Abstract
The past decade has shown a marked increase in the use of high-throughput assays in clinical research into human cancer, including acute myeloid leukemia (AML). In particular, genome-wide gene expression profiling (GEP) using DNA microarrays has been extensively used for improved understanding of the diagnosis, prognosis, and pathobiology of this heterogeneous disease. This review discusses the progress that has been made, places the technologic limitations in perspective, and highlights promising future avenues
Introduction
Acute myeloid leukemia (AML) is characterized by a maturation block and accumulation of myeloid progenitor cells.1,2 Clinically, it has been recognized as a heterogeneous disorder.1,2 Laboratory support for that notion has come from various directions. Chromosomal abnormalities and gene mutations are common in AML, many of which are apparent in particular subtypes.3,4
Classification of AML subtypes is clinically relevant, as particular abnormalities are associated with distinct clinical behavior.1 For instance, recurring reciprocal translocations t(15;17)(q22;q21), t(8;21)(q22;q22), or inv(16)(p13q22)/t(16)16(p13;q22), further abbreviated as t(15;17), t(8;21), and inv(16), respectively, predict favorable prognosis, whereas other chromosomal aberrations are associated with inferior outcome.1 Likewise, sequence mutations in certain genes are associated with either favorable or unfavorable response to treatment.4
Insight into cytogenetic and genetic aberrations is invaluable for diagnosis, and it may also allow for better understanding of the pathobiology. Furthermore, it may enable the development and application of specific treatment modalities targeted to underlying oncogenic abnormalities. The efficacy of such drugs as all-trans retinoic acid for the treatment of t(15;17) AML and imatinib for BCR-ABL–positive chronic myeloid leukemia offer well established examples.5
Despite great progress, much of the heterogeneity of AML remains to be resolved. A significant proportion of human AML appears cytogenetically and genetically normal, which implies that the underlying molecular abnormalities are still unknown. Furthermore, it is likely that in AMLs carrying recognizable aberrations additional hits await to be uncovered, as one lesion usually does not appear sufficient for full leukemic transformation.6
In recent years, DNA microarrays, together with the availability of the complete nucleotide sequence of the human genome, have spurred the search for abnormalities in cancer, including AML.7 The accessibility of these tools allows assessment of abnormalities and variations on a genome wide basis, covering various molecular levels.8 Gene expression profiling (GEP) is one of these technologies, in which DNA microarrays containing cDNAs or oligonucleotide probes are used to simultaneously measure levels of many different mRNA transcripts.7-10 In an early landmark study in 1999, Golub and colleagues were able to use GEP to discriminate a collection of AML from acute lymphoblastic leukemia (ALL) specimens.11 Their study suggested 3 important potential applications of GEP: class discovery, class prediction and class comparison. Class discovery refers to the identification of new subgroups while for class prediction one uses gene expression data to predict already defined subgroups. These 2 applications therefore have diagnostic implications. The third proposed application, class comparison, refers to the identification of genes that are deregulated in certain subgroups, and may address biologic questions. Given these proposed possibilities, investigators have used DNA microarrays to investigate gene expression in clinical AML samples in the past years.12-14
Now, some 10 years later, we shall discuss the results of a decade of experience with GEP as regards diagnosis, prognosis and biology of AML in the context of clinical studies. Is it possible to identify new clinically relevant subgroups of AML using GEP? Does GEP deserve a place in clinical diagnosis? Does GEP allow prediction of prognosis? And is it possible to extract new insights into the pathobiology of AML from clinical GEP data?
Can GEP identify new subgroups of AML?
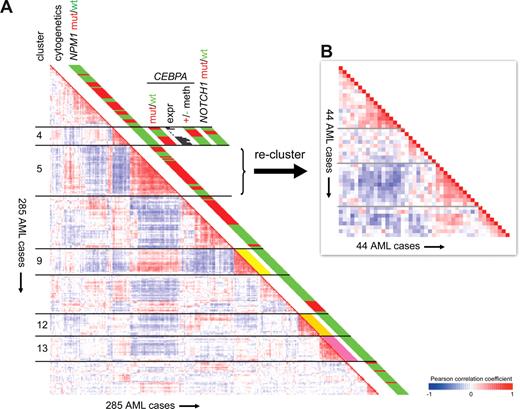
A straightforward way of using GEP is to compare expression profiles between cases of AML and to search for similarities and differences. In an unsupervised approach this is done in an unbiased way, ie without the use of external information such as mutations or karyotypic subtypes. This procedure is therefore representative of class discovery. The grouping of cases according to similar gene expression signatures is often referred to as clustering.15,16 The underlying assumption is that cases with the same gene expression profiles may carry the same genetic abnormality. Support for this hypothesis has come from observations, now confirmed by several research groups, that particular cytogenetic AML subtypes (eg, AMLs with t(8;21), t(15;17), and inv(16)) each share distinctive GEP profiles (Figure 1A).17-23 Likewise, mutations in CCAAT/enhancer binding protein alpha (CEBPA), and to a lesser extent nucleophosmin (NPM1) (Figure 1A), correlate with gene expression signatures that appear after unsupervised clustering.22,24
Summary of GEP findings in a cohort of 285 cases of AML. (A) A previous study of 285 cases of AML revealed 16 subgroups (clusters) of cases based on similarities in gene expression profiles.22 Pairwise correlations between these AML cases are shown on the left. The cells in the visualization are colored by Pearson correlation coefficient values, with deeper colors depicting higher positive (red) or negative (blue) correlations, as indicated by the scale bar. Five of the 16 clusters have been labeled as clusters 4, 5, 9, 12, and 13. One finding of the original study was the tight aggregation into distinct clusters of AML cases with cytogenetic abnormalities that predict good risk. For those cases, cytogenetic status is color-coded in the cytogenetics column: inv(16) is yellow, next to cluster 9; t(15;17) is orange, next to cluster 12; and t(8;21) is pink, next to cluster 13. A subsequent study in the same patient cohort identified NPM1 mutations in 95 of 285 cases. NPM1 mutational status is depicted next to each case (red indicates NPM1 mutant; green, NPM1 wild-type).24 The figure illustrates that NPM1 mutations were not randomly distributed over the 16 previously defined clusters, but enriched in several of them. Cluster 4 was found to associate with CEBPA mutations (red indicates CEBPA mutant). However, a subset of 6 patients in this cluster did not show any CEBPA mutation (green indicates CEBPA wild-type). It was found that these cases differed in their CEBPA mRNA expression as compared with the CEBPA mutant AMLs, as indicated by the histograms depicting signal intensity values for the CEBPA probe set on the microarray. In fact, whereas CEBPA mutant AMLs highly expressed CEBPA mRNA, expression was silenced in the cases lacking mutations. This silencing was associated with CEBPA DNA promoter hypermethylation (red indicates methylation; green, no methylation). In addition, NOTCH1 mutations were found as common characteristics of this subgroup (red indicates NOTCH1 mutation; green, NOTCH1 wild-type).31 (B) In the original analysis of 285 AML cases (panel A left), the 44 cases in cluster 5 aggregated very tightly, as indicated by the deep red colors representing positive Pearson correlation coefficients. Most of these 44 cases showed a monocytoid morphology (FAB-M4 or -M5).22 This raises the possibility that a significant part of the clustering effect was caused by specific up- or down-regulation of genes that are important in monocytic differentiation, resulting in a different signature than the remaining, mostly nonmonocytoid, cases of AML in the study. To answer whether gene expression profiling would enable identification of potential heterogeneity within this apparently homogeneous subgroup, in panel B the 44 cases were reclustered as an isolated cohort. For this analysis, only probe sets that showed a variable expression within these 44 AML cases were taken into account, as defined by a fold change of 3.5 to the mean in log2 scale in at least 1 case. The resulting cluster image shows that several potentially interesting subgroups can indeed be identified within these 44 AML cases, which have been indicated by gray lines.
Summary of GEP findings in a cohort of 285 cases of AML. (A) A previous study of 285 cases of AML revealed 16 subgroups (clusters) of cases based on similarities in gene expression profiles.22 Pairwise correlations between these AML cases are shown on the left. The cells in the visualization are colored by Pearson correlation coefficient values, with deeper colors depicting higher positive (red) or negative (blue) correlations, as indicated by the scale bar. Five of the 16 clusters have been labeled as clusters 4, 5, 9, 12, and 13. One finding of the original study was the tight aggregation into distinct clusters of AML cases with cytogenetic abnormalities that predict good risk. For those cases, cytogenetic status is color-coded in the cytogenetics column: inv(16) is yellow, next to cluster 9; t(15;17) is orange, next to cluster 12; and t(8;21) is pink, next to cluster 13. A subsequent study in the same patient cohort identified NPM1 mutations in 95 of 285 cases. NPM1 mutational status is depicted next to each case (red indicates NPM1 mutant; green, NPM1 wild-type).24 The figure illustrates that NPM1 mutations were not randomly distributed over the 16 previously defined clusters, but enriched in several of them. Cluster 4 was found to associate with CEBPA mutations (red indicates CEBPA mutant). However, a subset of 6 patients in this cluster did not show any CEBPA mutation (green indicates CEBPA wild-type). It was found that these cases differed in their CEBPA mRNA expression as compared with the CEBPA mutant AMLs, as indicated by the histograms depicting signal intensity values for the CEBPA probe set on the microarray. In fact, whereas CEBPA mutant AMLs highly expressed CEBPA mRNA, expression was silenced in the cases lacking mutations. This silencing was associated with CEBPA DNA promoter hypermethylation (red indicates methylation; green, no methylation). In addition, NOTCH1 mutations were found as common characteristics of this subgroup (red indicates NOTCH1 mutation; green, NOTCH1 wild-type).31 (B) In the original analysis of 285 AML cases (panel A left), the 44 cases in cluster 5 aggregated very tightly, as indicated by the deep red colors representing positive Pearson correlation coefficients. Most of these 44 cases showed a monocytoid morphology (FAB-M4 or -M5).22 This raises the possibility that a significant part of the clustering effect was caused by specific up- or down-regulation of genes that are important in monocytic differentiation, resulting in a different signature than the remaining, mostly nonmonocytoid, cases of AML in the study. To answer whether gene expression profiling would enable identification of potential heterogeneity within this apparently homogeneous subgroup, in panel B the 44 cases were reclustered as an isolated cohort. For this analysis, only probe sets that showed a variable expression within these 44 AML cases were taken into account, as defined by a fold change of 3.5 to the mean in log2 scale in at least 1 case. The resulting cluster image shows that several potentially interesting subgroups can indeed be identified within these 44 AML cases, which have been indicated by gray lines.
The potential of GEP to uncover new subgroups has been illustrated for several types of cancer, such as cutaneous malignant melanoma, diffuse large B-cell lymphoma, breast cancer, and acute lymphoblastic leukemia.25-28 In AML, several GEP cohort studies have been performed in the last few years. In many of those, new subgroups were discerned. In a cohort of 166 cases of AML, 2 subgroups of normal karyotypes with distinguishable expression profiles were identified.21 The investigators postulated that this subdivision of the normal karyotype group could be diagnostically relevant, but noted an association of the 2 subgroups with known factors: internal tandem duplications (ITDs) in fms-like tyrosine kinase 3 (FLT3) and FAB-M4 and M5 monocytic leukemias, respectively. In another dataset, of 285 AMLs, 16 subgroups were recognized, several of which lacked previously known denominators.22 A different study divided 170 cases of AML, mostly patients of older age lacking favorable cytogenetic features, into 6 subgroups, some of which appeared novel.29
Unsupervised analyses have also been performed to uncover heterogeneity within established AML subtypes. In a set of 166 AMLs, the core binding factor (CBF) AMLs, ie cases with t(8;21) or inv(16), each could be split into subgroups merely based on the GEP data, which was subsequently reproduced in another study.21 ,30 These observations suggested that CBF leukemias in terms of gene expression patterns may represent heterogeneous entities. In 130 cases of pediatric AML, heterogeneity within CBF leukemias was seen as well, particularly within AML inv(16).23
In each of the above studies, GEP revealed previously unrecognized heterogeneity of AML. But is this diversity relevant? As cluster algorithms, by definition, focus on similarities between cases, a newly identified group of AMLs does not necessarily have biologic or clinical importance. The biologic significance of a newly identified subgroup would be convincingly substantiated by the subsequent discovery of a related underlying defect or correlation with a characteristic clinical phenotype or treatment response. Alternatively, validation of a signature in one or more independent datasets would provide support that the novel subgroup is stable. Evidence indicating that the detection of a distinctive gene expression subtype indeed can lead to the discovery of a biologically and clinically significant subgroup has recently been demonstrated by the identification of a distinct form of leukemia characterized by epigenetic CEBPA silencing and an immature myeloid/T-lymphoid phenotype (Figure 1A).31 Importantly, these findings could be confirmed in an independent cohort of human AML. Studies like these show how complementing technologies can be used to make full use of the wealth of GEP data for subgroup discovery. Evidently, for successful subgroup discovery it is important to have access to sufficiently large series of cases that represent the variable subtypes of AML. Compatibility of platforms and data sharing through online repositories can facilitate the latter.32,33
Interpretation of the results of GEP studies for class discovery
While GEP has been demonstrated to offer robust and reproducible technology in the analysis of cohorts of AML,21-23 one should remain well aware of the factors that may affect the results. We discuss here some of the most important of those factors in the context of class discovery, although many will also affect class prediction and class comparison.
Differences in study design may determine the probability of the discovery of disease heterogeneity. Variations in the selection of study populations, in terms of size as well as demographic diversity, will have a direct impact on experimental results. Furthermore, various technical differences between studies may influence results, ranging from sample processing and mRNA isolation to microarray hybridization and analysis. Interstudy variations with regard to the bioinformatic and biostatistical approaches, which involve choices regarding data normalization, gene filtering, and clustering procedures, may exert marked effects on outcome of the analysis.10,34 Most cluster algorithms present results in a 2-dimensional way, in which slight changes in calculation may move samples from one cluster to another. Such analytical differences will most likely not have a major influence on subgroups with very distinctive signatures, but may have an effect on the identification of more subtle differences.
It is important to keep in mind that GEP based clustering is driven by similarities and differences in gene expression profiles between samples. The similarities could be caused by shared underlying genomic defects—which is what most researchers are primarily interested in—but could also be caused by factors that are not directly related to pathogenetic mechanisms, for example, similarities in maturation phenotype of the leukemias. This has indeed been observed in AML. In 2 relatively large studies, AMLs with monocytoid morphologies (FAB-M4-M5 leukemias) showed a tendency to aggregate according similar gene expression patterns.21,22 The differences in predominant maturation stage of the leukemias may also have influenced the aggregation of CEBPA mutant cases into 2 gene expression clusters, one cluster including cytologically more immature leukemias.22,31 Such pitfalls in the analysis of GEP for subgroup discovery may explain why the number of novel subtypes of AML reliably identified by GEP have remained relatively limited.
How can the potential of GEP for discovery of novel subgroups of AML be enhanced? In the search for pathogenetically relevant differences between leukemias, one would wish to avoid the interference of effects of phenotypic differences. One way of achieving this would be to restrict the analysis to relatively homogeneous populations. The feasibility of avoiding unwanted background differences that might obscure interesting pathogenetic differences in a selected predefined AML group was demonstrated by investigators who established 2 gene expression profiles of non–Down syndrome–associated acute megakaryoblastic leukemia.35 Similarly, it has been noted that distinct subgroups can be found within the preselected t(15;17) AML subtype.36 Likewise, the 44 AMLs of a previously established cluster that strongly associated with FAB-M4 and M5 leukemias exhibited notable internal heterogeneity when studied as an isolated population (Figure 1B). Studies dealing with purified progenitor cells will also be instrumental for exclusion of interfering transcriptional background. Accumulating evidence suggests the existence of leukemia stem cells.37,38 Profiling of those cells, instead of total blast populations, may enhance the possibilities of GEP for subgroup discovery. Focusing on the leukemic stem cell may also reveal stronger transcriptional profiles that may be buried only in subclones. However, this approach directly depends on the accepted definition of immunophenotypic markers of leukemic stem cells and suffers from the technical drawback of rare stem cell numbers. Another opportunity for class discovery lies in the application of improved analytical procedures. A notable example is the use of pathway oriented analyses, which allow the discovery of distorted functional networks of genes as opposed to gene-based approaches.39,40
Do genome-wide gene expression analyses deserve a place in clinical diagnosis?
Several techniques are currently used in the initial diagnosis of AML, including cytology, immunophenotyping, karyotyping, polymerase chain reaction (PCR), and fluorescence in situ hybridization (FISH). Because a GEP-based approach allows detection of many transcripts at the same time, it provides a transcriptional snap shot of the leukemia. Several research groups have investigated the possibility to define specific gene expression classifiers (also referred to as class predictors) for disease subtypes, eg a discriminative set of genes for AML with the t(8;21) translocation. This procedure of class prediction through the generation of classifiers differs from class discovery, as discussed in the first section, in one important aspect: it makes use of external information, such as absence or presence of t(8;21), to derive a signature that can subsequently be used for prediction of samples of leukemia of which the status is not known yet. This type of approach is, therefore, often referred to as supervised.
AMLs defined by t(8;21), AML inv(16) and acute promyelocytic leukemia with t(15;17) have consistently been found to be predictable using gene expression classifiers with almost 100% accuracy.17,22,23,41 For AMLs with 11q23 rearrangements involving the mixed lineage leukemia (MLL) gene, reported prediction accuracy in a study within several types of human leukemia was approximately 90%, while this was 95% in a study on pediatric AML.23,41 Efforts to derive gene expression classifiers for AMLs with other chromosomal abnormalities have as yet met little success. Thus, for instance AML with chromosomal abnormalities involving trisomy 8, complex karyotype and 3q appear not to be accurately predictable by GEP in representative cohorts of AML.20,21,41
Prediction of mutations in CEBPA and NPM1 has also been pursued.22,24 In one study, correct prediction of most NPM1 mutation positive cases was possible, albeit at the expense of a significant number of false positives, resulting in a positive predictive value of 70% to 75%.24 This would create a hurdle when GEP would be used for diagnostic purposes. For CEBPA mutations, prediction accuracy may be dependent on the type of mutation. While biallelic mutations, which are relatively frequent, appear to be predictable with a gene expression classifier with high positive and negative predictive values, heterozygous monoallelic mutations may not be accurately predicted.42
Likewise, abnormalities in signaling molecules such as FLT3 and RAS appear not to be readily predictable within diverse AML cohorts.22 This may not be too surprising taking into account their less direct role in transcriptional modulation. In some studies, reasonably successful prediction of FLT3-ITD status was achieved when cytogenetically normal AMLs were selectively analyzed.43,44 A recently reported classifier for FLT3-ITD mutation status in normal karyotype AML showed only a modest performance in predicting FLT3-ITD status in a validation cohort of 72 normal karyotype cases, with both a relatively high number of false positives and false negatives (sensitivity 73%, specificity 85%).45
Taken together, these observations suggest that, using current methodology, definition of GEP-based classifiers is only feasible for selected AML subtypes in which the underlying molecular abnormality is not too distantly involved in transcriptional modulation. Those abnormalities, involving t(8;21), inv(16), and t(15;17), can be identified by rapid and widely used methods such as PCR as well. This may raise the important question what the ultimate clinical utility of GEP classifiers will be. The particular value of GEP-based classification lies in its comprehensiveness (ie, the opportunity to perform many tests simultaneously), for instance using specifically designed diagnostic DNA microarrays. Evaluation of larger representative AML patient series will be needed to reveal whether it is possible to define additional classifiers for less frequent cytogenetic or molecular subtypes. Definition of such additional classifiers will enhance the attractiveness of GEP as a diagnostic assay.
Is genome-wide gene expression analysis useful for predicting prognosis?
Several attempts have been made to derive prognostic signatures for AML. This approach is similar to class prediction, as discussed in the previous section, but instead of subgroup status (eg, t(8;21)), outcome (eg, overall survival) is used as the end point to define a prognostic predictor.46,47
In one study, a set of 133 genes was demonstrated to predict survival among adults with normal karyotype AML.21 A second group of investigators converted this signature into a prognostic predictor and confirmed its prognostic ability as regards overall and disease-free survival in another series of normal karyotype AML.48 Because of differences in DNA microarray platforms, the investigators could only verify 81 of the original 133 genes, so that a complete validation of the original signature was not possible.34 A significant part of the prognostic effect was associated with FLT3-ITD mutations.48
For relapse of pediatric AML, a 2-gene predictor has been proposed.23 The predictive value of this indicator appeared to be modest in a small validation subset of the pediatric cohort as well as in a set of adult AML cases. The same investigators were not able to confirm the value of a prognostic signature for pediatric AML that had been proposed by a different group.49
While the above prognostic predictors were constructed without any a priori biologic assumption, a hypothesis-driven approach was used to construct a predictor consisting of 11 genes associated with a stem cell–like expression pattern. Those genes were chosen because of their relationship to BMI1 activation in a murine prostate cancer model and in human cancer samples.50 The predictor recognized adverse outcome in several types of human cancer including AML, but its value awaits independent evaluation.
Will these studies lead to the introduction of GEP-based prognostic tests for AML in clinical practice? Experience in the field of breast cancer and diffuse large B-cell lymphoma may exemplify the use of GEP-based prognostication.51-56 For breast cancer, 2 prognostic predictors were reported recently by independent groups.53,54 Initially these GEP-based predictors met some skepticism because of their minimal overlap in genes. Independent validation studies, however, subsequently confirmed prognostic value for both signatures, independent of other available prognostic markers.52,57 The very limited overlap in genes between the 2 predictors is most likely explained by the fact that in a typical GEP experiment, many transcripts are more or less similarly correlated to outcome. Consequently, several combinations of genes are equally informative for prognosis.58 The success of the breast cancer predictors has led to the recent approval by the Food and Drug Administration of a commercial test that is based on one of them.56,59
From currently available evidence it appears that GEP predictors for prognosis can probably be established for AML. This will have the advantage of a comprehensive prognostic test that can substitute several currently used prognostic markers. However, it is still necessary to better assess whether such GEP based predictors can add information over currently available cytogenetic and molecular markers for prognosis of AML.46,60 Validation of any prognostic signature in sufficiently large series will be necessary before accepting this as a solid and reliable indicator of prognosis.46,58,61-65
What can genome-wide gene expression analyses tell about the biology of AML?
An additional quality of genome-wide assessment of mRNA levels of tens of thousands of genes is that it may allow for the discovery of pathobiologic pathways. Unbiased genome-wide GEP in AML may identify critical downstream targets of known oncogenes or tumor suppressors or identify novel causative genetic abnormalities. This concept has been illustrated to be particularly promising in animal or cell line cancer models in which an oncogene of interest had been introduced. Recent examples include models for acute promyelocytic leukemia,66 mutant Cebpa67 and Mll-Af9.68 Indeed, these studies demonstrate that biologic information is captured within the thousands of transcripts measured.
Although the search for critical abnormalities in clinical AML sounds straightforward, in practice there may be nontrivial hurdles on the way to the discovery of genomic abnormalities playing a key role in the pathophysiology. There may be numerous differences in gene expression between AML cases, but only few of them may be truly disease pathogenesis-related. How to extract the biologically important transcriptional differences from the large amount of data? One possible approach is the supervised class comparison strategy, which typically involves 3 steps. First, a subgroup of AML of interest is defined. Next, the gene expression profiles of cases in this subgroup are compared with those of control cases, yielding a list of differentially expressed genes. And finally, a selection of several genes from that list is made for further study, based on their presumed biologic impact. For example, in one recent study, gene expression profiles from clinical acute promyelocytic leukemia samples with the t(11;17) translocation expressing both PLZF-RARA and the reciprocal fusion protein RARA-PLZF were compared with those from samples expressing the PLZF-RARA fusion only. This led to the identification of CRABP1 as a specific target of the reciprocal fusion product. CRABP1 was subsequently shown to play a functionally important role in retinoid resistance.69 Comparisons of FLT3 ITD and FLT3 TKD signatures to FLT3 wild-type signatures have led to the discovery of potentially relevant downstream targets as well, although these remain to be functionally validated.44
While these examples show the potential of this approach, the disadvantage of supervised comparisons with subsequent selection of candidate genes is restriction due to inherent selection bias. One way around the selection bias problem may be employment of pathway-oriented bioinformatic analyses. Using specialized software, it has become possible to investigate the differential expression of sets of genes known to function in the same biologic pathways or to identify genes that have frequently been linked in the literature.39,40,70 These analyses facilitate the identification of the most promising candidate genes and their selection for functional validation in either in vitro or in vivo model systems. At the same time, it should be kept in mind that relative levels of mRNA expression do not necessarily reflect biologic activity, as the latter may be highly dependent on other factors, such as posttranslational modifications.
Nevertheless, a recent report on a novel subgroup of immature leukemias with myeloid and T-lymphoid characteristics demonstrated the value of the application of biologic pathway analysis to clinical GEP data.31 In a clinical AML GEP study, these leukemias were found to carry expression profiles similar to AML cases with CEBPA mutations, but such mutations were not present (Figure 1A). Subsequent experiments elucidated the likely explanation for this phenomenon, as in the novel subgroup CEBPA expression was silenced, frequently through promoter hypermethylation. Pathway analysis then revealed that the leukemias carried both myeloid and T-lymphoid features. Mouse modeling subsequently demonstrated that lack of CEBPA expression induced the expression of certain T-cell genes in immature hematopoietic cells. Thus, the analysis of human AML GEP using pathway analysis in combination with experiments in a representative mouse model uncovered part of the pathobiology of a subgroup of leukemia characterized by a myeloid/T-lymphoid phenotype, CEBPA silencing, and, in fact, frequent NOTCH1 mutations.
Another strategy for distilling biologically significant genes from human AML gene expression data makes use of possible associations with large sets of genes involved in animal retroviral insertion leukemogenesis. A study that compared integration sites in retrovirally induced leukemias in mice with human AML datasets demonstrated that mouse cancer genes were frequently deregulated in the human AMLs.71 Moreover, pathway analysis defined several biologic networks that associated with particular AML subsets. Thus, comparisons of human AML GEP data with high-throughput results from dedicated experimental models provide valuable opportunities to identify candidate disease genes. As pointed out before, these experimental approaches could also utilize GEP of cell lines or animal models,25-27 or, alternatively, use techniques such as chromatin immunoprecipitation on DNA microarray chips (ChIP-chip),72 ChIP-sequencing73 and RNA interference libraries to uncover target genes. A successful example of the latter approach comes from diffuse large B-cell lymphoma, for which 2 previously GEP-defined disease subgroups were functionally investigated using an RNA interference library to search for specific targets inhibiting tumor growth.26,74
Another viable strategy is based on the correlation of putative disease genes from in vivo or in vitro models to specific human leukemia subtypes. The strength of this approach is that the gene choice is based on experimental data. This allows for a rapid correlation of basic research findings to human AML datasets: is transforming gene X, that induces a leukemic phenotype in an experimental mouse model, differentially expressed in particular forms of human AML? Using this approach, researchers have provided evidence that genes such as TrkA and Trib2, which are involved in murine leukemia, may also be engaged in t(8;21) AML and CEBPA-silenced AML, respectively.75,76
These studies demonstrate the abilities of GEP to resolve questions related to the biology of AML when combined with appropriate other experimental and analytical tools. At the same time, there are various other experimental options to pinpoint key genomic abnormalities through GEP in human AML samples.
Concluding remarks and future perspectives
Recent years have shown an increase in high-throughput applications apart from GEP.8 In this respect profiling of microRNA (miRNA) levels, chromosomal copy number changes, epigenetic modifications, and DNA sequencing offer interesting opportunities.
MiRNAs are small noncoding RNAs that play a role in transcriptional or posttranscriptional regulation of genes involved in numerous biologic processes, including differentiation and proliferation.77 Profiling of miRNA expression levels in AML cohorts has indicated that, similar to mRNA profiling, distinct miRNA signatures are associated with specific subgroups of AML.78-80 As a single miRNA can play a role in concomitant regulation of multiple genes, closer comparisons between miRNA and mRNA profiles may provide clues for significant defects.
Changes in expression levels of critical genes may be due to small DNA amplifications or deletions undetected by conventional cytogenetics. Platforms to study those small chromosomal aberrations include single nucleotide polymorphism (SNP) arrays and array-based comparative genomic hybridization (CGH).81,82 The power of the use of SNP arrays for this type of analysis was recently illustrated in a study of 242 cases of pediatric ALL.83 The investigators identified focal abnormalities in lymphocyte differentiation related genes in 40% of cases, including 30% in the PAX5 gene. As yet, only studies of limited size have been performed in AML.84-89 A study in which array CGH (bacterial artificial and P1-derived artificial chromosome clones) was used in a series of 60 cases of AML with complex karyotypes identified several recurrent lesions.90 It is not clear yet what the overall frequency and distribution of these genomic alterations in AML are. A notable observation from several SNP array–based studies is the relatively common occurrence of copy number–neutral loss of heterozygosity through segmental uniparental disomy.84,87,91 This phenomenon can lead to homozygous mutations or deletions of leukemia-related genes, including FLT3, WT1, RUNX1, and CEBPA.91,92
Variations in gene expression may also be caused by yet unknown gene sequence mutations. Whole genome sequencing is emerging as a means to address this issue on a global scale.93,94 A challenge in this regard will be to distinguish functionally relevant mutations from the abundant so-called passenger mutations—unimportant genetic changes caused by genomic instability of cancer cells—that will be picked up at the same time. Studies on tyrosine kinase abnormalities have underscored the need for validation of the biologic effects of novel mutations identified by high-throughput nucleotide sequencing.95,96
In addition to genetic alterations, epigenetic modifications, including DNA and histone methylation, play a pivotal role in gene regulation. Several platforms have been developed to study alterations in these mechanisms on a genome-wide scale.97,98 The first results of genome-wide CpG methylation profiling of AML cell lines and primary cells have now been reported, and it is likely that more extensive investigations will follow.99,100 Given the direct association between epigenetics and expression, much is to be expected from this field.
As these fields have just started to emerge, there is still a need for bioinformatics to keep up with the technical developments, and to develop software platforms that allow the integration of the massive amounts of data. It is likely that the combination of GEP with complementing technologies, such as those outlined above, will provide challenging opportunities to address questions that cannot be resolved by GEP alone.
Acknowledgments
We thank Dr Peter Valk for valuable comments on the manuscript.
This work was supported by the Dutch Cancer Society Koningin Wilhelmina Fonds (Amsterdam, The Netherlands) and by the National Institutes of Health (Bethesda, MD; CA118316).
National Institutes of Health
Authorship
Contribution: B.J.W., B.L., and R.D. wrote the paper.
Conflict-of-interest disclosure: R.D. and B.L. have declared ownership interests in Skyline, a spinoff company of Erasmus University Medical Center (Erasmus MC), held in a Special Purpose Foundation of Erasmus MC. B.J.W. declares no competing financial interests.
Correspondence: Bas Wouters or Ruud Delwel, Erasmus University Medical Center, Department of Hematology, rooms Ee1330B / Ee1342, PO Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: b.wouters@erasmusmc.nl or h.delwel@erasmusmc.nl.