The basic helix-loop-helix transcription factor Scl/Tal1 controls the development and subsequent differentiation of hematopoietic stem cells (HSCs). However, because few Scl target genes have been validated to date, the underlying mechanisms have remained largely unknown. In this study, we have used ChIP-Seq technology (coupling chromatin immunoprecipitation with deep sequencing) to generate a genome-wide catalog of Scl-binding events in a stem/progenitor cell line, followed by validation using primary fetal liver cells and comprehensive transgenic mouse assays. Transgenic analysis provided in vivo validation of multiple new direct Scl target genes and allowed us to reconstruct an in vivo validated network consisting of 17 factors and their respective regulatory elements. By coupling ChIP-Seq in model cell lines with in vivo transgenic validation and sophisticated bioinformatic analysis, we have identified a widely applicable strategy for the reconstruction of stem cell regulatory networks in which biologic material is otherwise limiting. Moreover, in addition to revealing multiple previously unrecognized links to known HSC regulators, as well as novel links to genes not previously implicated in HSC function, comprehensive transgenic analysis of regulatory elements provided substantial new insights into the transcriptional control of several important hematopoietic regulators, including Cbfa2t3h/Eto2, Cebpe, Nfe2, Zfpm1/Fog1, Erg, Mafk, Gfi1b, and Myb.
Introduction
Hematopoiesis has long served as a model system for adult stem cells, with many paradigms of stem cell biology first being established as a result of studying hematopoietic stem cells (HSCs). A large body of work over the past 25 years has established that transcription factors (TFs) play critical roles during the specification, maintenance, and/or differentiation of HSCs. However, the underlying mechanisms have remained largely obscure because of a lack of comprehensive data on target genes, as well as very limited information on the way key TFs interact to form the regulatory networks that control blood stem cell development and subsequent behavior.
The basic helix-loop-helix (bHLH) TF Scl (also known as Tal1) is required for the specification of HSCs as well as their subsequent differentiation into erythroid and megakaryocytic lineages.1,2 Scl-null embryos do not survive beyond embryonic day (E) 9.5 due to a complete absence of hematopoiesis,3,4 a more striking phenotype than seen with other important regulators of early hematopoiesis such as Runx1 or Gata2.5,–7 Moreover, together with its paralogue Lyl1, Scl was recently shown to be essential for the survival of adult HSCs, thus emphasizing critical functions for Scl at multiple stages of hematopoietic ontogeny.8 In addition to its pleiotropic roles in hematopoiesis, Scl is also required for vascular and central nervous system development.9,–11 Within the blood system, Scl is thought to be a key component of the regulatory networks controlling the specification and subsequent differentiation of HSCs.12,13 Studies on the transcriptional regulation of the murine Scl gene identified Ets and Gata factors as well as an autoregulatory loop as key upstream inputs.14,–16 However, to fully understand how Scl functions within hematopoietic regulatory networks, comprehensive information on downstream target genes will also be required. Scl has been found to regulate a handful of genes, including Gata1,17 Runx1,18 c-kit,19 and α-globin20 in different hematopoietic lineages. However, to date, no systematic genome-scale approach has been taken to interrogate Scl target genes at early developmental time points in which Scl function is critical.
Together with bHLH class I proteins, such as E47, Scl binds DNA as a heterodimer to the so-called E-box sequence motif CANNTG. In addition to its bHLH DNA-binding partners, Scl can interact with various proteins, including the lim-only protein Lmo2 and Gata factors Gata1/Gata2 in multimeric complexes.20,21 Scl/Gata-containing complexes are known to bind to a composite E-box/GATA motif present in many erythroid-specific regulatory elements.17,22 Moreover, it has been demonstrated that Gata sites alone can be sufficient to recruit Scl/Gata complexes to some regulatory elements,17 consistent with previous observations indicating that an Scl mutant unable to bind DNA can rescue most of the Scl phenotype observed in Scl−/− mice.23 However, without a genome-wide inventory of sequences bound by Scl in blood stem/progenitor cells, the relative contribution of the various modes of Scl binding toward controlling gene expression remains unclear.
The advent of deep-sequencing technology has made the genome-wide mapping of TF-binding events an attractive strategy for the identification of TF target genes and subsequent reconstruction of gene regulatory networks. Coupling chromatin immunoprecipitation (ChIP) with deep sequencing (the so-called ChIP-Seq approach) has recently been successful in delineating key aspects of the gene regulatory networks controlling pluripotency in embryonic stem cell lines.24 In this study, we have combined ChIP-Seq experiments in hematopoietic stem/progenitor cell lines with in vivo validation. This has resulted in the identification of 228 high-confidence–binding events of Scl. Transgenic analysis of 11 gene loci bound by Scl demonstrated that all contained Scl-bound regions with in vivo enhancer activity driving expression at key hematopoietic sites in transgenic mouse embryos, thus providing in vivo validation of an emerging transcriptional network controlled by Scl.
Methods
ChIP
Hematopoietic precursor cell-7 (HPC-7) cells were grown in stem cell factor,25 and ChIP assays were performed as previously described,26 using a polyclonal anti-Scl antibody.27 ChIP material was assayed by real-time polymerase chain reaction (PCR). For primer sequences, see Table S4 (available on the Blood website; see the Supplemental Materials link at the top of the online article). The Scl ChIP sample was amplified, divided in 2, and sequenced using the Illumina 1G genome analyzer following the manufacturer's instructions. Sequencing reads were mapped to the mouse reference genome using Illumina software and displayed as University of California Santa Cruz (UCSC) custom tracks. Raw sequence data have been submitted to the National Center for Biotechnology Information short sequence read archive to be accessed via the Gene Expression Omnibus portal (www.ncbi.nlm.nih.gov/geo; GEO record GSE15806).
Peak analysis
Two independent sequencing runs were processed separately to identify peaks of Scl binding using Findpeaks 3.128 and Illumina BeadStudio (www.illumina.com/pages.ilmn?ID = 265),29 respectively. Intersection of both approaches yielded 593 peaks that were further filtered using GALAXY (http://galaxyproject.org)30 to remove all regions with less than 50 bp alignable to the human genome. The final list of 228 Scl-binding events was obtained after removing a further 92 peaks located at the extreme ends of chromosomes in areas devoid of genes, but full of repeats, an artifact we have seen with other antibodies. The remaining peaks were allocated to genes whereby peaks within promoters and introns of genes were allocated to that gene. When determining which genes were the likely targets of Scl-binding events occurring in intergenic regions, we excluded flanking genes not expressed in HPC-7 cells or any primary hematopoietic blood lineages. There were only 2 instances in which both flanking genes were not expressed, and both 5′ and 3′ flanking genes were considered if both were expressed (see Table S3).31
Gene set enrichment analysis32,33 was used to determine overlaps with curated gene sets based on tissue origin or TF binding site overrepresentation. The Database for Annotation, Visualization, and Integrated Discovery (DAVID)34,35 tool was used to classify candidate target genes into functionally related categories in which significance of group classification was defined by enrichment scores based on Fisher exact statistics.
In vivo validation of Scl-bound regions
Scl-bound regions were PCR amplified from mouse genomic DNA and inserted into lacZ reporter plasmids (see Table S4 for PCR primers and Figure S1 for alignments). F0 transgenic mouse embryos were generated by pronuclear injection of lacZ reporter fragments and analyzed as described34 (for numbers of transgenic embryos, see Table S5). Expression of the transgene in fetal liver was confirmed in selected embryos by performing histologic sections, as described previously.36 All animal studies were performed according to United Kingdom Home Office guidelines with Home Office approval.
Whole-mount images were acquired using a Nikon Digital Sight DS-fL1 camera attached to a Nikon SM7800 microscope (Nikon, Kingston upon Thames, United Kingdom). Images of sections were acquired with the Zeiss AxioCom MRc5 camera attached to a Zeiss Axioscope2plus microscope (Carl Zeiss, Welwyn Garden City, United Kingdom) using Olympus UPlanApo 40×/0.85 numeric aperture (NA) and 100×/1.35 NA objectives. Axio Vision Rel version 4.3.1.0 software (Carl Zeiss) was used for acquisition of digital images, which were processed using Adobe Photoshop and Adobe Illustrator (Adobe Systems, San Jose, CA).
Motif analysis of Scl-bound regions
The 228 Scl-bound sequences were separately scanned with a consensus-based and 2 position-specific scoring matrices (PSSMs)–based libraries of known motifs to detect putative binding sites. An International Union of Pure and Applied Chemistry library of motifs of TFs known to play essential roles in blood development was used37 in the consensus-based approach. For the PSSM-based searches, the JASPAR Core database containing 138 matrices38,39 and a subset of TRANSFAC release 10.4 (506 matrices of human and mouse origin)40 were used with the program matrix scan from RSAT following the protocol described previously.41 The statistical significance of the matching sites in Scl-bound sequences was estimated by z-score analysis against a negative background of promoter sequences of an average length of 400 nucleotides (the average length of the Scl-bound regions). To select an appropriate negative background, the BloodExpress database31 was interrogated for genes that were specifically not expressed in mouse blood progenitor cells (long-term and short-term HSCs). This search returned a nonredundant list of 3021 ENSEMBL genes. The 400 bases immediately upstream of the transcription start sites of each of these genes were extracted, and 1000 sets of 228 background sequences each were set to constitute a negative background distribution. The result of the pattern matching analysis using the 3 libraries is provided in Table S2.
Results
A genome-wide catalog of Scl-binding events
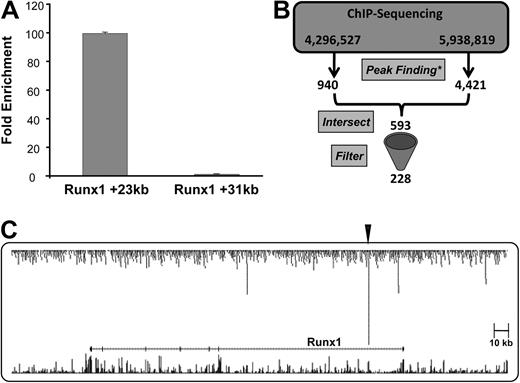
ChIP-Seq technology was used to generate a genome-scale catalog of sequences bound by Scl in the model cell line HPC-7, which requires stem cell factor for growth and has multilineage differentiation capacity.25 Chromatin from 20 million cells was immunoprecipitated using a Scl antibody and the resulting ChIP material tested by quantitative real-time PCR. We had shown previously that the Runx1 intron 1 +23 enhancer was bound by Scl in the aorta-gonad-mesonephros region and fetal liver,18 and now show that this region was also highly enriched in HPC-7 cells (Figure 1A).
The Scl ChIP material was divided in 2 and then analyzed by ultra-high throughput sequencing using the Illumina genome analyzer in 2 separate runs yielding 4.3 million and 5.9 million mappable reads, respectively. Visualization of the raw data across the Runx1 locus showed that ChIP-Seq reproduced the PCR results with highly specific enrichment of the Runx1 +23 region (Figure 1C), thus suggesting that the ChIP-Seq datasets should be a rich source of novel Scl target regions. To identify these regions, the 2 separate runs were processed individually and analyzed using 2 different peak prediction tools based on the premise that independent analysis would reduce the number of false-positive predictions. Even though the raw data were highly reproducible for the 2 runs, the different peak prediction algorithms generated overlapping, yet not identical lists of peaks due to the differences between the 2 individual algorithms (see “Peak analysis”). To focus subsequent analysis on those regions with the highest likelihood to represent real binding events, the intersection of the 2 datasets was therefore obtained and further filtered, as outlined in “Peak analysis,” resulting in a list of 228 Scl-binding events in HPC-7 cells (Figure 1B and Table S1). It is important to emphasize that the stringent filtering methods used in this study to analyze the current Scl ChIP-seq dataset were designed to maximize the identification of high-confidence–binding events with likely biologic function. However, using such stringent methods at very low false discovery rates, some true binding events are likely to have been missed. We have, therefore, made the raw dataset available to the wider scientific community for their own analyses (see “ChIP”). In any case though, given that the nonrepeat portion of the mouse genome contains more than 7.5 million E-box motifs, even at low stringency only a very small proportion of all these potential sites will be classified as occupied by Scl.
ChIP-Seq analysis recovers known Scl-binding events. (A) Real-time PCR analysis of ChIP assays in HPC-7 cells performed with Scl antibody. Levels of enrichment were normalized to IgG and compared with a negative control region (Runx1 +31kb). (B) Schematic diagram of the intersection and filtering of the ChIP-Seq data (* relevant peak-finding software, either Findpeaks 3.1 or Illumina BeadStudio). (C) Representation of combined ChIP-Seq sequencing reads (raw data) across the Runx1 locus, showing significant binding of Scl at the +23kb region, highlighted by the arrowhead. Sequencing reads are displayed as a custom track on the UCSC genome browser together with the UCSC known genes and vertebrate multiz alignment and conservation tracks.
ChIP-Seq analysis recovers known Scl-binding events. (A) Real-time PCR analysis of ChIP assays in HPC-7 cells performed with Scl antibody. Levels of enrichment were normalized to IgG and compared with a negative control region (Runx1 +31kb). (B) Schematic diagram of the intersection and filtering of the ChIP-Seq data (* relevant peak-finding software, either Findpeaks 3.1 or Illumina BeadStudio). (C) Representation of combined ChIP-Seq sequencing reads (raw data) across the Runx1 locus, showing significant binding of Scl at the +23kb region, highlighted by the arrowhead. Sequencing reads are displayed as a custom track on the UCSC genome browser together with the UCSC known genes and vertebrate multiz alignment and conservation tracks.
Candidate Scl target genes are highly enriched for TFs and proteins involved in signal transduction
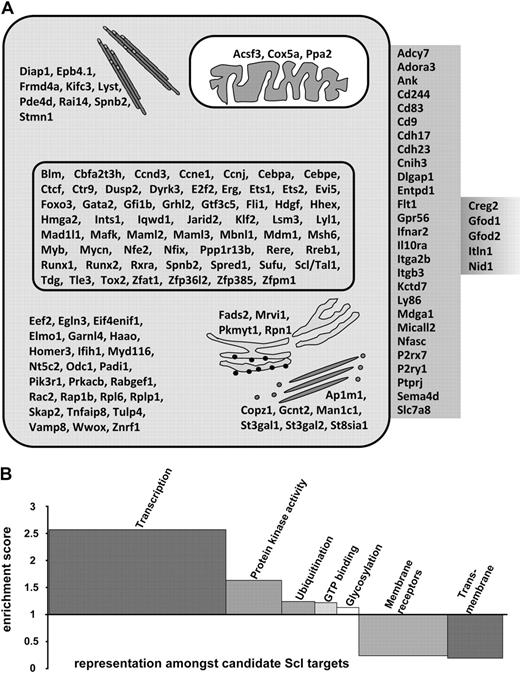
Scl-bound regions were mapped to their nearest gene (see “Peak analysis”) to obtain an initial list of candidate Scl target genes in HPC-7 cells (Figure 2). This list contained the 4 previously known HSC targets of Scl (Scl itself, Gata2, Fli-1, and Runx1).13,18,42 In addition, we observed Scl binding to several other regulatory elements known to be active in hematopoietic stem/progenitor cells, but not previously known to be targets of Scl (Hhex +1, Lyl-1 promoter, Vav1 intron1). The fact that known targets of Scl in HSCs (such as the Runx1 +23 element), as well as known enhancers active in HSCs, but not known to be controlled by Scl, were identified in this screen suggested that our curated list of 228 binding events was likely to contain a significant number of key building blocks of stem/progenitor cell regulatory network.
Candidate Scl target genes. (A) Diagram of identified Scl target genes and relative location within cellular compartments; cytoskeleton, mitochondrion, nucleus, cytoplasm, endoplasmic reticulum, Golgi, plasma membrane, and extracellular region/secreted proteins. (B) Representative grouping of gene ontology of candidate Scl target genes. Candidate target genes were classified into functionally related categories using the DAVID analysis tool.32,33 Significance of group classification was defined by enrichment scores based on Fisher exact statistics. Overrepresented categories are greater than 1, whereas underrepresented categories are smaller than 1.
Candidate Scl target genes. (A) Diagram of identified Scl target genes and relative location within cellular compartments; cytoskeleton, mitochondrion, nucleus, cytoplasm, endoplasmic reticulum, Golgi, plasma membrane, and extracellular region/secreted proteins. (B) Representative grouping of gene ontology of candidate Scl target genes. Candidate target genes were classified into functionally related categories using the DAVID analysis tool.32,33 Significance of group classification was defined by enrichment scores based on Fisher exact statistics. Overrepresented categories are greater than 1, whereas underrepresented categories are smaller than 1.
Gene set enrichment analysis32,33 indicated that our curated list of Scl target genes was highly enriched for hematopoietic TFs (most overrepresented category with a P value of 6.76−13). A more general analysis of gene ontology identified 5 overrepresented and 2 underrepresented functional categories (Figure 2B). Of note, the 2 most highly overrepresented categories were “transcription” and “signaling,” consistent with the known biologic function of Scl as a powerful developmental regulator. Taken together, this analysis suggested that our curated list of 228 Scl-bound regions is a rich source of key nodes in the hematopoietic regulatory networks serving to connect other important regulators with Scl.
In vivo occupancy in primary cells
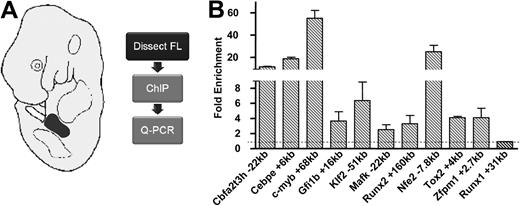
Reconstruction of developmental regulatory networks requires validation of key regulatory interactions in primary tissue. The limiting amount of primary material available negates the use of ChIP-Seq to analyze TF occupancy in blood stem/progenitor cells. We therefore elected to validate Scl-binding events seen in HPC-7 cells by performing ChIP assays, followed by quantitative PCR using material from E11.5 fetal liver, as a rich source of immature hematopoietic progenitors. We focused validation of Scl-bound regions on candidate elements in TF gene loci, because direct control of other TF genes by Scl would constitute the prime means of information flow through HSC networks. From the list of 228 peaks, we identified 40 TF genes that were expressed in blood stem/progenitor cells. For fetal liver ChIP validation, we focused our analysis on TFs known to be important for blood development, but not previously implicated as Scl target genes (see Figure S2 for expression levels), as well as the High Mobility Group box factor Tox2, which was chosen due to its expression in E8.5 yolk sac and E11.5 FL (data not shown). Validating functional interactions between previously unconnected regulators not only constitutes the first and essential step toward building wider transcriptional hierarchies and networks, but also provides a potentially powerful route toward identifying major regulatory elements involved in regulating expression in stem/progenitor cells. Specific primers were designed for the bound regions within different TF loci (Cbfa2t3h, Cebpe, c-myb, Gfi1b, Klf2, Mafk, Nfe2, Runx2, Tox2, and Zfpm1). All the regions tested showed significant levels of enrichment compared with a negative control region (Figure 3). Binding of Scl to these same regions was also verified in an independent ChIP sample from HPC-7 cells, again showing significant binding to all regions tested (Figure S3). Taken together, real-time PCR validation provided crucial evidence of novel Scl targets in vivo and confirmed that the HPC-7 cell line represents a suitable in vitro model in which true Scl binding sites important for hematopoietic regulatory networks may be found.
Confirmation of novel targets of Scl binding in vivo in primary tissue. (A) Fetal livers were dissected from E11.5 wild-type embryos, and chromatin was prepared and immunoprecipitated using an antibody against Scl. Enriched DNA was then analyzed by quantitative real-time PCR. (B) Real-time PCR analysis of ChIP assays in E11.5 fetal liver (FL) validates Scl binding to previously unknown TF targets of Scl. Fold enrichment levels were normalized to IgG and compared with a negative control region (Runx1 +31kb).
Confirmation of novel targets of Scl binding in vivo in primary tissue. (A) Fetal livers were dissected from E11.5 wild-type embryos, and chromatin was prepared and immunoprecipitated using an antibody against Scl. Enriched DNA was then analyzed by quantitative real-time PCR. (B) Real-time PCR analysis of ChIP assays in E11.5 fetal liver (FL) validates Scl binding to previously unknown TF targets of Scl. Fold enrichment levels were normalized to IgG and compared with a negative control region (Runx1 +31kb).
Transgenic validation of Scl-bound candidate enhancers
Regulatory elements represent the building blocks of transcriptional regulatory networks. To further our understanding of HSC networks, it was therefore important to demonstrate that Scl-bound regions identified in this study represent bona fide regulatory elements. Whereas promoter/enhancer elements can be assayed in cell lines, only transgenic analysis can identify true in vivo activities. For transgenic analysis, candidate regulatory elements are inserted into lacZ reporter constructs containing basal promoters, which on their own are largely silent and only show rare and ectopic staining in a small minority of transgenic embryos. For transgenic embryos that carry candidate regulatory elements fused to the minimal promoter-lacZ cassette, tissue-specific activity of these elements is revealed through consistent lacZ expression in 1 or more tissues.43 Given that putative activity of candidate elements is assessed based on comparison with control transgenic embryos, the choice of negative control constructs is critical. We have shown previously that the simian virus 40 (SV40) minimal promoter serves as a valid negative control when assessing candidate regulatory elements at midgestation for potential activity in the Scl expression domain (eg, midbrain and fetal liver; see Table S5).36
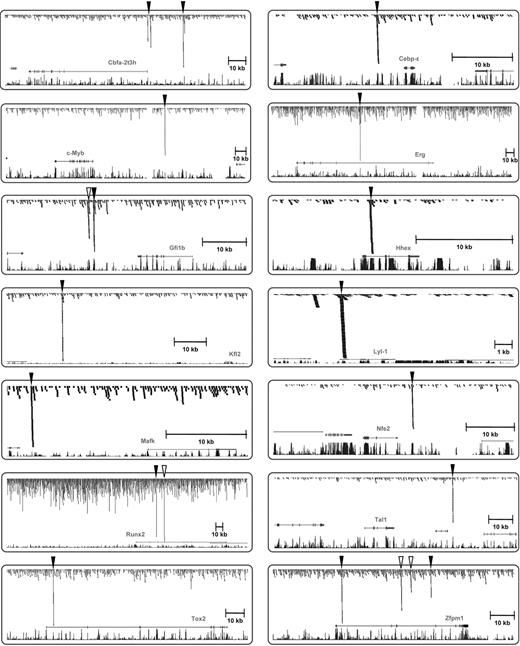
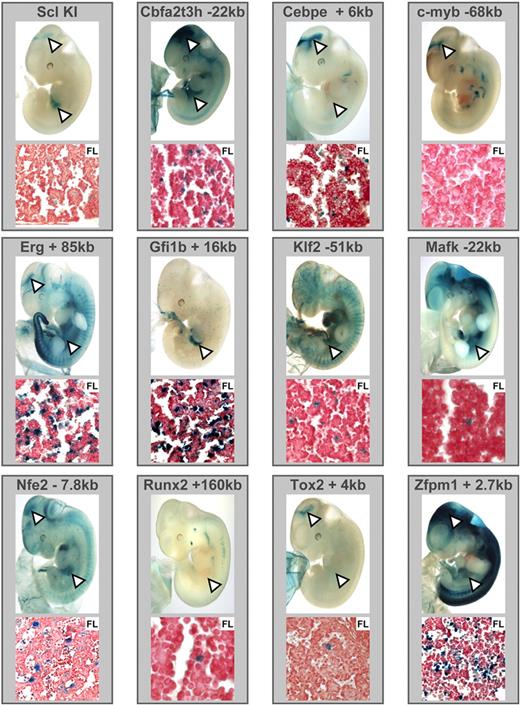
As outlined above, the Lyl-1 promoter and Hhex + 1 enhancer were known to be active in fetal liver hematopoietic stem/progenitor cells in transgenic mice, but only our genome-wide ChIP-Seq study showed that these 2 regions were in fact bound by Scl (Figure 4). To investigate whether Scl-bound regions from the additional 11 TF gene loci shown in Figure 4 also corresponded to hematopoietic enhancers, we generated lacZ reporter constructs for all Scl-bound regions from those 11 TF gene loci by inserting candidate enhancer regions into a reporter construct in which lacZ is driven by the neutral SV40 minimal promoter.36,46 A total of 59 transgenic mice was generated by microinjection, and in vivo activity of lacZ reporter constructs was assessed at midgestation using the colorimetric lacZ substrate X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactoside; see Table S5 and Figure S1 for alignments). Staining patterns of whole-mount embryos and histologic sections were compared with E11.5 Scl lacZ knockin embryos, which show characteristic staining in the midbrain and fetal liver hematopoietic cells as well as rare endothelial and circulating blood cells (Figure 5). Remarkably, Scl-bound regions from all 11 TF gene loci directed expression to fetal liver and/or midbrain, suggesting that our screen for Scl-bound regions represents a highly reliable way of identifying bona fide elements controlled by Scl in vivo (see Figure S4 for array of staining patterns seen for the Gfi1b +16 kb construct). With the exception of the Myb −68 kb region, which showed expression in the midbrain, but may require interaction with other elements in the Myb locus to provide hematopoietic expression, all other elements showed fetal liver activity with the Cbfa2t3h −22 kb, Cebpe +6 kb, Erg +85 kb, Nfe2 −7 kb, Tox2 +4 kb, and Zfpm1 +2.7 kb regions showing activity in both midbrain and fetal liver, whereas the Gfi1b +16 kb, Klf2 −51 kb, Mafk −22 kb, and Runx2 +160 kb regions only targeted fetal liver (representative embryos shown in Figure 5). Several of the Scl-bound enhancers showed staining outside the Scl expression domain when tested in transgenic mice. This is most likely due to either additional motifs in the elements responding to the TFs in these tissues or bHLH factors other than Scl operating through the Scl binding site. Importantly, transgenic validation provided direct in vivo validation of multiple new direct links in the transcriptional regulatory networks controlling early hematopoiesis (Figure 6A).
Novel binding targets of Scl within 11 TF gene loci. Combined ChIP-Seq raw data were displayed on UCSC genome browser as for Figure 1C for the Scl locus, as well as 13 newly identified TF targets of Scl. Significant peaks of Scl binding are highlighted with arrowheads. Open arrowheads were not functionally validated, and filled arrowheads were functionally validated in vivo in this study except for Hhex and Lyl-1, which had already been tested in transgenic mice.42,43
Novel binding targets of Scl within 11 TF gene loci. Combined ChIP-Seq raw data were displayed on UCSC genome browser as for Figure 1C for the Scl locus, as well as 13 newly identified TF targets of Scl. Significant peaks of Scl binding are highlighted with arrowheads. Open arrowheads were not functionally validated, and filled arrowheads were functionally validated in vivo in this study except for Hhex and Lyl-1, which had already been tested in transgenic mice.42,43
Scl-bound regions within TF loci function as enhancers in transgenic mice and display characteristic Scl knockin staining patterns. DNA fragments corresponding to the Scl-bound regions were cloned into a LacZ reporter plasmid and used to generate transgenic F0 embryos. A Scl LacZ knockin embryo is shown as a reference for the characteristic staining pattern (see text). (Note that the LacZ sequence used to create the Scl knockin mice differs from the one used in transgenic reporters; hence, the more punctuate staining with the knockin.) E11.5 X-Gal–stained whole-mount embryos and paraffin sections of fetal liver are shown. White arrowheads highlight midbrain and fetal liver staining.
Scl-bound regions within TF loci function as enhancers in transgenic mice and display characteristic Scl knockin staining patterns. DNA fragments corresponding to the Scl-bound regions were cloned into a LacZ reporter plasmid and used to generate transgenic F0 embryos. A Scl LacZ knockin embryo is shown as a reference for the characteristic staining pattern (see text). (Note that the LacZ sequence used to create the Scl knockin mice differs from the one used in transgenic reporters; hence, the more punctuate staining with the knockin.) E11.5 X-Gal–stained whole-mount embryos and paraffin sections of fetal liver are shown. White arrowheads highlight midbrain and fetal liver staining.
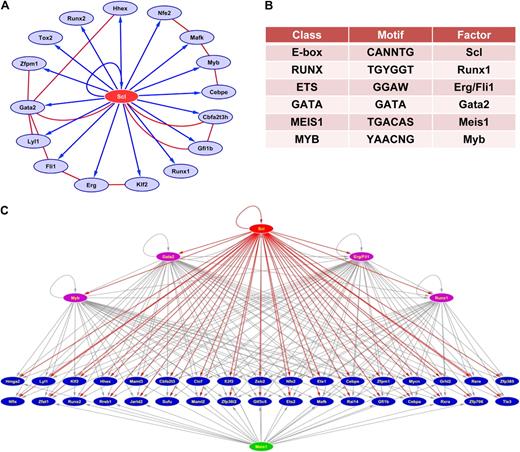
Reconstruction of Scl-centric regulatory networks. (A) In vivo validated core network. Blue arrows indicate activation through Scl-bound enhancers validated in transgenic mice. Red lines indicate protein-protein interactions curated from the literature.58,–60 (B) Motif overrepresentation analysis of 228 Scl-bound regions shows overrepresentation of multiple consensus binding sites associated with stem cell regulators (for full results, see Table S2). (C) Extended Scl-centric TF network with Scl at the top and Scl targets involved in transcription regulation underneath. Red arrows correspond to Scl controlling target genes through elements found to be occupied by Scl in the current study. Gray arrows indicate inferred links based on the presence of overrepresented motifs from (B). Factors with inferred links are shown in purple if they are Scl targets, in green if they are not.
Reconstruction of Scl-centric regulatory networks. (A) In vivo validated core network. Blue arrows indicate activation through Scl-bound enhancers validated in transgenic mice. Red lines indicate protein-protein interactions curated from the literature.58,–60 (B) Motif overrepresentation analysis of 228 Scl-bound regions shows overrepresentation of multiple consensus binding sites associated with stem cell regulators (for full results, see Table S2). (C) Extended Scl-centric TF network with Scl at the top and Scl targets involved in transcription regulation underneath. Red arrows correspond to Scl controlling target genes through elements found to be occupied by Scl in the current study. Gray arrows indicate inferred links based on the presence of overrepresented motifs from (B). Factors with inferred links are shown in purple if they are Scl targets, in green if they are not.
An emerging regulatory network controlling early embryonic hematopoiesis
Transgenic validation suggested that a large proportion of the 228 Scl-bound regions identified in this study represents regulatory elements active in vivo during early embryonic development. We therefore decided to explore whether analysis of DNA motif content of this unique dataset would allow us to gain new insights into the regulatory networks controlled by Scl. Like other bHLH TFs, Scl is known to bind to the E-box consensus site CANNTG, but has also been shown to be recruited to DNA via Gata factors bound to the GATA site.20,21 To identify putative TF binding sites that might be overrepresented, motif overrepresentation analysis was performed on the 228 Scl-bound regions using the 138 position frequency matrices from the JASPAR Core database,38,39 the 506 Transfac Matrices,40 and an in-house library of 54 blood development–specific consensus motifs.37 Motif overrepresentation was calculated against a negative background of promoter regions from genes not expressed in mouse blood stem/progenitor cells (see “Motif analysis of Scl-bound regions”).
The first and second most overrepresented motifs in overrepresentation analysis (z-scores of 13.6 and 11.9, respectively) corresponded to the E-box and GATA motifs, with 210 of 228 Scl-bound regions containing at least 1 E-box and 212 regions containing at least 1 GATA site (see Figure 6B and Table S2 for full results). This suggested that unlike with some other recent genome-scale surveys of TF binding, the majority of Scl-bound regions contained motifs consistent with the previously recognized major modes of Scl binding to DNA. The remaining overrepresented motifs contained binding sites for several known regulators of blood stem cells, such as Meis1, Erg/Fli-1, and Runx1, suggesting that Scl collaborates with these factors in controlling the activity of blood stem cell enhancers. Interestingly, several of these factors (Runx1, Fli-1, Erg, Gata2, and Myb) were identified in this study as direct Scl targets, whereas Meis1 was not. This suggests that there is an interface between a tightly knit Scl-centered core network interacting with several other regulatory pathways such as Meis1. Figure 6C shows a representation of the emerging transcriptional regulatory network connecting Scl with 39 other genes involved in transcriptional regulation together with inferred regulatory interactions based on motif content. This analysis indicated that the Scl targets Gata2, Erg/Fli-1, Myb, and Runx1 are likely to participate in the control of significant subsets of Scl targets because the regions bound by Scl also contained consensus binding sites for these factors. Moreover, Meis1, which was not identified as a high-confidence Scl target, is also predicted to play a role in the control of multiple Scl targets, thus providing potential crosstalk between the Scl core network and other potential hubs of stem/progenitor regulatory networks. To begin to corroborate the predictive power of regulatory interactions inferred on the basis of motif content, we examined whether the Scl-bound regions of the gene loci shown in Figure 6C were also bound by Gata2. Thirty-nine of 40 Scl-bound regions contained at least 1 GATA site, and 32 of these showed strong Gata2 binding (see Figure S6 and Table S6), thus highlighting the potential power of integrated bioinformatic analysis of ChIP-Seq data.
Discussion
At the molecular level, development can be characterized as a journey through progressive regulatory states defined by the dynamic changes in gene expression patterns during differentiation cascades from multipotent cells to mature progeny. The primary control of gene expression patterns is exerted by TFs, and it is therefore no surprise that TFs have emerged as some of the most powerful developmental regulators. However, to understand the underlying mechanisms, it will be important to identify downstream target genes as well as gain insight into combinatorial TF interactions that form the building blocks of wider transcriptional regulatory networks. In this study, we have used a combination of ChIP-Seq, transgenic reporter assays, and bioinformatic analysis to define target genes and transcriptional network interactions for the blood stem cell master regulator Scl.
An emerging core network of embryonic hematopoietic development centered around Scl
Even though the function of Scl as a key regulator of blood stem cell development was established more than 15 years ago, only 3 Scl target genes (Runx1, Gata2, Fli-1) have been validated in vivo during early embryonic hematopoiesis.13,18,42 The current study adds more than 200 candidate target genes with transgenic validation of 11 new elements as well as the realization that 2 previously characterized elements are bound by Scl (Lyl-1 promoter and Hhex +1 enhancer, respectively).44,45 This has allowed us to generate a fully in vivo validated core network with Scl at the center surrounded by 16 target gene TFs connected via 16 tissue-specific regulatory elements, all of which have been validated in transgenic mice (Figure 6A and previously published data13,18,42,44,45 ). A closer examination of this emerging core network provides several interesting observations.
Data mining of protein-protein interactions curated from the literature shows extensive protein-protein interactions within this network, several of which involve the Scl protein itself. This observation suggests that there are multiple feedback loops at the level of protein complexes, and it will be important to decipher which complexes act on which Scl-bound regions. Moreover, this observation is highly reminiscent of the embryonic stem cell pluripotency network being defined both at the level of protein/DNA and protein/protein interactions.47,48
The emerging network contains several factors thought to predominantly act as transcriptional repressors, such as Cbfat2t3h, Gfi1b, and Hhex. Of particular interest may be the observation that Cbfat2t3h is a direct Scl target because it is known to form a complex with Scl protein, which functions to repress transcription.49,–51 Although it is at this stage unclear whether Scl/Cbfat2t3h complexes have a restricted binding specificity to only a subset of target regions bound by Scl, our observation suggests that negative feedback may be built into Scl-mediated control of downstream target genes. In terms of regulatory network architecture, this also provides the necessary design elements to have abundant so-called incoherent feed-forward loops (gene A turns on genes B and C, in which C is a repressor that will in turn repress gene B). Incoherent feed-forward loops create bursts of gene expression that may be important if Scl plays a role in the response of stem cells to external stimuli such as cytokines or stem cell niche signals.
Although several of the Scl targets identified by ChIP-Seq were previously known to be important for hematopoietic stem/progenitor cell function, most of these had not been implicated as direct downstream targets of Scl. Moreover, by functionally validating the Scl-bound regions in vivo in transgenic mice, we have defined new regulatory elements for 11 gene loci, which previously had not been studied using transgenic mouse assays. By validating all these elements involved in controlling early hematopoietic expression, the current study therefore provides substantial new insights into the transcriptional control of multiple important hematopoietic regulators, such as Cbfa2t3h, Cebpe, Nfe2, Zfpm1, Erg, Mafk, Gfi1b, and Myb. Of note, 8 of 11 of the new enhancers validated in this study showed neuronal expression in the region of the midbrain known to express Scl. Analysis of conditional Scl null mice has identified a critical function during neuronal development. Knockout mice have been generated for 7 of the 8 genes with neuronal enhancer activity, but a conditional brain-specific knockout has only been described for c-myb, which like Scl was found to be critical for neuronal development.52
Known target genes of Scl in more mature erythroid and megakaryocyte cells such as Gata1,53 Lmo2,54 α- and β-globin,20,55 as well as NF-E2 promoter,56 respectively, were not bound by Scl in the HPC-7 cell line. We also did not observe binding to the Runx3 promoter, which previously showed low-level Scl binding in fetal liver and no binding in E8.5 yolk sac,18 suggesting potential binding of Scl within subpopulations of hematopoietic progenitor cells. Absence of binding events characteristic for mature cell types not only provides molecular validation of HPC-7 as a progenitor model cell line, but also underlines the dynamic nature of regulatory networks with Scl likely to control overlapping, yet distinct sets of downstream target genes at different stages of hematopoietic maturation. Moreover, it raises the issue as to which factors or complexes are responsible for opening up regions bound by Scl later in development, and therefore, underscores the importance of deciphering the processes by which so-called pioneer factors57,58 open up chromatin, as this is likely to be one of the key mechanisms causing remodelling of network topology, and thus initiate transition through successive regulatory states.
Beyond TF centric networks
Reconstruction of regulatory networks has largely focused on either transcriptional regulatory networks or signaling networks. Transcriptional network models describe the sum of TF and target gene interactions, whereas signaling networks focus on protein-protein interactions in which one protein is generally the substrate for a posttranslational modification mediated by enzymatic activity of the other protein (eg, a phosphorylation event mediated by a protein kinase). At least partly due to largely different technologies used for their analysis (eg, bulk measurements for TF networks and single-cell measurements for signaling), there is very limited information on how these networks interface, even though it is clear that cellular behavior is controlled by both. It was therefore interesting to see that after transcriptional regulation, gene ontology categories associated with signaling were the second most enriched categories among the candidate Scl target genes identified in this study.
Pathway analysis demonstrated that components of the mitogen-activated protein (MAP) kinase and focal adhesion kinase pathways were overrepresented among our list of candidate Scl target genes (see Figure S5). The MAP kinase cascade operates downstream of several receptor kinases important in blood stem and progenitor cells, such as Flt3 and c-kit. Moreover, the focal adhesion kinase pathway transmits both growth factor and extracellular matrix signals via growth factor receptors and integrins, respectively.59 It is therefore tempting to speculate that by controlling the expression levels of multiple components of signal transduction pathways, Scl might be able to modulate the interaction of hematopoietic stem 1 progenitor cells (HSPCs) with their niche.
Complementary approaches permit network reconstruction in rare stem/progenitor populations
New postgenomic tools are revolutionizing our ability to map TF binding sites, and therefore reconstruct regulatory networks. However, these techniques currently require large amounts of biologic material prohibiting their use in many stem cell settings in which cell numbers are limiting. Because regulatory elements represent the building blocks for transcriptional regulatory networks, their identification and subsequent characterization will remain a key activity of any program of work directed toward the reconstruction of regulatory networks. In this study, we show that genome-scale ChIP-Seq surveys in carefully chosen in vitro models can be used to define the connectivity of stem cell regulators such as Scl when combined with appropriate in vivo validation such as transgenic assays. Moreover, bioinformatic analysis of newly identified regulatory elements demonstrated highly significant overrepresentation of the binding sites of other HSC regulators within Scl-bound regions, thus opening up new avenues into deciphering combinatorial regulatory codes. Whereas complete functional validation of inferred network connections will depend on the availability of suitable antibodies, preliminary analysis showed that 83% of predicted Gata2-regulated elements are bound in vivo by Gata2, thus underlining the power of comprehensive bioinformatics analysis. In conclusion, this study not only provides many new candidate regulators of HSPCs acting downstream of Scl, but also may serve as a blueprint for deciphering regulatory networks in other developmental/stem cell systems in which the amounts of biologic material are limiting.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Cheng-Eng Ang and Richard Dixon (Medical Solution), as well as Yongjun Zhao and Martin Hirst (British Columbia Cancer Agency Genome Sciences Center, Vancouver, BC) for expert managing of Illumina sequencing runs; Duncan Odom and Dominic Schmidt for advice on processing samples for ChIP-Seq; and John Pimanda for critically reading the manuscript.
This work was supported by the Leukaemia Research Fund, London, United Kingdom: Leukemia & Lymphoma Foundation, New York, NY; Cancer Research UK, London, United Kingdom; Newton Trust, Cambridge, United Kingdom; United Kingdom Medical Research Council, London, United Kingdom; and Cambridge Cancer Center, Cambridge, United Kingdom.
Authorship
Contribution: N.K.W., D.M.-S., and R.J. performed research, analyzed data, and wrote the paper; S.K., N.B., S.D.F., F.C.-N., M.A.D., and I.J.D. performed research; J.F. and X.-H.S. contributed essential reagents; A.J.B. designed research; S.A.T. analyzed data and wrote the paper; and B.G. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Berthold Göttgens, University of Cambridge, Department of Haematology, Cambridge Institute for Medical Research, Hills Rd, Cambridge, CB2 0XY, United Kingdom; e-mail: bg200@cam.ac.uk.