Abstract
AHI-1 is an oncogene often targeted by provirus insertional mutagenesis in murine leukemias and lymphomas. Aberrant expression of human AHI-1 occurs in cutaneous T-cell lymphoma (CTCL) cells and in CD4+CD7− Sezary cells from patients with Sezary syndrome. Stable knockdown of AHI-1 using retroviral-mediated RNA interference in CTCL cells inhibits their transforming activity in vitro and in vivo. To identify genes involved in AHI-1–mediated transformation, microarray analysis was performed to identify differentially expressed genes in AHI-1–suppressed CTCL cells. Fifteen up-regulated and 6 down-regulated genes were identified and confirmed by quantitative reverse transcription-polymerase chain reaction. Seven were further confirmed in a microarray analysis of CD4+CD7− Sezary cells from Sezary syndrome patients. HCK and BIN1 emerged as new candidate cooperative genes, with differential protein expression, which correlates with observed transcript changes. Interestingly, changes in HCK phosphorylation and biologic response to its inhibitor, dasatinib, were observed in AHI-1–suppressed or –overexpressed cells. The tumor suppressor BIN1 physically interacts with MYC in CTCL cells, which also exhibit differential MYC protein expression. In addition, aberrant expression of alternative splicing forms of BIN1 was observed in primary and transformed CTCL cells. These findings indicate that HCK and BIN1 may play critical roles in AHI-1–mediated leukemic transformation of human CTCL cells.
Introduction
Cutaneous T-cell lymphomas (CTCLs) are lymphoproliferative disorders characterized by the homing of malignant T cells to the surface of the skin.1-3 As a heterogeneous group, CTCLs are the most common, with an incidence rate of approximately 1500 to 2000 new cases annually in the United States.4 The 2 main types of CTCLs are mycosis fungoides (MF) and its leukemic variant, Sezary syndrome (SS), which together represent approximately 65% to 70% of all CTCL cases.1,4,5 SS is a very aggressive disease with high mortality and a 5-year survival rate of only 24%.6 SS patients present with erythroderma, general lymphadenopathy, and Sezary cells in the skin, lymph nodes, and peripheral blood.4,6,7 Sezary cells are circulating malignant mature memory CD4+ CD45RO+ T cells with atypical cerebriform nuclei; these cells often lose expression of CD7 and CD26.3,4,6,7 The development of Sezary cells denotes a poor prognosis, and treatments for SS are generally disappointing. To date, little is known about the molecular nature of the lymphoma cells in SS and MF, although gene expression studies on MF and SS primary samples have recently been conducted to gain insight into the pathogenesis of these 2 diseases, with the hope of identifying diagnostic and therapeutic target molecules.3,4,8 However, the precise genetic pathogenesis of these diseases still needs to be determined
The novel oncogene Abelson helper integration site-1 (Ahi-1) was identified by provirus insertional mutagenesis in various murine leukemias and lymphomas.9 Murine Ahi-1 encodes a modular protein with several Src homology 3 (SH3) binding sites (PxxP), an SH3 domain, and 7 tryptophan-aspartic acid 40 (WD40) repeat domains.9 Further, there are many potential phosphorylation sites, including 2 tyrosine kinase phosphorylation sites, an amino acid–rich domain, and 3 potential PEST sequences.9,10 These domains and motifs are known to be important mediators of protein-protein interactions.9,11 Interestingly, human AHI-1 contains an additional N-terminal coiled coil domain, absent in mouse Ahi-1, a domain frequently involved in intramolecular and intermolecular interactions and in homo- and heterodimerization.12,13 There appear to be at least 3 human isoforms of AHI-1, which differ in their C-terminus sequence.9,14
The oncogenic potential of AHI-1 is further illustrated by highly deregulated expression in several human leukemic cell lines of myeloid, B-cell and T-cell origin, suggesting that aberrant expression of AHI-1 contributes to leukemia and lymphoma development.14 The most striking up-regulation of AHI-1 occurs in the CTCL cell lines, Hut 78 (∼ 40-fold), derived from blood of a patient with SS, and Hut 102 (∼ 30-fold), derived from blood of a patient with MF.14 Moreover, aberrant expression of AHI-1, at RNA and protein levels, has been detected in primary CD4+CD7− Sezary cells.11 We have recently demonstrated that the lymphomagenic activity of Hut 78 cells is directly dependent on expression of AHI-1 by stably knocking down endogenous AHI-1 by retroviral-mediated RNA interference in Hut 78 cells, which inhibited their transforming activity in vitro and in vivo.11 Although there is substantial evidence that Ahi-1/AHI-1 can act as an oncogene in hematopoietic cells, the specific molecular mechanisms of this transformation have yet to be determined.
To elucidate molecular mechanisms of AHI-1–mediated leukemic transformation, a microarray analysis has been performed in CTCL cells with stable AHI-1 suppression to identify differentially expressed genes as potential AHI-1 cooperative genes. Several identified genes were confirmed in CTCL cells and in Sezary cell-enriched populations (CD4+CD7− cells) of SS patient samples and further assessed for differential expression and phosphorylation at the protein level. HCK (hemapoietic cell kinase) and BIN1 (bridging integrator 1) have emerged as candidate cooperative genes that could be functionally connected to AHI-1 in CTCL.
Methods
Human primary cells
Peripheral blood from 6 SS patients at diagnosis (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) and 5 normal persons was obtained from the Department of Dermatology, University of British Columbia.11 Informed consent was obtained in accordance with the Declaration of Helsinki, and the procedures used were approved by the Research Ethics Board of the University of British Columbia. All patients were screened for human T-cell leukemia virus-1 status and were negative. Leukemic Sezary cells and normal blood cells were purified by negative selection with monoclonal antibodies directed against granulocytes, B cells, and CD8+ and CD7+ T cells using a Rosette Sep kit (StemCell Technologies, Vancouver, BC) as previously described.11 Cell purity was verified by fluorescence-activated cell sorter, with more than 90% purity by immune phenotyping (CD4+CD7−) obtained from patient samples studied as described.11
Cell culture and lentiviral transduction of Hut 78 cells
Hut 78 cells (transduced and untransduced) were cultured in RPMI 1640 medium with 10% fetal bovine serum, 100 U/mL penicillin, 0.1 mg/mL streptomycin, and 10−4 M β-mercaptoethanol (StemCell Technologies). An EF1α-AHI-1-IRES-YFP lentiviral vector was constructed as previously described.15 Lentiviral production was performed, Hut 78 cells were transduced, and YFP+ transduced cells were fluorescence-activated cell sorter-purified as described.15
Colony-forming cell assay
Colony-forming cell (CFC) assays were performed in fetal calf serum-containing methylcellulose cultures (H4230; StemCell Technologies) in the presence or absence of dasatinib (DS; 10-100 nM) and imatinib (IM; 1-10 μM). Colony counts were performed, using standard scoring criteria, on the basis of the ability to produce colonies containing a minimum of 20 cells after 12 to 14 days of incubation.
RNA isolation
RNA was isolated by pelleting 105 to 106 cells and washing them in 10 mL Dulbecco phosphate-buffered saline (PBS; StemCell Technologies). Washed cells were repelleted, and the Absolutely RNA Miniprep Kit (Stratagene, La Jolla, CA) was used to purify total RNA using the procedure described for tissue-culture cells grown in suspension. RNA yield was quantified by measuring the optical density at 260 nm and 280 nm using a Nanodrop ND-100 Spectrophotometer (Thermo Fischer Scientific, Wilmington, DE).
Microarray analysis
A total of 1 μg purified total RNA from parental Hut 78, Hut 78 RPG (Hut 78 cells transduced with an empty vector), Hut 78/sh4 bulk (AHI-1–suppressed cells), and Hut 78/sh4 clone 1 cells (AHI-1–suppressed clonal cells) was used for microarray analysis (Michael Smith Genome Sciences Center, British Columbia Cancer Agency). Controls consisted of 2 RNA samples of Hut 78 cells and 3 RNA samples of Hut 78 RPG cells. Experimental samples included 3 RNA samples of Hut 78/sh4 bulk cells and 3 RNA samples of Hut 78/sh4 clone 1 cells. The Affymetrix GeneChip Human Genome U133 Plus 2.0 Array was used to determine differential gene expression between the 2 groups (Affymetrix, Santa Clara, CA). Data were analyzed by DNA-Chip Analyzer (dChip) and Linear Model for Microarray Data (Limma) software programs to identify significantly differentially expressed genes.16,17
The microarray data have been deposited in the GEO database under accession number GSE14746. All data obtained from the microarray analysis have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14746).
Quantitative reverse transcription–polymerase chain reaction
RNA (0.1-0.5 μg) was reverse transcribed in a 25-μL reaction using random primers (1 μg; Invitrogen, Burlington, ON) and SuperScript III Reverse Transcriptase (Invitrogen), as described.18 Polymerase chain reaction (PCR) was performed using 12.5 μL Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 1 μL cDNA, 1 μL 20 μM gene specific primer (Invitrogen; Table S2), and 10.5 μL water, for a total reaction volume of 25 μL. Quantification of gene expression was performed using the 7500 Real Time PCR System (Applied Biosystems). The thermal profile of the reaction was: 50°C for 2 minutes, 95°C for 10 minutes, and 45 or 50 cycles of 95°C for 15 seconds followed by 60°C for 1 minute. The 7500 Real Time PCR System Software was used for data analyses (Applied Biosystems).
Cell lysate
Cells were pelleted and washed with PBS and placed at −70°C to freeze dry. The pellet was resuspended in an appropriate volume of lysis buffer (106 cells/30 μL lysis buffer), and the mixture was incubated for 1 hour on a rotator at 4°C. The lysis buffer consisted of 1 mL PSB, 10 μL phenylmethylsulfonyl fluoride (Sigma-Aldrich, Oakville, ON), 50 μL protease inhibitor cocktail (Sigma-Aldrich), and 50 μL NP-40 Alternative, Protein Grade Detergent (Calbiochem, San Diego, CA). After incubation, lysed cells were centrifuged at 9000g for 10 minutes at 4°C, and the supernatant was harvested and stored at −70°C.
Protein lysate quantification
Bradford assays were used to determine lysate concentration by generation of a standard curve with bovine serum albumin (Bio-Rad, Mississauga, ON) using concentrations ranging from 50 to 1000 ng/μL. Samples were diluted to 1/20× to ensure a reading in the linear range of the standard curve, and 20 μL of each diluted sample and standard was aliquoted into an untreated 96-well plate. Next, 200 μL Bio-Rad Protein Assay Dye Reagent was mixed in each well and the plate incubated at room temperature for at least 5 minutes. The absorbance of samples/standards was measured at 630 nm using the ELx808 Absorbance Microplate Reader (BioTek Instruments, Winooski, VT).
Western blotting and immunoprecipitation
Protein expression was assessed by Western blotting with the NuPAGE Novex Bis-Tris Gel Electrophoresis system (Invitrogen). Samples were prepared using 20 μg protein lysate, 2.5 μL NuPAGE LDS sample buffer (4×), 1.0 μL NuPAGE Reducing Agent (10×), and up to 6.5 μL deionized water for a final volume of 10 μL (Invitrogen). Samples were heated at 70°C for 10 minutes and loaded into a NuPAGE Novex 4% to 12% Bis-Tris Gel 1.0 mm, 10 well (Invitrogen) along with the PageRuler Prestained Protein Ladder (Fermentas, Burlington, ON). Using the XCell Surelock Mini-cell and NuPAGE MOPS SDS Running Buffer (Invitrogen), the gel was run under reducing conditions at 200 V for 50 minutes. Proteins were then transferred from the gel onto Immobilon-P polyvinylidene fluoride 0.45-μm membrane (Millipore, Billerica, MA) using NuPAGE Transfer Buffer (Invitrogen) in the XCell II Blot Module (Invitrogen) at 30 V for 1 hour. The membrane was next dried, blocked in Tris-buffered saline Tween 20 (TBST) with 5% skim milk for 1 hour at room temperature, and washed for 5 minutes (2 times) with TBST. The membrane was then incubated with primary antibody overnight at 4°C, washed for 10 minutes (3 times) with TBST, and incubated with secondary antibody for 1 hour at room temperature, followed again by 10-minute washes (3 times) with TBST (conditions used for various primary and secondary antibodies are listed in Table S3). Western Lightning Western Blot Chemiluminescence Reagent Plus and KODAK BioMax XAR autoradiography film (PerkinElmer Life and Analytical Sciences, Waltham, MA) were used to image the membrane with the Fuji RGII x-ray film processor. Relative protein expression was determined by densitometry using the image analysis program ImageQuant, version 5.2 (Molecular Dynamics, Sunnyvale CA). For immunoprecipitation, protein extracts were diluted to approximately 1 μg protein/μL with PBS containing proteinase inhibitors (250 μg total protein), and specific antibodies were added and incubated at 4°C overnight. The immunocomplexes were then captured by protein G bead slurry, separated from the beads, and Western blotting performed as previously described.15
Peptide competition
Peptide competition experiments were performed on the HCK primary antibody (sc-72; Santa Cruz Biotechnology, Santa Cruz, CA) using the HCK epitope (sc-72 P; Santa Cruz Biotechnology) and a random peptide, AHI-1 (Imgenex, San Diego, CA). Neutralization of the primary antibody was achieved by incubating 10 μL of 0.2 μg/μL HCK antibody with either 50 μL of 0.2 μg/μL HCK blocking peptide and 440 μL PBS, or 20 μL of 0.5 μg/μL AHI-1 peptide and 470 μL PBS. The final 500-μL reaction volume was incubated overnight at 4°C while gently shaking and was subsequently diluted with 9.5 mL of TBST to a final primary antibody concentration of 1:1000. Neutralized antibody was then used as described in the Western blotting procedure.
Cloning and sequencing of PCR products
PCR was performed with 1.0 μL of 10 mM dNTP, 2.0 μL of 20 μM BIN1 specific primer (Table S2), 5.0 μL of 10× PCR buffer, 1.5 μL of 50 mM MgCl2, 1.0 μL Platinum Taq DNA polymerase, 2.0 μL cDNA,and 37.5 μL water mixed together for a total reaction volume of 50 μL (Invitrogen). The thermal profile of the reaction was: 95°C for 5 minutes, 35 cycles of 94°C for 30 seconds, 66°C for 30 seconds, and 72°C for 45 seconds, followed by 72°C for 10 minutes. PCR products were analyzed on a 2% Agarose SFR (Amresco, Solon, OH) TAE gel at 90 V for 2.5 hours. The resulting bands were excised and purified using the MinElute Gel Extraction Kit (QIAGEN, Mississauga, ON). The extracted PCR products were then ligated into the pCR2.1 vector using the TOPO TA Cloning Kit (Invitrogen). This was followed by transformation into the MAX Efficiency DH5 αT1 Phage Resistant One Shot Competent Escherichia coli (Invitrogen), which were plated on 100 μg/mL ampicillin LB selection plates with 40 μL of 20 μg/μL X-gal. Positive colonies were expanded in liquid culture, and plasmids were purified with the GeneJET Plasmid Miniprep Kit (Fermentas). Both M13 PCR and digestion using HindIII and EcoRV restriction enzymes (Invitrogen) were subsequently used to verify the positive colonies, and those clones that successfully confirmed were then sequenced (McGill University and Genome Québec Innovation Center, Montreal, QC).
Statistical analysis
Data analysis used 2-sample Student t tests to determine the statistical significance of the results. The 2 arrays compared consisted of the Hut 78 and Hut 78 RPG data versus the Hut 78/sh4 bulk and Hut 78/sh4 clone 1 data. The analysis included 2 tails and assumed unequal variance because the datasets were relatively small (N = 6, 8, 10, or 12).
Results
Microarray identification of differentially expressed genes in human CTCL cells with suppression of AHI-1
To identify genes involved in AHI-1–mediated leukemic transformation, a microarray experiment compared parental Hut 78 cells with those with a stable knockdown of all AHI-1 isoforms. AHI-1 knockdown cells were previously generated using a short hairpin transcript (sh4) derived from an N-terminal 19 nucleotide sequence of AHI-1 in a modified retroviral vector (RPG), which was transduced into Hut 78 cells.11 Gene expression of parental Hut 78 cells and Hut 78 cells transduced with empty RPG vector (Hut 78 RPG) was compared with a bulk population (Hut 78/sh4) and a clonal population (Hut 78/sh4 clone 1) of AHI-1–suppressed cells. The Affymetrix GeneChip Human Genome U133 Plus 2.0 Array was used to evaluate gene expression of 6 RNA samples from AHI-1–suppressed cells (Hut 78/sh4) against 5 control samples, and the resulting pooled data were analyzed by 2 independent software programs (Limma and dChip). Differentially expressed genes were identified using the threshold values of a fold change of 2 or less or 2 or more and a P value of .01 or lower.
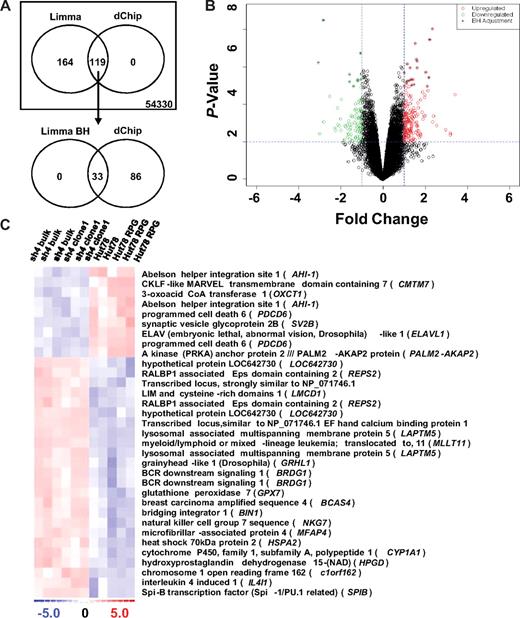
Limma analysis initially listed 283 differentially expressed probe sets (218 genes) that were further refined to a list of 33 (27 genes, Figure 1A,C). Refinement occurred by performing the Benjamini and Hochberg (BH) adjustment on the P values to correct for discovery of false positives (Figure 1).19 Using the same threshold values, the dChip analysis identified 119 probe sets (95 genes) with differential expression, with identified genes overlapping those detected in the initial Limma analysis (Figure 1A,B). Notably, significant down-regulation of AHI-1 itself (P < .001) was confirmed by microarray analysis in all 6 Hut 78/sh4 RNA samples studied (Figure 1C).
Differentially expressed genes in AHI-1–suppressed cells identified through microarray analysis. (A) Venn diagram of differentially expressed probes selected by both Limma and dChip analyses. The Affymetrix GeneChip Human Genome U133 Plus 2.0 Array screened 54 330 probe sets using the threshold values of a fold change of 2 or higher and 2 or lower and a P value of .01 or less for differential expression. Initial Limma analysis selected 283 differentially expressed probe sets, which was further refined to 33 with the BH P value adjustment. Importantly, the 119 probes sets identified by dChip analysis completely overlapped the initial Limma results. (B) Volcano plot of differentially expressed probes selected by Limma analysis (red represents up-regulated; green, down-regulated; and +, after BH adjustment). (C) Heat map of differentially expressed genes from the refined Limma (BH) analysis. Up-regulated (red) and down-regulated (blue) genes are defined by comparing the pooled gene expression of Hut 78/sh4 bulk and Hut 78/sh4 clone 1 samples with Hut 78 and Hut 78 RPG samples.
Differentially expressed genes in AHI-1–suppressed cells identified through microarray analysis. (A) Venn diagram of differentially expressed probes selected by both Limma and dChip analyses. The Affymetrix GeneChip Human Genome U133 Plus 2.0 Array screened 54 330 probe sets using the threshold values of a fold change of 2 or higher and 2 or lower and a P value of .01 or less for differential expression. Initial Limma analysis selected 283 differentially expressed probe sets, which was further refined to 33 with the BH P value adjustment. Importantly, the 119 probes sets identified by dChip analysis completely overlapped the initial Limma results. (B) Volcano plot of differentially expressed probes selected by Limma analysis (red represents up-regulated; green, down-regulated; and +, after BH adjustment). (C) Heat map of differentially expressed genes from the refined Limma (BH) analysis. Up-regulated (red) and down-regulated (blue) genes are defined by comparing the pooled gene expression of Hut 78/sh4 bulk and Hut 78/sh4 clone 1 samples with Hut 78 and Hut 78 RPG samples.
Quantitative RT-PCR validation of microarray data in human CTCL cells with suppression of AHI-1
Differentially expressed genes identified by both the Limma BH (27 genes) and dChip (95 genes) analyses were carefully reviewed to select genes with connections to SS, CTCL, or other cancers; roles in cell cycle, proliferation, or apoptosis; or specificity to hematopoietic cells and particularly T cells, and the most significantly up and down-regulated genes. In addition, because AHI-1 has several domains involved in protein interactions, genes with complementary structural domains were also of interest. After evaluation, 15 genes up-regulated and 6 genes down-regulated in AHI-1–suppressed Hut 78/sh4 cells, compared with control Hut 78 cells, were selected for validation by quantitative RT-PCR (Table 1). Up- and down-regulated genes were defined by their average pooled expression levels in Hut 78/sh4 bulk and Hut 78/sh4 clone 1 cells compared with their average pooled expression levels in Hut 78 and Hut 78 RPG cells. Quantitative RT-PCR validation was performed on 4 technical replicates using the same RNA samples provided for microarray analysis. First, suppression of AHI-1 was verified using N-terminal and C-terminal primer sets with P values of 1.01 × 10−5 and 2.84 × 10−6, respectively. Next, the 21 selected genes were quantified. Differential expression of the 21 genes was validated, with P less than .05, ranging from 1.97 × 10−10 to 6.55 × 10−3 (Table 1). The only gene with a P value greater than .05 was BRDG1 at 5.88 × 10−2. Interestingly, functional grouping of the differentially expressed genes in AHI-1–suppressed cells showed several involved in signal transduction (BRDG1, HCK, and REPS2), cell-cycle control (CCNG2, CDKN1C, and PDCD6), cell proliferation and differentiation (BIN1 and MLLT11), and mRNA stability (ELAVL1). In addition, our observed deregulated expression of NKG7, HCK, and CDKN1C has recently been documented in both SS and MF patient samples.8,20,21 Furthermore, candidate tumor suppressor genes (BIN1 and CDKN1C) were found to be highly elevated in AHI-1–suppressed cells.
Quantitative RT-PCR validation of the 21 differentially expressed genes identified by microarray
| Gene . | Microarray, fold change . | P . | Quantitative RT-PCR, fold change . | P . |
|---|---|---|---|---|
| Down-regulated | ||||
| AHI-1 | 0.44 | ≤ .01 | 0.27 | 2.84 × 10−6 |
| PALM2-AKAP2 | 0.12 | ≤ .01 | 0.03 | 3.92 × 10−3 |
| SV2B | 0.33 | ≤ .01 | 0.14 | 6.45 × 10−3 |
| CMTM7 | 0.34 | ≤ .01 | 0.18 | 2.06 × 10−3 |
| PDCD6 | 0.38 | ≤ .01 | 0.41 | 1.51 × 10−5 |
| OXCT1 | 0.44 | ≤ .01 | 0.27 | 7.11 × 10−4 |
| ELAVL1 | 0.48 | ≤ .01 | 0.29 | 6.59 × 10−5 |
| Up-regulated | ||||
| IL1RN* | 10.78 | ≤ .01 | 4.97 | 2.27 × 10−4 |
| CDKN1C* | 5.31 | ≤ .01 | 3.55 | 6.70 × 10−4 |
| LMCD1 | 5.09 | ≤ .01 | 3.23 | 3.25 × 10−6 |
| LAPTM5 | 4.64 | ≤ .01 | 3.22 | 6.73 × 10−6 |
| MLLT11 | 4.41 | ≤ .01 | 3.33 | 1.97 × 10−10 |
| NKG7 | 4.30 | ≤ .01 | 2.63 | 1.12 × 10−5 |
| GRHL1 | 4.11 | ≤ .01 | 1.98 | 4.20 × 10−4 |
| HCK* | 3.92 | ≤ .01 | 1.63 | 1.84 × 10−3 |
| SPIB | 2.79 | ≤ .01 | 1.87 | 2.45 × 10−5 |
| CCNG2* | 2.73 | ≤ .01 | 1.44 | 6.55 × 10−3 |
| IL4I1 | 2.69 | ≤ .01 | 1.90 | 6.41 × 10−5 |
| BIN1 | 2.59 | ≤ .01 | 1.82 | 1.17 × 10−3 |
| BRDG1 | 2.52 | ≤ .01 | 1.37 | 5.81 × 10−2 |
| REPS2 | 2.46 | ≤ .01 | 6.65 | 3.51 × 10−4 |
| BCAS4 | 2.24 | ≤ .01 | 1.94 | 1.52 × 10−5 |
| Gene . | Microarray, fold change . | P . | Quantitative RT-PCR, fold change . | P . |
|---|---|---|---|---|
| Down-regulated | ||||
| AHI-1 | 0.44 | ≤ .01 | 0.27 | 2.84 × 10−6 |
| PALM2-AKAP2 | 0.12 | ≤ .01 | 0.03 | 3.92 × 10−3 |
| SV2B | 0.33 | ≤ .01 | 0.14 | 6.45 × 10−3 |
| CMTM7 | 0.34 | ≤ .01 | 0.18 | 2.06 × 10−3 |
| PDCD6 | 0.38 | ≤ .01 | 0.41 | 1.51 × 10−5 |
| OXCT1 | 0.44 | ≤ .01 | 0.27 | 7.11 × 10−4 |
| ELAVL1 | 0.48 | ≤ .01 | 0.29 | 6.59 × 10−5 |
| Up-regulated | ||||
| IL1RN* | 10.78 | ≤ .01 | 4.97 | 2.27 × 10−4 |
| CDKN1C* | 5.31 | ≤ .01 | 3.55 | 6.70 × 10−4 |
| LMCD1 | 5.09 | ≤ .01 | 3.23 | 3.25 × 10−6 |
| LAPTM5 | 4.64 | ≤ .01 | 3.22 | 6.73 × 10−6 |
| MLLT11 | 4.41 | ≤ .01 | 3.33 | 1.97 × 10−10 |
| NKG7 | 4.30 | ≤ .01 | 2.63 | 1.12 × 10−5 |
| GRHL1 | 4.11 | ≤ .01 | 1.98 | 4.20 × 10−4 |
| HCK* | 3.92 | ≤ .01 | 1.63 | 1.84 × 10−3 |
| SPIB | 2.79 | ≤ .01 | 1.87 | 2.45 × 10−5 |
| CCNG2* | 2.73 | ≤ .01 | 1.44 | 6.55 × 10−3 |
| IL4I1 | 2.69 | ≤ .01 | 1.90 | 6.41 × 10−5 |
| BIN1 | 2.59 | ≤ .01 | 1.82 | 1.17 × 10−3 |
| BRDG1 | 2.52 | ≤ .01 | 1.37 | 5.81 × 10−2 |
| REPS2 | 2.46 | ≤ .01 | 6.65 | 3.51 × 10−4 |
| BCAS4 | 2.24 | ≤ .01 | 1.94 | 1.52 × 10−5 |
Genes identified in dChip but not in refined Limma BH analysis.
Several genes were selected for comprehensive validation in 2 additional biologic replicates of Hut 78 and Hut 78–transduced cells. In each biologic replicate, defined by an independent RNA isolation, quantitative RT-PCR expression analysis began again with verification of AHI-1 suppression, followed by expression analyses of HCK, BIN1, CDKN1C, ELAVL1, PDCD6, and REPS2. Differential expression of these genes was confirmed in biologic replicates (data not shown).
Correlations of candidate differentially expressed genes in Sezary cell–enriched population in primary Sezary syndrome samples
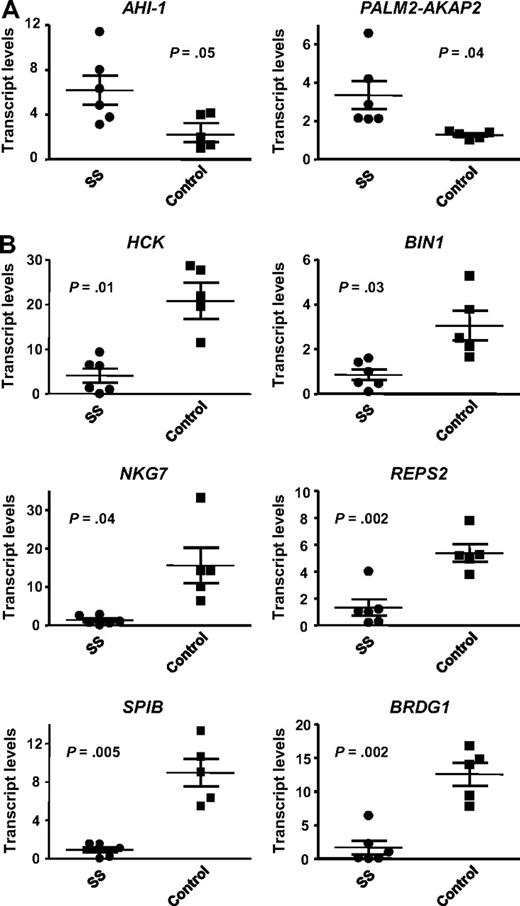
To determine whether the observed expression changes in Hut 78 cell lines held clinical relevance, several differentially expressed genes were screened in primary SS samples. As shown in Table S1, the 6 patient sample studied all had typical SS presentations with erythroderma, lymphadenopathy, leukemic Sezary cells in the peripheral blood ( > 5%-75%), and high degree of phenotypic loss of CD7 (68%-94%) and CD26 expression. Gene selection was based on previous documented links to SS and promising results in a recent microarray analysis of CD4+CD7− Sezary cell–enriched population from these SS patients (Y. Zhou, unpublished results July 2008). Quantitative RT-PCR analysis was performed to evaluate the expression of AHI-1, PALM2-AKAP2, HCK, BIN1, NKG7, REPS2, SPIB, BRDG1, and ELAVL1 in CD4+CD7− cells isolated from the 6 SS patient samples compared with 5 normal controls. Genes that were found to be down-regulated in AHI-1–suppressed Hut 78/sh4 cell lines were expected to be up-regulated in SS patients compared with controls, and vice versa. All the genes, with the exception of ELAVL1 (data not shown), demonstrated up- or down-regulation, which corresponded to the differential expression observed in the Hut 78 cell lines (Figure 2). Finding of expression correlations between primary SS samples and AHI-1–suppressed Hut 78 cell lines suggests that the identified differentially expressed genes may be important in the pathogenesis of SS.
Quantitative RT-PCR validation of 8 differentially expressed genes in CD4+CD7− Sezary cells from SS patients. The expression of several genes identified in the microarray comparing AHI-1–suppressed Hut 78/sh4 cells with Hut 78 control cells were evaluated by quantitative RT-PCR in both CD4+CD7− primary leukemic cells from 6 SS patients and 5 CD4+ normal controls. (A) Validation of up-regulated genes in primary SS samples compared with normal controls: AHI-1 and PALM2-AKAP2. (B) Validation of down-regulated genes in primary SS samples compared with normal controls: HCK, BIN1, NKG7, REPS2, SPIB, and BRDG1. Values shown are the mean ± SEM. The P values presented are the result of 2-sample t tests comparing the SS patient expression data with that of normal controls.
Quantitative RT-PCR validation of 8 differentially expressed genes in CD4+CD7− Sezary cells from SS patients. The expression of several genes identified in the microarray comparing AHI-1–suppressed Hut 78/sh4 cells with Hut 78 control cells were evaluated by quantitative RT-PCR in both CD4+CD7− primary leukemic cells from 6 SS patients and 5 CD4+ normal controls. (A) Validation of up-regulated genes in primary SS samples compared with normal controls: AHI-1 and PALM2-AKAP2. (B) Validation of down-regulated genes in primary SS samples compared with normal controls: HCK, BIN1, NKG7, REPS2, SPIB, and BRDG1. Values shown are the mean ± SEM. The P values presented are the result of 2-sample t tests comparing the SS patient expression data with that of normal controls.
Identification of differential protein expression and phosphorylation of HCK in AHI-1–suppressed and –overexpressed CTCL cells
We next assessed whether changes observed in mRNA translated to the protein level, which could imply a biologic or functional connection to AHI-1. Eight genes were selected based on differential expression in primary SS patient samples and transformed CTCL cells, intriguing function and structure, convincing biologic connections, and antibody availability. An additional motive for selection of IL1RN and PALM2-AKAP2 was that they were the most significantly up- and down-regulated genes, respectively, that resulted from AHI-1 suppression. Conscious of the fact that correlations between mRNA and protein expression are not inherently strong, it was not completely surprising to next discover that only 2 of the 8 selected genes, HCK and BIN1, demonstrated differential protein expression by Western blot analysis. There were no changes detected in CDKN1C, ELAVL1, IL1RN, PALM2-AKAP2, PDCD6, and REPS2 protein expression (data not shown).
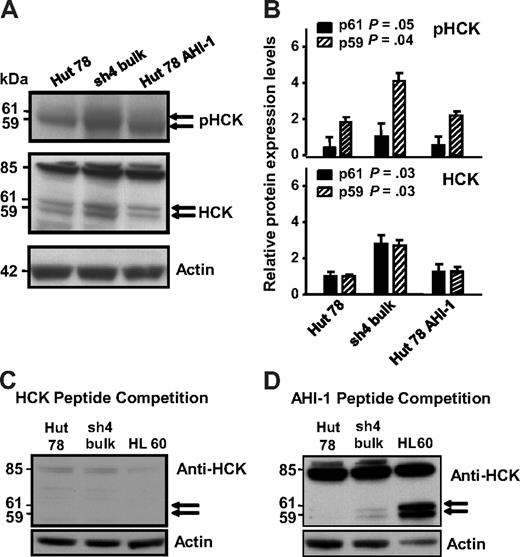
HCK, a Src family tyrosine kinase, showed significant up-regulation in AHI-1–suppressed cells compared with control cells, correlating with earlier mRNA expression data (Figure 3A middle panel, lane 1 compared with lane 2, and 3B). Two isoforms of HCK have been reported to be generated by alternative translation: HCK (p59HCK) and HCK (p61HCK).22 Distinct up-regulation of HCK was observed in a double band at approximately 60 kDa, indicating that these 2 isoforms became up-regulated in AHI-1–suppressed cells (P = .03 for both p59 and p61; Figure 3B). In addition, HCK competition experiments with primary antibody confirmed the observed expression to be specific to HCK (Figure 3C,D). There was also a strong 85-kDa band evident in the both Hut 78 cells and in a positive control HL-60 cell line. Although this band has not been previously described, HCK peptide competition experiments confirmed that this band was specific to the HCK antibody (Figure 3C, compared with a control peptide competition in Figure 3D). Interestingly, phosphorylation of HCK also increased in AHI-1–suppressed cells compared with control cells (Figure 3A top panel).
Differential protein expression and phosphorylation of HCK in AHI-1–suppressed and –overexpressed CTCL cells. (A) Up-regulation of 2 isoforms of HCK (p61 and p59) was evident in AHI-1–suppressed Hut 78/sh4 cells compared with the control Hut 78 cells. Reduced HCK protein expression and phosphorylation were observed when AHI-1 was overexpressed in Hut 78 cells. In addition, an uncharacterized 85-kDa band was also observed. (B) Quantification of the differential expression and phosphorylation of HCK for both the p61HCK and p59HCK isoforms relative to actin. (C) Peptide competition experiment in which the HCK primary antibody was incubated with 5-fold of its cognate epitope before use in the Western blot. Dissipation of the doublet bands (p61 and p59) and the uncharacterized 85-kDa band indicated that these results were specific for the HCK antibody. (D) Additional peptide competition experiment in which the HCK antibody was incubated with 5-fold of a random peptide (AHI-1) to show that the previous competition experiment was a direct result of saturation of the HCK antibody. Values shown are the mean ± SEM. The P values presented are the result of 2-sample t tests comparing the expression in cells with AHI-1 suppression with that of Hut 78 controls.
Differential protein expression and phosphorylation of HCK in AHI-1–suppressed and –overexpressed CTCL cells. (A) Up-regulation of 2 isoforms of HCK (p61 and p59) was evident in AHI-1–suppressed Hut 78/sh4 cells compared with the control Hut 78 cells. Reduced HCK protein expression and phosphorylation were observed when AHI-1 was overexpressed in Hut 78 cells. In addition, an uncharacterized 85-kDa band was also observed. (B) Quantification of the differential expression and phosphorylation of HCK for both the p61HCK and p59HCK isoforms relative to actin. (C) Peptide competition experiment in which the HCK primary antibody was incubated with 5-fold of its cognate epitope before use in the Western blot. Dissipation of the doublet bands (p61 and p59) and the uncharacterized 85-kDa band indicated that these results were specific for the HCK antibody. (D) Additional peptide competition experiment in which the HCK antibody was incubated with 5-fold of a random peptide (AHI-1) to show that the previous competition experiment was a direct result of saturation of the HCK antibody. Values shown are the mean ± SEM. The P values presented are the result of 2-sample t tests comparing the expression in cells with AHI-1 suppression with that of Hut 78 controls.
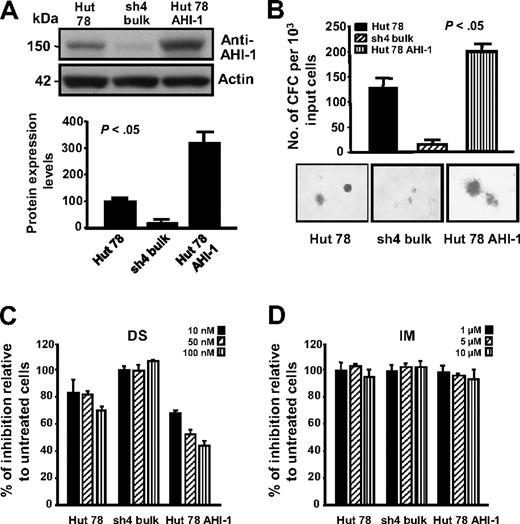
To further investigate specific cooperative roles of AHI-1 and HCK in CTCL cell transformation, overexpression of AHI-1 by transduction of EF1α-AHI-1-IRES-YFP lentivirus in Hut 78 cells was performed and confirmed (Figure 4A). Interestingly, changes in phosphorylation levels of HCK were also observed in AHI-1–suppressed and –overexpressed Hut 78 cells (Figure 3A top panel). Strikingly, overexpression of human AHI-1 in Hut 78 cells resulted in sharply increased growth factor (GF) independence in semisolid cultures, producing more CFCs compared with control cells (∼ 2-fold; P < .05; Figure 4B). In contrast, knockdown of AHI-1 expression (sh4 cells) reduced GF-independent colony-forming ability as we previously demonstrated (Figure 4B).11 In addition, AHI-1–overexpressed cells showed a significant reduction in CFC output in a dose-dependent fashion in response to DS,23 a small molecular inhibitor of Src family tyrosine kinases (Figure 4C). Slightly increased CFC numbers were observed in AHI-1–suppressed cells in response to DS. Of note, control Hut 78 cells also responded to DS treatment, suggesting that HCK itself may play a role in CTCL cell transformation. This is further supported by treatment of the same cells with IM,24 a specific inhibitor for ABL and c-kit tyrosine kinases, where no changes in CFC output were observed (Figure 4D).
Knockdown and overexpression of AHI-1 in CTCL cells mediate their GF-independent colony-forming ability and their response to Src tyrosine kinase inhibitor, DS. (A) Western analysis of AHI-1 protein expression in control Hut 78 cells, AHI-1–suppressed cells (sh4 bulk), and AHI-1–overexpressed cells (Hut 78 AHI-1). The bottom panel shows quantification of AHI-1 protein expression relative to actin. (B) The number of GF-independent CFC colonies produced and their appearance in semisolid cultures in the same transduced cells. (C,D) CFCs produced in semisolid cultures in response to DS and IM. Values shown are the mean ± SEM. The P values presented are the result of 2-sample t tests comparing the results in cells with AHI-1 suppression and overexpression with that of Hut 78 controls.
Knockdown and overexpression of AHI-1 in CTCL cells mediate their GF-independent colony-forming ability and their response to Src tyrosine kinase inhibitor, DS. (A) Western analysis of AHI-1 protein expression in control Hut 78 cells, AHI-1–suppressed cells (sh4 bulk), and AHI-1–overexpressed cells (Hut 78 AHI-1). The bottom panel shows quantification of AHI-1 protein expression relative to actin. (B) The number of GF-independent CFC colonies produced and their appearance in semisolid cultures in the same transduced cells. (C,D) CFCs produced in semisolid cultures in response to DS and IM. Values shown are the mean ± SEM. The P values presented are the result of 2-sample t tests comparing the results in cells with AHI-1 suppression and overexpression with that of Hut 78 controls.
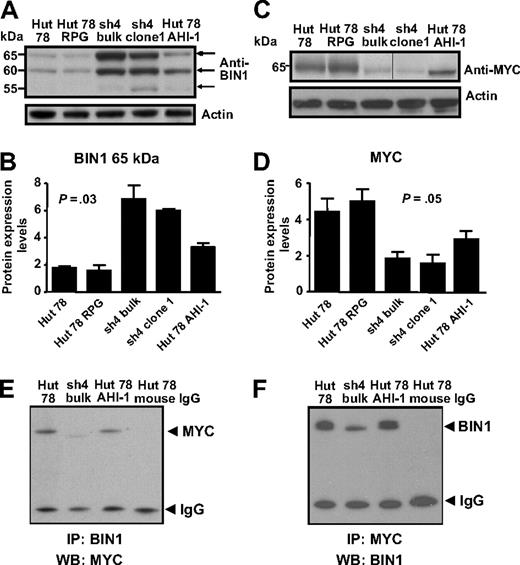
Identification of differential protein expression of BIN in AHI-1–suppressed and –overexpressed CTCL cells and its physical interaction with MYC
BIN1, a tumor suppressor gene, also showed up-regulation in AHI-1–suppressed cells compared with control cells (Figure 5A,B). Again, this correlated well with the previous mRNA expression data. Of the 3 isoforms observed (65, 60, and 55 kDa), up-regulation was most evident at 65 and 60 kDa. Interestingly, BIN1 expression was significantly reduced when AHI-1 was overexpressed in Hut 78 cells (Figure 5A,B), indicating that mediating AHI-1 expression by suppression or overexpression of AHI-1 changes BIN1 expression levels in CTCL cells.
Differential protein expression of BIN1 and MYC in AHI-1–suppressed and –overexpressed CTCL cells. (A) Up-regulation of BIN1 was evident in AHI-1–suppressed Hut 78/sh4 cell lines. Of the 3 isoforms detected at 65, 60, and 55 kDa, the most significant up-regulation was observed in the 65- and 60-kDa isoforms. Reduced BIN1 protein expression was also observed when AHI-1 was overexpressed in Hut 78 cells (Hut 78 AHI-1). (B) Quantification of the differential expression of the 65-kDa isoform of BIN1 relative to actin. (C) Changes in MYC expression were observed in AHI-1–suppressed Hut 78/sh4 cell lines and AHI-1–overexpressed cells compared with the control cells (Hut 78 and Hut 78 RPG). Vertical line has been inserted to indicate a repositioned gel lane. (D) Quantification of the differential expression of MYC relative to actin. (E,F) Protein lysates from control Hut 78 cells, AHI-1–suppressed cells (sh4 bulk), and AHI-1–overexpressed cells were used to immunoprecipitate BIN1 (E) and MYC (F), and proteins were detected by Western blotting with antibodies to MYC (E) and BIN1 (F) as indicated. Values shown are the mean plus or minus SEM. The P values presented are the result of 2-sample t tests comparing the AHI-1–suppressed expression data with that of Hut 78 controls.
Differential protein expression of BIN1 and MYC in AHI-1–suppressed and –overexpressed CTCL cells. (A) Up-regulation of BIN1 was evident in AHI-1–suppressed Hut 78/sh4 cell lines. Of the 3 isoforms detected at 65, 60, and 55 kDa, the most significant up-regulation was observed in the 65- and 60-kDa isoforms. Reduced BIN1 protein expression was also observed when AHI-1 was overexpressed in Hut 78 cells (Hut 78 AHI-1). (B) Quantification of the differential expression of the 65-kDa isoform of BIN1 relative to actin. (C) Changes in MYC expression were observed in AHI-1–suppressed Hut 78/sh4 cell lines and AHI-1–overexpressed cells compared with the control cells (Hut 78 and Hut 78 RPG). Vertical line has been inserted to indicate a repositioned gel lane. (D) Quantification of the differential expression of MYC relative to actin. (E,F) Protein lysates from control Hut 78 cells, AHI-1–suppressed cells (sh4 bulk), and AHI-1–overexpressed cells were used to immunoprecipitate BIN1 (E) and MYC (F), and proteins were detected by Western blotting with antibodies to MYC (E) and BIN1 (F) as indicated. Values shown are the mean plus or minus SEM. The P values presented are the result of 2-sample t tests comparing the AHI-1–suppressed expression data with that of Hut 78 controls.
Because BIN1 is known to interact with MYC to inhibit malignant cell transformation,25-27 MYC was also investigated in CTCL cells, although no significant changes in its mRNA expression appeared in the microarray results. However, noticeable down-regulation and up-regulation of MYC protein were observed in the AHI-1–suppressed and –overexpressed cells, and the connection to BIN1 suggests that this could be of functional significance (Figure 5C,D).
To determine whether a physical interaction between BIN1 and MYC occurs in human CTCL cells, coimmunoprecipitation experiments were performed. Direct interaction between BIN1 and MYC at endogenous levels was detected by a specific MYC antibody in transduced Hut 78 cells after immunoprecipitation with a BIN1 antibody, with a reduced band of MYC in AHI-1–suppressed cells (Figure 5E). This interaction was not found in control antibody-coimmunoprecipitated Hut 78 cells. This was confirmed by detection of BIN1 in the same cells after IP with a specific antibody to MYC (Figure 5F).
Characterization of BIN1 isoforms in primary and transformed CTCL cells
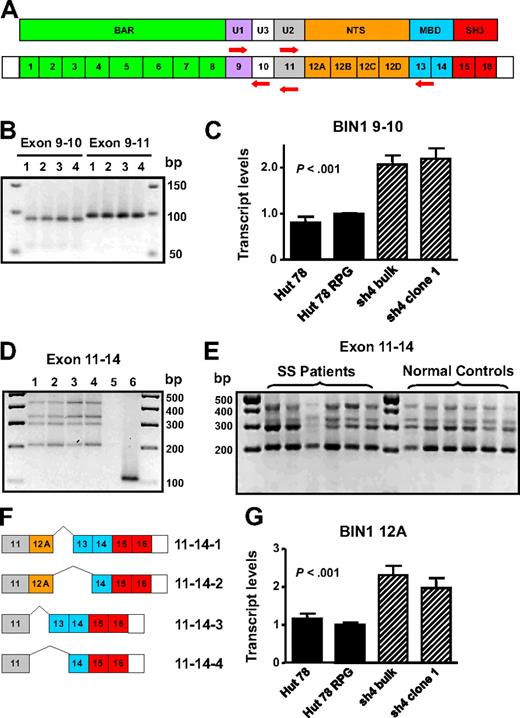
Detection of BIN1 up-regulation and down-regulation as a result of AHI-1 suppression and overexpression was intriguing because certain isoforms of this gene have been described to have tumor suppressor properties.25,28,29 In addition, BIN1 attenuation is frequently described in several cancers,30-37 and aberrant splicing of BIN1 yields isoforms unable to bind MYC to inhibit malignant transformation in melanoma cells and primary neuroblastoma cells.27,30,35 Therefore, characterization of the BIN1 isoforms in Hut 78-transduced cells was performed to illuminate how AHI-1 expression may mediate expression changes and possibly generate aberrantly spliced isoforms of BIN1. Based on previously described isoforms, the regions of the gene susceptible to differential splicing are exons 10, 12A-D, and 13.28,30,38 Therefore, RT-PCR exon-specific primers for these segments of BIN1 were designed to elucidate the presence or absence of these exons (Figure 6A).
Characterization of BIN1 isoforms in primary and CTCL cells. (A) Schematic diagram of protein domains and exon organization of BIN1 and the specific primers sets generated to detect exons 10, 12A-D, and 13: BIN1 9/10, BIN1 9/11, and BIN1 11/14. BAR indicates BIN1/Amphiphysin/RVS167-related; U, unique; NTS, neural tissue specific; MBD, MYC-binding domain; and SH3, Src homology 3. (B) Yield of both BIN1 9/10 and BIN1 9/11 PCR products by RT-PCR indicates differential splicing of exon 10. Lane 1 indicates Hut 78; lane 2, Hut 78 RPG; lane 3, Hut 78/sh4 bulk; lane 4, Hut 78/sh4 clone 1. (C) Quantitative RT-PCR using the BIN1 9/10 primer set demonstrated up-regulation of BIN1 transcripts containing exon 10 in AHI-1–suppressed cells compared with the control Hut 78 cells. (D) Four distinct RT-PCR products were identified and cloned in Hut 78 cell lines using BIN1 11/14 primers. Lane 1 indicates Hut 78; lane 2, Hut 78 RPG; lane 3, Hut 78/sh4 bulk; lane 4, Hut 78/sh4 clone 1; lane 5, negative control (no RNA); lane 6, positive control (Hut 78 RPG with GAPDH primer set). (E) RT-PCR revealed that the 4 BIN1 11/14 RT-PCR products identified in the Hut 78-transduced cells were also present in both CD4+CD7− SS patients samples and in normal controls. (F) Schematic diagram of the differential splicing of exons 12A and 13 characterized in both the Hut 78 cell lines and primary samples. (G) Quantitative RT-PCR using a BIN1 12A primer set demonstrated up-regulation of BIN1 transcripts containing exon 12A in AHI-1–suppressed cells compared with the control Hut 78 cells. Values shown are the mean plus or minus SEM. The P values presented are the result of 2-sample t tests comparing the AHI-1–suppressed expression data to that of Hut 78 controls.
Characterization of BIN1 isoforms in primary and CTCL cells. (A) Schematic diagram of protein domains and exon organization of BIN1 and the specific primers sets generated to detect exons 10, 12A-D, and 13: BIN1 9/10, BIN1 9/11, and BIN1 11/14. BAR indicates BIN1/Amphiphysin/RVS167-related; U, unique; NTS, neural tissue specific; MBD, MYC-binding domain; and SH3, Src homology 3. (B) Yield of both BIN1 9/10 and BIN1 9/11 PCR products by RT-PCR indicates differential splicing of exon 10. Lane 1 indicates Hut 78; lane 2, Hut 78 RPG; lane 3, Hut 78/sh4 bulk; lane 4, Hut 78/sh4 clone 1. (C) Quantitative RT-PCR using the BIN1 9/10 primer set demonstrated up-regulation of BIN1 transcripts containing exon 10 in AHI-1–suppressed cells compared with the control Hut 78 cells. (D) Four distinct RT-PCR products were identified and cloned in Hut 78 cell lines using BIN1 11/14 primers. Lane 1 indicates Hut 78; lane 2, Hut 78 RPG; lane 3, Hut 78/sh4 bulk; lane 4, Hut 78/sh4 clone 1; lane 5, negative control (no RNA); lane 6, positive control (Hut 78 RPG with GAPDH primer set). (E) RT-PCR revealed that the 4 BIN1 11/14 RT-PCR products identified in the Hut 78-transduced cells were also present in both CD4+CD7− SS patients samples and in normal controls. (F) Schematic diagram of the differential splicing of exons 12A and 13 characterized in both the Hut 78 cell lines and primary samples. (G) Quantitative RT-PCR using a BIN1 12A primer set demonstrated up-regulation of BIN1 transcripts containing exon 12A in AHI-1–suppressed cells compared with the control Hut 78 cells. Values shown are the mean plus or minus SEM. The P values presented are the result of 2-sample t tests comparing the AHI-1–suppressed expression data to that of Hut 78 controls.
To detect exon 10, reportedly restricted to muscle tissue, quantitative RT-PCR analysis was performed.28 PCR products generated from BIN1 9/10 and BIN1 9/11 primer sets suggested that this exon was differentially spliced and that transcripts containing exon 10 were rare (Figure 6B). Cloning and sequencing of several PCR products confirmed differential splicing of exon 10 in these cells. Quantitative RT-PCR analysis also demonstrated up-regulation of BIN1 transcripts containing exon 10 in AHI-1–suppressed cells compared with Hut 78 controls (Figure 6C). To detect exons 12A-D and 13, RT-PCR was performed with the BIN1 11/14 primer set. Results revealed 4 distinct RNA isoforms, which were cloned and sequenced to confirm alternative splicing of exons 12A and 13 (Figure 6D,F). Interestingly, in both CD4+CD7− cells from SS patient samples and similarly isolated cells from normal controls, these 4 isoforms were also present, in addition to transcripts with exon 10 (Figure 6E; and data not shown). Quantitative RT-PCR analysis using a primer set specific for BIN1 exon 12A demonstrated up-regulation of transcripts containing this exon in AHI-1–suppressed cells compared with control cells (Figure 6G).
Discussion
Microarray analysis identified several differentially expressed genes in AHI-1–suppressed Hut 78/sh4 cells compared with control Hut 78 cells. These results were validated by quantitative RT-PCR, and several genes were found to have clinical relevance in SS patient samples, suggesting that these genes with a connection to AHI-1 may be involved the pathogenesis of SS. Further investigation of 8 genes revealed that only 2, HCK and BIN1, demonstrated differential expression at the protein level, a typical expression discrepancy.39-41 For example, investigations in yeast revealed that correlations between protein and mRNA were insufficient for prediction of protein expression from quantitative mRNA data, with projected correlation for all yeast proteins at less than 0.4.39 This expression discordance demonstrates that, although many genes are under transcriptional control, several are more tightly regulated by translational control and/or posttranslational modifications.
HCK was found to be up-regulated in AHI-1–suppressed cells at mRNA and protein levels, including up-regulation of its phosphorylation level. As a member of the Src family tyrosine kinases, HCK has a kinase domain, an SH2 domain, and an SH3 domain.42 Alternative translation of the transcript generates 2 isoforms, p59HCK and p61HCK, both up-regulated in AHI-1–suppressed cells.22 Expression of HCK is reportedly restricted to hematopoietic cells with predominant expression in myeloid lineage cells and B lymphocytes.43,44 Commonly, constitutive activation of Src family kinases contributes to transformation of a cell, and this oncogenic potential of HCK has been demonstrated in Philadelphia chromosome-positive (Ph+) leukemias and lymphomas.42,45 However, there is also evidence of potential tumor suppressor properties for HCK in Ph− leukemias and other cancers.46 Recently, aberrant methylation of the CpG island in the HCK promoter was detected in 13 of 23 hematopoietic and 8 of 10 nonhematopoietic cancer cell lines but not in normal controls.46 There are also 2 independent reports that mouse Hck can trigger caspase-mediated apoptosis, supporting the tumor suppressor hypothesis.44,47 Because changes in HCK protein and phosphorylation were observed in the AHI-1–suppressed and –overexpressed cells, HCK could be a critical player in a signaling cascade in CTCL cells to regulate caspase-mediated apoptosis. Alternatively, AHI-1 may directly affect HCK expression by regulation of specific genes involved in its specific transcription and translational control, or instead may modulate methylation of the HCK promoter. Interestingly, suppression of activities of Src family kinases, including HCK activity, by DS resulted in reduced or increased GF-independent growth of AHI-1–overexpressed or –suppressed cells in a dose-dependent fashion (Figure 4C), whereas no changes were triggered in the same cells by IM, a non-Src inhibitor. Although it is not clear whether DS may also inhibit tyrosine kinase activity of other members of Src family that may contribute to biologic changes observed in AHI-1–overexpressed or –suppressed cells, these results strongly suggest that HCK may be an important signaling molecule and potential target in AHI-1–mediated CTCL cell transformation.
BIN1 expression was also up-regulated or down-regulated in AHI-1–suppressed or –overexpressed cells, respectively. This protein contains an N-terminal BIN1/Amphiphysin/RVS167–related domain important in nuclear localization, an internal neural tissue-specific domain (NTS) thought to have endocytic functions in neural isoforms, a MYC-binding domain, and a C-terminal SH3 domain (Figure 6A).26-28 As a nucleocytosolic adapter protein, elucidation of the specific function of BIN1 is challenging because there are more than 10 splice isoforms, each with unique roles, specific cellular localization, and tissue specificity.28,34 Nuclear isoforms of BIN1 act as tumor suppressors, physically interacting with and inhibiting the transforming activity of MYC.25,26 BIN1 also inhibits cell proliferation and activates cell death processes through both MYC-dependent and MYC-independent mechanisms.26,48,49 In addition, both loss of heterozygosity and attenuated BIN1 expression have been described in breast cancer, prostate cancer, and malignant melanoma.35-37,48 Moreover, aberrant splicing of an NTS exon (12A) in BIN1 transcripts has been described in both primary neuroblastoma cells and melanoma cell lines.30,35 Inclusion of the 12A exon has recently been shown to abolish the ability of BIN1 to bind MYC.27
Characterization of BIN1 isoforms in hematopoietic cells and T cells has not been previously described, and our results in Hut 78 cells indicate that BIN1 splice isoforms tightly regulate the inclusion of exon 10, a rare transcript, and exhibit alternative splicing of exons 12A and 13. The unusual presence of exon 10, thought to be a muscle-specific exon, may indicate a general function in hematopoietic cells, or more specifically in T cells, or a role in leukemic cells because exon 10 up-regulation was observed in AHI-1–suppressed cells compared with the control cells (Figure 6C). Importantly, 2 of the 4 BIN1 isoforms characterized included exon 12A, which is specifically associated with MYC interaction.26,30 Of note, although MYC did not demonstrate any significant differential mRNA expression in the microarray analysis, down-regulation of the protein was observed in AHI-1–suppressed cells (Figure 5C); this can be mediated when AHI-1 is overexpressed in Hut 78 cells.
Because up-regulation of BIN1 is observed in AHI-1–suppressed cells and down-regulation of BIN1 is found in SS patient samples (Figures 2, 5), AHI-1 may directly or indirectly inhibit expression of BIN1 to enhance its oncogenic transformation, possibly through MYC. This could occur by inhibiting transcription of BIN1 or manipulating genes involved in both its mRNA and/or protein stability. Alternatively, AHI-1 may alter splicing of BIN1 to yield perturbed expression of isoforms, which incorrectly express exon 12A to abolish MYC binding and tumor suppressor properties. Recently, the SF2/ASF splicing factor was reported to manipulate splicing regulation and increase inclusion of exon 12A in BIN1 transcripts.50 Although the expression patterns and localization of these specific isoforms in Hut 78 cells with and without AHI-1 suppression need to be further elucidated, these results strongly suggest that AHI-1 may cooperate with BIN1 in the loss of its tumor suppressor activity through its interacting oncoprotein MYC to mediate cellular proliferation and apoptosis control of human CTCL cells. Indeed, we have now demonstrated a direct physical interaction between BIN1 and MYC at endogenous levels in transduced CTCL cells, supporting a cooperative role of AHI-1 and BIN1-MYC interaction complex in CTCL cell transformation.
In conclusion, microarray analysis has identified several differentially expressed genes involved in AHI-1–mediated leukemic transformation in human CTCL cells. In particular, HCK and BIN1 have emerged as new candidate cooperative genes that could be biologically connected to AHI-1 in CTCL. Importantly, evaluation of HCK, BIN1, and other gene expression patterns in primary Sezary cell–enriched population (CD4+CD7− cells) from SS patients revealed correlating expression changes to the microarray results. These results indicate that the differentially expressed genes identified through AHI-1 suppression may have significance in SS and other human CTCLs and thus could help elucidate the pathogenesis of these diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank K. Saw and M. Hale for their excellent technical assistance and Novartis for imatinib and Bristol-Myers Squibb for dasatinib.
This work was supported by grants from the Leukemia & Lymphoma Society of Canada, the Galloway Leukemia Research Seed Funding from the University of British Columbia, and in part by the Cancer Research Society (X.J.). E.K. was a recipient of the Medical Genetics Graduate Entrance Scholarship from the University of British Columbia. A.R. was a recipient of a British Columbia Cancer Studentship. X.J. is a Michael Smith Foundation for Heath Research Scholar.
Authorship
Contribution: E.K., A.R., L.L.Z., and S.E. designed and performed the experiments and analyzed the data; H.Q. assisted in microarray data analysis; M.-w.S. and Y.Z. provided clinical data and RNA samples from patients with Sezary syndrome and performed and analyzed microarray data on the clinical Sezary cell samples; X.J. developed the concept of the study and designed and supervised the experiments; E.K., A.R., Y.Z., and X.J. wrote the manuscript, and all other authors commented on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiaoyan Jiang, Terry Fox Laboratory, British Columbia Cancer Agency, 675 West 10th Ave, Vancouver, BC V5Z 1L3, Canada; e-mail: xjiang@bccrc.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal