood (PB) samples were analyzed from 617 younger adult patients (16-60 years) with CN-AML. Patients had been entered onto 3 consecutive multicenter AMLSG treatment trials: AML HD93 (n = 77),19 AML HD98-A (n = 268; NCT00146120),20 and AMLSG 07-04 (n = 272; NCT00151242)21 ; 499 patients had de novo AML, 60 had secondary AML after myelodysplastic syndrome, 12 had treatment-related AML, and in 46 patients this information was missing. For the present study, the only criterion used to include patients was the availability of a BM or PB sample from diagnosis for gene mutation analysis. All patients gave informed consent for both treatment and genetic analysis in accordance with the Declaration of Helsinki; all studies were approved by the institutional review board of the University of Ulm. All patients received intensive double-induction and consolidation therapy. All 3 trials included a genetic randomization for allogeneic transplantation from a matched related donor. Details of the treatment plans, including dosages of cytotoxic drugs, are given in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Cytogenetic and molecular genetic analyses
Pretreatment samples from all patients were studied centrally by chromosome banding analysis. To improve the accuracy of cytogenetic diagnosis, specimens were also analyzed by fluorescence in situ hybridization or polymerase chain reaction (PCR) for the presence of the recurring gene fusions RUNX1-RUNX1T1, CBFB-MYH11, MLL-MLLT3, and PML-RARA.4
Molecular diagnostics included mutation analyses of the FLT3 (ITDs and tyrosine kinase domain [TKD] mutations at codon D835 and I836), NPM1, CEBPA, MLL (partial tandem duplication [PTDs]), and NRAS genes.4
Analysis of WT1 mutations
Genomic DNA was isolated from mononuclear cells stored at −80°C using the All Prep Kit (QIAGEN, Hilden, Germany) according to the manufacturer's recommendations. PCR of the whole coding region of the WT1 gene (exons 1-10) was carried out using intron-exon flanking primer pairs (Table S1). Because of GC-rich sequences of exon 1, Hot Star Plus polymerase (QIAGEN) was used for DNA amplification; for the remaining exons 2 to 10, Platinum Taq polymerase (Invitrogen, Karlsruhe, Germany) was used. The total reaction volume of 25 μL contained approximately 100 ng DNA, 10 pmol of each primer, deoxynucleoside triphosphates (10 mM each), and Hot Star Plus polymerase or Platinum Taq polymerase with supplied buffers. DNA was amplified using the following PCR conditions: 95°C for 10 minutes, 40 cycles of 95°C for 10 seconds, 57°C for 30 seconds, and 72°C for 30 seconds; and finally 72°C for 7 minutes. PCR products were purified by standard methods and subsequently sequenced with M13 forward primer using the ABI Ready Reaction Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Darmstadt, Germany).
In addition, PB samples from 38 healthy volunteers were analyzed for the presence of WT1 polymorphisms. In 26 of the 78 mutated AML cases, DNA obtained from buccal swabs or remission material from patients in complete remission was available for WT1 germline mutation analysis.
Statistical analyses
The median follow-up for survival was calculated according to the method of Korn.22 The definition of CR followed the recommended criteria.23 OS endpoints, measured from entry into one of the prospective studies, were death (failure) and alive at last follow-up (censored).23 Relapse-free survival (RFS) endpoints, measured from the date of documented CR, were relapse (failure), death in CR (failure), and alive in CR at last follow-up (censored).23 Comparisons between patient characteristics (covariates) were performed by Kruskal-Wallis test for continuous variables and by Fisher exact test for categorical variables. A multivariable logistic model was used to analyze associations between presenting features and response to induction therapy. The Kaplan-Meier method was used to estimate the distribution of RFS and OS. Confidence interval estimation for the survival curves was based on the cumulative hazard function using Greenwood's formula for the standard error (SE) estimation.24 A Cox model was used to identify prognostic variables.25 Missing data were estimated using a multiple-imputation technique using predictive mean matching with n equals 50 for multiple imputations.26 All statistical analyses were performed with the use of the R package (version 2.0-12) of the R statistical software platform (version 2.4.1).27
Results
WT1 mutations
Overall, 87 WT1 mutations were identified in 78 of the 617 (12.6%) patients (Table S2). Mutations clustered in exon 7 (n = 60) and exon 9 (n = 13) but also occurred in exons 1 (n = 1), 2 (n = 3), 3 (n = 5), and 8 (n = 5) (Figure 1). In exon 7, most of the mutations were frameshift mutations as a result of insertions (n = 39) or deletions (n = 10), ranging in length from 1 to 28 bp; 11 mutations were substitutions, in 2 of them, serine was replaced by a stop codon (S312X); 3 were synonymous mutations and 6 were missense mutations. In exon 9, most mutations (n = 11) were substitutions, with 3 of them causing a stop codon (R390X); the other 8 were missense mutations; 2 patients had an insertion leading to a frameshift. Fourteen mutations did not affect exon 7 and 9: one missense substitution in exon 1; 3 substitutions in exon 2 (2 cases with a stop codon, one missense mutation); one substitution (with a stop codon), one deletion and 3 insertions in exon 3; and 4 substitutions (one with a stop codon, 3 missense mutations) and one insertion in exon 8. Types of exon 7 and 9 mutations were similar to those recently described by King-Underwood et al,6,15,16 Virappane et al,28 and Paschka et al.29 The mutation P181S in exon 2 was previously described by Köhler et al as a germline missense mutation in a male patient with ambiguous genitalia, normal testosterone production, absence of kidney disease, and associated heart defect.30 Three mutations (2 polymorphisms and 1 insertion, which resulted in a truncated protein) in exon 1 have already been described by King-Underwood earlier,6 but these were different from mutation P13T found by us. Seventy-two patients had heterozygous and 6 patients had homozygous mutations. In all 6 patients, both alleles carried the same mutation, a finding that is consistent with acquired uniparental disomy. Data from single nucleotide polymorphism analysis were available in 2 of the 6 patients revealing acquired 11p uniparental disomy in both cases (data not shown). Three patients exhibited 2 mutations, 2 with mutations in exon 3 and exon 7, and 1 patient with mutations in exon 7 and exon 9.
None of the 38 PB samples obtained from healthy volunteers revealed WT1 sequence variations compared with the WT1 wild-type sequence (NCBI; cDNA NM_0024426; www.ncbi.org). No WT1 germline mutations were detected in the 26 cases in which DNA from buccal swabs, PB, or BM samples in CR was available.
Association with other molecular markers
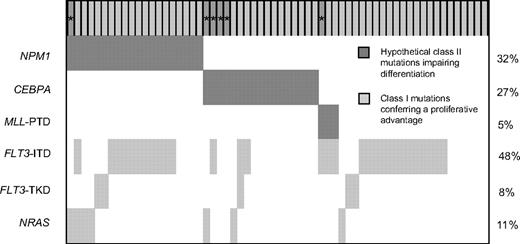
WT1 mutations (WT1mut) were significantly associated with the presence of FLT3-ITD (P < .001; marker constellation analyzed in 612 patients) and CEBPA mutations (P = .004; marker constellation analyzed in 552 patients); 41 of 78 (53%) WT1mut patients also harbored a FLT3-ITD compared with 171 of 534 (32%) WT1wt patients, and 17 of 72 (24%) WT1mut patients had additional CEBPA mutations compared with 51 of 480 (11%) WT1wt patients. The median of the FLT3-ITD mutant/wild-type ratio in WT1mut patients was 0.53 (range, 0.02-4.10) compared with 0.51 in WT1wt patients (range, 0.006-14.2, Wilcoxon test, P = .65). In contrast, there was an inverse relationship between the presence of WT1 mutations and NPM1 mutations: 24 of 75 (32%) WT1mut patients had NPM1 mutation, whereas 286 of 523 (55%) WT1wt patients had an additional NPM1 mutation (P < .001). There was no significant association with regard to FLT3-TKD mutations (P = .66), MLL-PTD (P = .64), or NRAS mutations (P = .30; Figure 2).
Frequencies and distribution of the WT1, NPM1, CEBPA, MLL, FLT3, and NRAS mutations. For WT1 mutations, each patient is represented by a gray column. Dark gray columns marked with * represent homozygous mutations. ITD denotes internal tandem duplication; PTD, partial tandem duplication; and TKD, mutation of the tyrosine kinase domain.
Frequencies and distribution of the WT1, NPM1, CEBPA, MLL, FLT3, and NRAS mutations. For WT1 mutations, each patient is represented by a gray column. Dark gray columns marked with * represent homozygous mutations. ITD denotes internal tandem duplication; PTD, partial tandem duplication; and TKD, mutation of the tyrosine kinase domain.
Patient characteristics by WT1 mutation status
WT1 mutations were significantly associated with younger age (P = .007), higher lactate dehydrogenase (LDH) serum levels (P = .007), and higher PB blast counts (P = .01). Of note, LDH and PB blast counts were significantly associated with the combined mutation status WT1mut/FLT3-ITDpos, in that highest values were present in double-positive cases (median LDH level, 755.5 U/L; median blast count, 71%) followed by cases harboring FLT3-ITD (559.5 U/L; 55%) or WT1mut (422 U/L; 35%); lowest values were found in patients with the WT1wt/FLT3-ITDneg genotype (362 U/L; 27%; P = .007 and P = .02 for overall comparison, respectively). Patients' characteristics according to WT1 and FLT3-ITD mutation status are given in Table 1.
Patient characteristics according to the WT1 and FLT3-ITD mutation status
| Characteristic . | WT1wt/FLT3-ITDneg . | WT1mut/FLT3-ITDneg . | WT1wt/FLT3-ITDpos . | WT1mut/FLT3-ITDpos . | P . |
|---|---|---|---|---|---|
| Sex, no. female/no. male | 178/185 | 23/14 | 104/67 | 22/19 | .05 |
| Type of AML, n* | .35 | ||||
| De novo | 288 | 29 | 146 | 33 | |
| s-AML | 43 | 3 | 10 | 2 | |
| t-AML | 9 | 0 | 3 | 0 | |
| Missing | 23 | 5 | 12 | 6 | |
| Median age, y (range) | 49.4 (16-60.9) | 41.9 (23-59.8) | 47.2 (18.3-60.9) | 46.4 (19.4-58.8) | .007 |
| Median WBC count, ×109/L (range) | 13.5 (0.2-369) | 13.9 (0.2-369) | 39.2 (0.2-345) | 30.1 (1.1-220) | .22 |
| Median PB blasts, % (range) | 27 (0-99) | 35 (0-98) | 55 (0-100) | 71 (1-96) | .01 |
| Median BM blasts, % (range) | 70 (0-100) | 72 (7-96) | 87 (2-100) | 88 (20-100) | .14 |
| Median LDH, U/L (range) | 362 (89-2371) | 422 (131-4566) | 559.5 (122-4091) | 755.5 (184-2992) | .007 |
| Characteristic . | WT1wt/FLT3-ITDneg . | WT1mut/FLT3-ITDneg . | WT1wt/FLT3-ITDpos . | WT1mut/FLT3-ITDpos . | P . |
|---|---|---|---|---|---|
| Sex, no. female/no. male | 178/185 | 23/14 | 104/67 | 22/19 | .05 |
| Type of AML, n* | .35 | ||||
| De novo | 288 | 29 | 146 | 33 | |
| s-AML | 43 | 3 | 10 | 2 | |
| t-AML | 9 | 0 | 3 | 0 | |
| Missing | 23 | 5 | 12 | 6 | |
| Median age, y (range) | 49.4 (16-60.9) | 41.9 (23-59.8) | 47.2 (18.3-60.9) | 46.4 (19.4-58.8) | .007 |
| Median WBC count, ×109/L (range) | 13.5 (0.2-369) | 13.9 (0.2-369) | 39.2 (0.2-345) | 30.1 (1.1-220) | .22 |
| Median PB blasts, % (range) | 27 (0-99) | 35 (0-98) | 55 (0-100) | 71 (1-96) | .01 |
| Median BM blasts, % (range) | 70 (0-100) | 72 (7-96) | 87 (2-100) | 88 (20-100) | .14 |
| Median LDH, U/L (range) | 362 (89-2371) | 422 (131-4566) | 559.5 (122-4091) | 755.5 (184-2992) | .007 |
WBC denotes white blood cell; PB, peripheral blood; BM, bone marrow; LDH, lactate dehydrogenase (normal value < 250 U/L); s-AML, secondary AML; and t-AML, therapy-associated AML.
Status on FLT3-ITD missing in 3 de novo AML and in 2 secondary AML after myelodysplastic syndrome.
Response to induction therapy
In univariable analysis, there was no difference in the rates of CR (77% vs 79%), refractory disease (RD; 19% vs 14%), and early death (4% vs 7%) between WT1mut and WT1wt patients, respectively.
Based on the previous observation that the concurrent presence of WT1 mutation and FLT3-ITD may be associated with induction failure5 and based on our findings showing that patients with the genotype WT1mut/FLT3-ITDpos appeared to have a high proliferative activity indicated by high blood blast counts and high LDH serum levels, a subset analysis was performed exploring the impact of an additional FLT3-ITD in patients with WT1 mutations. Patients with the genotype WT1mut/FLT3-ITDpos (n = 40) had a significantly lower CR rate (63%) and a significantly higher RD rate (33%) compared with patients (n = 37) with WT1 mutations in the absence of FLT3-ITD (92% and 5%; P = .003 and P = .003, respectively). Patients with the WT1wt/FLT3-ITDpos (n = 169) and WT1wt/FLT3-ITDneg (n = 357) genotypes had comparable CR rates (78% and 79%, respectively) and RD rates (6.5% and 8%, respectively; missing values for response evaluation n = 9).
As a consequence of these findings, we performed an explorative multivariable logistic regression analysis, including the variables FLT3-ITD and WT1mut as single markers as well as with an interaction term. This model revealed CEBPAmut (odds ratio [OR], 2.67; P = .02) and NPM1mut (OR, 2.13; P = .001) to be significantly associated with response to induction therapy, whereas logarithm of white blood cell (WBC; OR, 0.63; P = .009) and the genotype WT1mut with concurrent FLT3-ITDpos (OR 0.19; P = .03) were significantly associated with induction failure.
Survival analysis
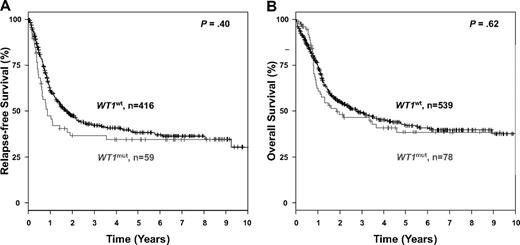
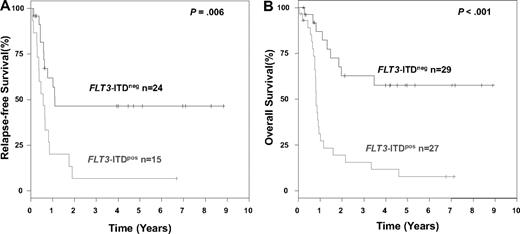
The median follow-up time for survival was 44 months. In univariable analysis, there was no difference in RFS (P = .40) and OS (P = .62) between the WT1mut and the WT1wt groups (Figure 3). Results for survival were very similar when only exon 7 and 9 mutations were considered, or when only de novo AML were included in statistical analyses. When considering only those patients who received either repetitive cycles of high-dose cytarabine or autologous stem cell transplantation for consolidation and excluding those who received allogeneic transplantation, again, no difference in RFS (P = .23) or OS was found (P = .46). Subset analysis according to the FLT3-ITD status (excluding patients who received an allogeneic transplantation) revealed a significantly inferior RFS (P = .006) and OS (P < .001) for patients with the genotype WT1mut/FLT3-ITDpos compared with patients with the genotype WT1mut/FLT3-ITDneg (Figure 4).
Kaplan-Meier survival estimates, according to WT1 mutation status. Data are shown for relapse-free survival (A) and overall survival (B).
Kaplan-Meier survival estimates, according to WT1 mutation status. Data are shown for relapse-free survival (A) and overall survival (B).
Kaplan-Meier survival estimates for patients with WT1 mutation, according to FLT3-ITD mutation status. Data are shown for relapse-free survival (A) and overall survival (B). Patients after allogeneic stem cell transplantation have been excluded from this analysis.
Kaplan-Meier survival estimates for patients with WT1 mutation, according to FLT3-ITD mutation status. Data are shown for relapse-free survival (A) and overall survival (B). Patients after allogeneic stem cell transplantation have been excluded from this analysis.
Multivariable analysis on the endpoints RFS and OS revealed a significant impact of the genotypes NPM1mut/FLT3-ITDneg (hazard ratio [HR], 0.45 and 0.52, respectively), CEBPAmut (HR, 0.51 and 0.39, respectively), MLL-PTD (HR, 1.57 only for RFS), age (10 years difference, HR 1.2 and 1.3), and logarithm of WBC (HR 1.55 and 1.58, respectively). WT1 mutations had no significant impact.
Discussion
WT1 mutations in acute leukemia were indeed reported more than 10 years ago,6 but it is only recently that these mutations received attention again.5 In this study, we assessed the incidence and prognostic significance of WT1 mutations in a very large cohort of younger adult patients with CN-AML who were prospectively enrolled into 3 AMLSG protocols.
WT1 mutations were identified in 78 of the 617 (12.6%) patients. This percentage of cases is slightly higher than the 10% and 10.7% frequencies that were recently reported in the studies by Virappane et al28 and Paschka et al,29 respectively (Table 2). The difference may be explained by the fact that in our study the entire WT1 coding sequence was analyzed, and mutations other than those in exons 7 or 9 were identified.
Comparison of the WT1 mutation studies performed by AMLSG, CALGB, and MRC
| . | AMLSG . | CALGB . | MRC . |
|---|---|---|---|
| Patients, n | 617 | 196 | 470 |
| Type of AML | De novo, s-AML, t-AML | De novo | 94% de novo AML |
| Age, y | 16-60 | 18-60 | 15-60; 13 patients > 60 |
| Median follow-up time, mo | 33 | 50.4 | 122.4 (range, 27-213) |
| Trials | AML HD93, AML HD98-A, AMLSG 07-04 | 9621, 19808 | MRC10, MRC12 |
| Exons | 1-10 | 7 and 9 | 7 and 9 |
| No. of mutated patients | 78/617 (13%) | 21/196 (11%) | 47/470 (10%) |
| Associated mutations (WT1mut) | FLT3-ITD (P = .001) CEBPAmut (P = .004) NPM1mut (inverse, P = .001) | FLT3-ITD (P = .06) | FLT3-ITD (P = .08) NPM1mut (inverse, P = .05) |
| Clinical characteristics (WT1mut) | ↓ age (P = .007) ↑ LDH serum levels (P = .007) ↑ PB blast counts (P = .01) | ↑ WBC counts (P = .01) | No difference in age, sex, type of AML, WBC count |
| Response to induction (WT1mut) | No difference in rate of CR, RD, and ED | No difference in rate of CR | Inferior response to induction therapy (P = .02) |
| Survival analysis (WT1mut) | |||
| Univariable analysis | No difference in RFS and OS | ↓ DFS (P < .001), ↓ OS (P < .001) | ↓ RFS (P = .005), ↓ OS (P = .007) |
| Multivariable analysis | No difference in RFS and OS | ↓ DFS (P = .009), ↓ OS (P < .001) | ↓ RFS (P < .02), ↓ OS (P < .04) |
| Explorative subset analysis | WT1mut/FLT3-ITDneg: ↑ CR (92%), ↓ RD (5%) WT1mut/FLT3-ITDpos: ↓ CR (63%), ↑ RD (33%) ↓ RFS (P = .006), ↓ OS (P < .001) | Not performed | Not performed |
| Cumulative dosages of high-dose cytarabine | 18-54 g/m2 | 13-25 g/m2 | 1-8 g/m2 |
| . | AMLSG . | CALGB . | MRC . |
|---|---|---|---|
| Patients, n | 617 | 196 | 470 |
| Type of AML | De novo, s-AML, t-AML | De novo | 94% de novo AML |
| Age, y | 16-60 | 18-60 | 15-60; 13 patients > 60 |
| Median follow-up time, mo | 33 | 50.4 | 122.4 (range, 27-213) |
| Trials | AML HD93, AML HD98-A, AMLSG 07-04 | 9621, 19808 | MRC10, MRC12 |
| Exons | 1-10 | 7 and 9 | 7 and 9 |
| No. of mutated patients | 78/617 (13%) | 21/196 (11%) | 47/470 (10%) |
| Associated mutations (WT1mut) | FLT3-ITD (P = .001) CEBPAmut (P = .004) NPM1mut (inverse, P = .001) | FLT3-ITD (P = .06) | FLT3-ITD (P = .08) NPM1mut (inverse, P = .05) |
| Clinical characteristics (WT1mut) | ↓ age (P = .007) ↑ LDH serum levels (P = .007) ↑ PB blast counts (P = .01) | ↑ WBC counts (P = .01) | No difference in age, sex, type of AML, WBC count |
| Response to induction (WT1mut) | No difference in rate of CR, RD, and ED | No difference in rate of CR | Inferior response to induction therapy (P = .02) |
| Survival analysis (WT1mut) | |||
| Univariable analysis | No difference in RFS and OS | ↓ DFS (P < .001), ↓ OS (P < .001) | ↓ RFS (P = .005), ↓ OS (P = .007) |
| Multivariable analysis | No difference in RFS and OS | ↓ DFS (P = .009), ↓ OS (P < .001) | ↓ RFS (P < .02), ↓ OS (P < .04) |
| Explorative subset analysis | WT1mut/FLT3-ITDneg: ↑ CR (92%), ↓ RD (5%) WT1mut/FLT3-ITDpos: ↓ CR (63%), ↑ RD (33%) ↓ RFS (P = .006), ↓ OS (P < .001) | Not performed | Not performed |
| Cumulative dosages of high-dose cytarabine | 18-54 g/m2 | 13-25 g/m2 | 1-8 g/m2 |
AMLSG denotes German-Austrian AML Study Group; CALGB, Cancer and Leukemia Group B; MRC, Medical Research Council; s-AML, secondary AML; t-AML, therapy-associated AML; WBC, white blood cell; LDH, lactate dehydrogenase; PB, peripheral blood; CR, complete remission; RD, refractory disease; ED, early death; RFS, relapse-free survival; OS, overall survival; and DFS, disease-free survival.
WT1 mutations in our study were significantly associated with younger age, higher LDH serum levels, and higher PB blast counts, with the latter 2 factors probably reflecting a higher proliferative activity of these leukemias. In the study by Cancer and Leukemia Group B (CALGB),29 there was an association of WT1 mutations with higher WBC counts; however, no association with surrogate markers for proliferation was found in the Medical Research Council (MRC) study.28 In accordance with the findings by Summers et al,5 in our study the presence of WT1 mutations was significantly associated with FLT3-ITD; in addition, we found that, among hypothetical class II mutations, CEBPA mutations, but not NPM1 mutations, were significantly associated with WT1 mutations. In both the MRC and CALGB studies, there also was a higher frequency of FLT3-ITD among WT1 mutated cases; however, this did not reach statistical significance (Table 2).
Of note, in our study, we did not observe any prognostic impact of WT1 mutations on achievement of CR, RFS, and OS in either univariable or multivariable analysis. This finding is in marked contrast to the data by CALGB and by MRC that evaluated the prognostic significance of WT1 mutations in 196 and 470 younger adults with CN-AML, respectively.28,29 With regard to response to induction therapy, in the study by CALGB, there was also no difference in CR rates between patients with or without WT1 mutations; whereas in the study by MRC, patients with WT1 mutations had a significantly inferior CR rate because of a higher rate of resistant disease. In the study by CALGB, patients with WT1 mutations had inferior DFS and OS; the 3-year OS rate was 10% and 56% for patients with and without WT1 mutations, respectively, and in multivariable analysis, WT1 mutation independently predicted for DFS and OS. Similarly, in the study by MRC, patients with WT1 mutations had inferior RFS and OS; the 5-year OS rate was 26% and 47% for patients with and without WT1 mutations, respectively; in multivariable analysis, WT1 mutation was an independent adverse prognostic factor.
The observation by Summers et al5 that the presence of a FLT3-ITD in addition to WT1 mutation may be associated with induction failure and inferior outcome prompted us to perform an exploratory subset analysis taking into account the FLT3-ITD status. Of note, comparing the laboratory characteristics among the various genotypes, the genotype WT1mut/FLT3-ITDpos was associated with highest serum LDH levels and highest blood blast counts, suggesting that the 2 mutations cooperate and confer a high proliferative capacity to these leukemias. In accordance with the findings by Summers et al,5 the WT1mut/FLT3-ITDpos genotype was significantly associated with induction failure and inferior RFS and OS (Figure 4). This effect could only be shown for response to induction in multivariable analysis and for RFS and OS in univariable analysis.
The marked differences between our data and those by CALGB and MRC remain elusive. One possible explanation is that the discrepancies may be a consequence of differences in treatment. Indeed, comparing the treatment protocols, the most striking difference was in the cumulative dosages of high-dose cytarabine given for consolidation treatment. Both MRC 10 and MRC 12 protocols used relatively low cumulative doses of high-dose cytarabine (∼ 6 g/m2 and ∼ 1-8 g/m2, respectively). Similarly, in CALGB protocol 9621 (dose escalation study of daunorubicin and etoposide) and protocol 19808, the cumulative dosages of high-dose cytarabine ranged between 13 g/m2 and 25 g/m2. In contrast, in our treatment protocols, the dosages of high-dose cytarabine ranged between 18 g/m2 and 54 g/m2, with the majority of patients receiving more than 36 g/m2 (Table 2). These data would suggest that the negative impact of WT1 mutations reported by others may be overcome by the use of repetitive cycles of high-dose cytarabine, especially in the subgroup of patients with an FLT3-ITDneg genotype. Of note, similar differential treatment effects of various dosages of cytarabine have previously been reported for other genotypic subsets of AML, ie, for AML with RUNX1-RUNX1T1 and CBFB-MYH11 gene fusions31 and, more recently, for AML with RAS mutations.32 Thus, it will be important to prospectively evaluate the impact of WT1 mutations with respect to different treatments, in particular to the use of repetitive cycles of high-dose cytarabine for consolidation treatment.
In accordance with previous studies, the majority of WT1 mutations in our study were frameshift mutations occurring in exon 7, followed by single amino acid substitutions in exon 9; whereas frameshift mutations in exon 9 were rare and have been identified in only 2 of our patients. Because exon 7 encodes the first Zn finger of WT1, mutations would result in truncated proteins lacking the 4 Zn fingers that contain functional domains, such as the DNA-binding portion, the nuclear localization signal and the binding domains for interacting proteins. Exon 9 frameshift mutations are predicted to lack zinc fingers 3 and 4; exon 9 substitutions, in particular mutations at codon D396 and H397, would also be expected to impair DNA binding and have been shown to destabilize the helix of Zn finger 3.33 The missense mutation R390X in exon 9 has previously been reported as somatic as well as germline WT1 mutation in WT.14,34,35 Beyond the mutation hotspot regions in exons 7 and 9, we found mutations in exons 1, 2, 3, and 8. Exon 8 frameshift mutations are predicted to result in truncated proteins lacking the second Zn finger; the frameshift mutations in exon 2 and 3 would result in truncated proteins lacking the trans-repression and the trans-activation domain localized at exon 1 to 4 as well as the 4 zinc fingers. Mutations in exon 1 have been reported previously and would lead to an almost complete lack of the protein from the affected allele.6 Nevertheless, it is unknown whether the various WT1 mutation types that so far have been described in AML also have different functional consequences and therefore differ in their impact on clinical outcome. Of note, there are marked differences in WT1 mutation types between the distinct disease entities. In AML, the mutations predominantly cluster in exon 7 (mostly frameshift mutations resulting from insertions or deletions) and 9 (mostly substitutions); whereas in patients with WAGR, Denys-Drash syndrome, or Frasier syndrome, the majority of WT1 mutations are point mutations located either in Zn finger 2 and 3 (exon 8 and 9) or in cases with Frasier syndrome in intron 9. In WT, large deletions or intragenic mutations involving especially exons 7 and 9 have been found. These distinct mutation types suggest different pathogenic mechanism of mutant WT1 in differentiating hematopoietic cells and cells of the genitourinary system.
In conclusion, in our large cohort of patients with CN-AML, the presence of WT1 mutation per se did not have an impact on treatment outcome; however, the WT1mut/FLT3-ITDpos genotype appeared to be associated with worse clinical course. Testing for WT1 mutations should become part of the molecular risk assessment in future clinical trials to resolve the discrepancies observed among the different studies and to further evaluate whether the use of repetitive cycles of high-dose cytarabine may indeed overcome the strong negative prognostic impact reported in the studies by MRC and CALGB.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all members of the German-Austrian AML Study Group for their participation in this study and for providing patient samples.
This work was supported in part by the Network of Competence Acute and Chronic Leukemias (grant 01GI9981) and IPD-Meta-Analysis (grant 01KG0605), a model-based hierarchical prognostic system for adult patients with acute myeloid leukemia, the Bundesministerium für Bildung und Forschung, Germany, the Deutsche José Carreras Leukämie-Stiftung (R 08/23v), and the Else Kröner-Fresenius-Stiftung (P38/05//A49/05//F03).
Authorship
Contribution: V.I.G., H.D., and K.D. designed research, analyzed data, and wrote the paper; R.F.S. analyzed data and critically reviewed the paper; S.M. and A.B. performed experiments; and L.B., A.C., B.S., J.K., and A.G. contributed intellectually and critically reviewed the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the German-Austrian AML Study Group (AMLSG) appears in the Appendix, available online on the Blood website.
Correspondence: Konstanze Döhner, Department of Internal Medicine III, University Hospital of Ulm, Albert-Einstein-Allee 23, 89081 Ulm, Germany; e-mail: konstanze.doehner@uniklinik-ulm.de.