Abstract
Class IIa histone deacetylases (HDACs) are signal-responsive regulators of gene expression involved in vascular homeostasis. To investigate the differential role of class IIa HDACs for the regulation of angiogenesis, we used siRNA to specifically suppress the individual HDAC isoenzymes. Silencing of HDAC5 exhibited a unique pro-angiogenic effect evidenced by increased endothelial cell migration, sprouting, and tube formation. Consistently, overexpression of HDAC5 decreased sprout formation, indicating that HDAC5 is a negative regulator of angiogenesis. The antiangiogenic activity of HDAC5 was independent of myocyte enhancer factor-2 binding and its deacetylase activity but required a nuclear localization indicating that HDAC5 might affect the transcriptional regulation of gene expression. To identify putative HDAC5 targets, we performed microarray expression analysis. Silencing of HDAC5 increased the expression of fibroblast growth factor 2 (FGF2) and angiogenic guidance factors, including Slit2. Antagonization of FGF2 or Slit2 reduced sprout induction in response to HDAC5 siRNA. Chromatin immunoprecipitation assays demonstrate that HDAC5 binds to the promoter of FGF2 and Slit2. In summary, HDAC5 represses angiogenic genes, such as FGF2 and Slit2, which causally contribute to capillary-like sprouting of endothelial cells. The derepression of angiogenic genes by HDAC5 inactivation may provide a useful therapeutic target for induction of angiogenesis.
Introduction
Histone deacetylases (HDACs) are cofactors for the regulation of gene transcription. Human HDACs are grouped into 3 classes based on their similarity to known yeast factors: class I HDACs are similar to the yeast transcriptional repressor yRPD3, class II HDACs to yHDA1, and class III HDACs to ySIR2.1 Nonselective inhibitors of class I and class II HDACs reduce tube formation of endothelial cells in vitro,2 inhibit postnatal neovascularization in response to hypoxia,3 and block tumor angiogenesis.4 Moreover, the enzymatic activity of class I and class II histone deacetylases is essential for endothelial commitment of progenitor cells.5 Mice deficient for the class IIa HDAC isoform HDAC7 fail to maintain proper endothelial cell-cell adhesion and have a leaky, rupture-prone vasculature, leading to embryonic lethality.6 Recently, it has been shown that members of class IIa HDACs are modulators of angiogenesis in vitro.7–10 In particular, protein kinase D–dependent phosphorylation of HDAC5 and HDAC7 plays an important role in vascular endothelial growth factor (VEGF)–induced angiogenesis in vitro.9,10 However, beyond the characterization of HDAC7 as an essential regulator of embryonic blood vessel development, the individual functions of class IIa HDAC isoforms during postnatal angiogenesis and the underlying mechanisms remain poorly defined.
Angiogenesis requires the migration of endothelial cells from preexisting vessels.11 Paracrine secretion of angiogenic growth factors creates a milieu that regulates directed movement of endothelial cells.12 Among these growth factors, fibroblast growth factor 2 (FGF2) released from endothelial cells potently stimulate migration and capillary-like growth of these cells via an autocrine action.13–16 Beyond growth factors, so-called axon guidance factors known for their role in neuronal wiring have recently been identified as essential regulators of capillary sprout formation.17 These factors provide guidance cues for the directional movement of specialized endothelial “tip” cells at the forefront of navigating blood vessels similar to axonal growth cones.18 Guidance factors are families of surface membrane receptors and their ligands that can be secreted (ie, Slit2) or membrane-bound ligands (ie, EphrinB2). After ligand-receptor interaction, both partners enable bidirectional communication between cells, which includes reverse signaling via the membrane-bound ligand to the ligand-bearing cell in addition to the outside-in signaling that arises from ligand-mediated activation of the receptor.19 Signaling via the guidance factor EphB4 is crucial for both endothelial cell migration20,21 and the formation of capillary-like structures.22 Likewise, Slit2 acts as molecular guidance cue in cellular migration, which is mediated by its interaction with one of 4 different roundabout (robo) receptors. Depending on the differentiation stage and the individual robo receptor subtype involved, Slit2 can act as a chemorepellent or attractant.23 In a model of tumor angiogenesis, Slit2 attracts endothelial cells and promotes tube formation, and its interaction with Robo1 is essential for tumor vascularization.24
Because angiogenic activation of endothelial cells to migrate and to form sprouts is associated with characteristic changes in gene expression profiles,25 the identification of upstream regulatory mechanisms that coordinate pro-angiogenic activation is crucial. Here, we assessed the role of individual HDAC isoenzymes for endothelial cell migration and capillary-like sprout-forming capacity and explored the role of HDAC5 as an essential repressor of a gene expression pattern required for angiogenic functions in endothelial cells.
Methods
Cell culture
Pooled human umbilical vein endothelial cells (HUVECs) were purchased from Lonza Verviers (Verviers, Belgium) and cultured in endothelial basal medium (EBM; Lonza Verviers SPRL) supplemented with hydrocortisone, bovine brain extract, gentamicin, amphotericin B, epidermal growth factor, and 10% fetal calf serum (Invitrogen, Karlsruhe, Germany) until the third passage. After detachment with trypsin, cells were grown in 6-cm culture dishes for at least 18 hours. Neutralizing FGF2 antibody (clone bFM-1) was bought from Upstate Biotechnology (Lake Placid, NY). RoboN and R5 were kindly donated by Jian-Guo Geng (Division of Hematology, Oncology and Transplantation, University of Minnesota Medical School, Minneapolis, MN).
Plasmids and transfection
HUVECs (3.5 × 105 cells/6-cm well) were grown to 60% to 70% confluence and then transfected with 3 μg plasmid DNA. Suk-Chul Bae kindly provided the wild-type human HDAC5 construct (HDAC5 in pCS4-3myc) as described previously.26 The HDAC5 S259/498A construct was engineered by stepwise site-directed mutagenesis. The HDAC5 Δmyocyte enhancer factor-2 (MEF2) mutant was constructed by site-directed mutagenesis as described for the homologous isoenzyme HDAC4.27 Eric N. Olson (Department of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX) kindly provided the HDAC5 1-767aa construct.28 Constitutively active PKD1 and HDAC7 wt were kindly provided by Franck Dequiedt (Cellular and Molecular Biology Unit, FUSAGx, Gembloux, Belgium).29 pcDNA3.1-myc-His served as empty vector control (mock). Transfection was performed using Superfect transfection reagent (QIAGEN, Hilden, Germany) according to the manufacturer's protocol.
RNA interference
For siRNA-mediated silencing, HUVECs were grown to 60% to 70% confluence and transfected with GeneTrans II (MoBiTec, Göttingen, Germany) according to the manufacturer's protocol. siRNAs were synthesized by Eurogentec (Cologne, Germany) or QIAGEN targeting the sequences given in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). All other siRNA oligonucleotide sequences are available on request.
Western blot analysis
For Western blot analysis, HUVECs were lysed with radio-immunoprecipitation assay (RIPA) buffer (ready-to-use solution containing 150 mM NaCl, 1.0% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 50 mM Tris, pH 8.0; Sigma-Aldrich, Munich, Germany) for 20 minutes on ice. After centrifugation for 15 minutes at 20 000g (4°C), the protein content of the samples was determined according to the Bradford method. For immunoprecipitation, cells were lysed with RIPA buffer for 15 minutes on ice. Equal amounts of protein were loaded onto SDS-polyacrylamide gels and blotted onto polyvinylidene difluoride membranes. Western blots were performed using antibodies directed against HDAC5 (Cell Signaling Technology, Danvers, MA), topoisomerase Ia (Santa Cruz Biotechnology, Heidelberg, Germany), c-Myc (Santa Cruz Biotechnology), MEF2 (Santa Cruz Biotechnology), protein kinase D (Cell Signaling Technology), or tubulin-α (Lab Vision, Fremont, CA).
Spheroid-based angiogenesis assay
Endothelial cell spheroids of defined cell number were generated as described previously.30 Further details are provided in Document S1.
Scratched wound assay
Migration of HUVECs was detected using a “scratched wound assay.”31 Further details are provided in Document S1.
Cell-cycle analysis
Twenty-four hours after transfection, adherent cells were incubated with bromodeoxyuridine (BrdU; 10 μM). Cells were detached with trypsin, washed in phosphate-buffered saline, and incubated with 20 μL anti–BrdU-fluorescein isothiocyanate for 20 minutes and with 2.5 μL 7-amino-actinomycin D for 15 minutes according to the manufacturer's protocol (BrdU Flow Kit; BD Biosciences). Analysis was performed using a FACS SCAN flow cytometer and CellQuest software (BD Biosciences).
MTT viability assay
Assessment of cell viability was performed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. At 24 hours and 48 hours after transfection, 0.5 mg/mL MTT was added to each well, and cells were incubated for 4 hours at 37°C. Cells were washed with phosphate-buffered saline and lysed 30 minutes at room temperature with lysis buffer (40 nM HCl in isopropanol). Absorbance was photometrically measured at 550 nm.
Tube formation assay
Twenty-four hours after transfection, HUVECs (5 × 104) were cultured in a 12-well plate (Greiner Bio-One, Frickenhausen, Germany) coated with 200 μL Matrigel Basement Membrane Matrix (BD Biosciences). Tube length was quantified after 24 hours by measuring the cumulative tube length in 5 random microscopic fields with a computer-assisted microscope using the program KS300 3.0 (Carl Zeiss, Jena, Germany).
Matrigel plug assay
HUVECs transfected with siRNA against HDAC5 or scrambled siRNA were suspended in Matrigel, which was then subcutaneously injected into the backs of 8- to 12-week-old nude mice.32 After 7 days, blood vessel infiltration in Matrigel plugs was quantified by analysis of hematoxylin and eosin staining using microscopy. To analyze perfused capillaries, 200 μL fluorescein isothiocyanate–conjugated lectin (1 mg/mL; Sigma-Aldrich) was injected intravenously 30 minutes before the mice were killed. To visualize capillaries, plug sections were stained with an antibody against mouse CD31. Nuclei were stained with 4,6-diamidino-2-phenylindole. For hemoglobin analysis, the Matrigel plug was removed after 7 days and homogenized in 130 μL deionized water. After centrifugation, the supernatant was used in the Drabkin assay (Sigma-Aldrich) to measure hemoglobin concentration. Stock solutions of hemoglobin are used to generate a standard curve. Results are expressed relative to total protein in the supernatant. Animal studies were performed with the permission of the State of Hesse, Regierungspraesidium Darmstadt, and conform to the Guide for the Care and Use of Laboratory Animals.
RT-PCR
Total RNA was isolated using the RNeasy Mini Kit (QIAGEN); 2 μg RNA from each sample was reverse-transcribed (RT) into cDNA and subjected to conventional PCR using the oligonucleotide primers summarized in Table S2. Semiquantitative analysis was performed densitometrically using the Scion Image software (version 4.0.2; Scion, Frederick, MD).
Real-time PCR (Lightcycler)
Total RNA was isolated using the RNeasy Mini Kit; 2 μg RNA from each sample was reverse-transcribed into cDNA and subjected to real-time PCR using the LightCycler FastStart DNA MasterPlus SYBR Green I Kit (Roche Applied Science, Indianapolis, IN). Lightcycler reactions were performed with the LightCycler 1.2 System and LightCycler 3.5 quantification software (Roche Applied Science). Oligonucleotides used for real-time PCR are summarized in Table S3. All other oligonucleotide sequences are available on request.
Oligonucleotide array
Total RNA was isolated from 3 independently transfected HUVECs; 10 μg total RNA was hybridized to the HG U133 Plus 2.0 microarray (Affymetrix, Santa Clara, CA). The standard protocol used for sample preparation and microarray processing is available from Affymetrix. To detect differentially expressed genes, data analysis was performed as described previously.33 The microarray data have been deposited into the GEO database under accession number GSE15499.34
Boyden chamber migration
Transwell membranes (8 μm; Corning Life Sciences, Schiphol-Rijk, The Netherlands) were coated on both sides with fibronectin (2.5 μg/mL; Sigma-Aldrich) overnight at 4°C; 600 μL serum-free EBM with 1% bovine serum albumin containing VEGF, FGF2, or 600 μL conditioned medium of siRNA-transfected HUVECs were placed in the lower chambers. Then 5 × 104 HUVECs in 100 μL serum-free EBM containing 1% bovine serum albumin were incubated in the upper chamber at 37°C in 5% CO2 for 5 hours. Cells remaining on the upper surface of the filters were mechanically removed, and HUVECs that had migrated to the lower surface were fixed with 4% formaldehyde and counted in 5 fields using a fluorescence microscope (Axiovert 100; Carl Zeiss).
ChIP
HUVECs were harvested by scraping and cross-linked for 5 minutes by directly adding 1% formaldehyde to the culture medium. The fixed cells were washed and lysed for 5 minutes on ice (50 mM Tris pH 8.0, 2 mM ethylenediaminetetraacetic acid, 0.1% NP-40, 10% glycerol) to obtain cell nuclei. The nuclei were pelleted and lysed for another 5 minutes on ice (50 mM Tris pH 8.0, 5 mM ethylenediaminetetraacetic acid, 0.1% SDS). The chromatin was sheared by sonication (Branson Sonifier 450; Branson Ultrasonics, Danbury, CT). Sheared chromatin samples were taken as input control or used for immunoprecipitation with an antibody directed against c-Myc (Santa Cruz Biotechnology). After immunoprecipitation, the samples were washed 5 times and subjected to either Western blot analysis or PCR using primer pairs directed against promoter regions within the FGF2 or Slit2 genes (Table S4).
Statistical analysis
Data are mean plus or minus SEM. Two treatment groups were compared with the independent samples t test or Mann-Whitney U test, and 3 or more groups by one-way analysis of variance followed by post hoc analysis adjusted with a least significant difference correction for multiple comparisons (SPSS, Chicago, IL). Results were considered statistically significant when P value was less than .05.
Results
Effects of HDAC isoenzymes on in vitro angiogenesis
To investigate the individual contribution of class IIa HDAC isoenzymes to angiogenic functions of endothelial cells in vitro, we transfected HUVECs with siRNA oligonucleotides targeting the individual HDAC isoenzymes (Table S1). siRNA transfection caused a significant suppression of individual HDAC isoenzymes after 24 hours (Figures 1A, S1A). Silencing of the isoforms HDAC7 and HDAC9 blocked endothelial sprouting in a 3-dimensional spheroid assay and endothelial cell migration in a scratched wound assay (Figure 1B,C). In contrast, surprisingly, siRNA directed against HDAC5 profoundly increased sprout length, the number of branch points, and the number of sprouts per spheroid (Figures 1B, S2) and stimulated endothelial cell migration (Figure 1C). Because HDACs may affect cell cycle progression and cell death,35 we measured proliferation by BrdU incorporation and viability by MTT assay. However, transfection of siRNA against HDAC5, HDAC7, or HDAC9 did not significantly affect endothelial cell proliferation (Figure 1D). In addition, knockdown of HDAC5 and HDAC9 did not affect viability, whereas siRNA against HDAC7 slightly but significantly decreased viability after 48 hours (Figure 1E). To further investigate toxic effects of the siRNA transfection, we used 2 additional siRNA oligonucleotides (siRNA I and II) against HDAC5, HDAC7, and HDAC9. As shown in Figure S1, HDAC5 and HDAC9 siRNA did not affect endothelial cell viability, whereas siRNA against HDAC7 (siRNA II) significantly reduced cell viability after 48 hours. These data indicate that, among the class II HDACs, HDAC5 exhibits a specific and unique antiangiogenic activity without affecting proliferation or survival.
Role of class IIa HDAC isoenzymes for angiogenic function and cell-cycle progression in endothelial cells. (A-F) Effects of siRNA against individual class IIa HDAC isoenzymes in HUVECs on (A) HDAC mRNA expression as measured by quantitative PCR. Data (delta Ct) are mean ± SEM (n = 3, *P < .05 vs scrambled). (B) Capillary-like sprout formation from spheroid cultures (n ≥ 4; *P < .05). (C) Scratched wound endothelial cell migration (n ≥ 3; *P < .05). (D) Proliferation rate measured by flow cytometric detection of BrdU-labeled HUVECs in the S-phase (n = 7-11). (E) Viability measured by MTT assay (n = 3). (F) Tube formation in a Matrigel assay (n = 6; *P < .05). (G-K) Biochemical and histologic analysis of Matrigel plugs containing scrambled versus HDAC5 siRNA-transfected HUVECs at day 7 after subcutaneous implantation (n ≥ 3 per group). (G) Representative hematoxylin and eosin–stained plug sections. Pictures were taken using an Axiovert 100M microscope, an AxioCam camera, a Plan-NEOFLUAR 10×/0.30∞/0.17 objective, and the AxioVision Rel. 4.6.3 Sp1 software (Carl Zeiss). (H) Number of invaded cells per hematoxylin and eosin–stained plug section (*P < .05). (I) Number of CD31+ structures per plug section. (J) Number of lectin+ structures per plug section. (K) Hemoglobin content of Matrigel plugs (* P < .05).
Role of class IIa HDAC isoenzymes for angiogenic function and cell-cycle progression in endothelial cells. (A-F) Effects of siRNA against individual class IIa HDAC isoenzymes in HUVECs on (A) HDAC mRNA expression as measured by quantitative PCR. Data (delta Ct) are mean ± SEM (n = 3, *P < .05 vs scrambled). (B) Capillary-like sprout formation from spheroid cultures (n ≥ 4; *P < .05). (C) Scratched wound endothelial cell migration (n ≥ 3; *P < .05). (D) Proliferation rate measured by flow cytometric detection of BrdU-labeled HUVECs in the S-phase (n = 7-11). (E) Viability measured by MTT assay (n = 3). (F) Tube formation in a Matrigel assay (n = 6; *P < .05). (G-K) Biochemical and histologic analysis of Matrigel plugs containing scrambled versus HDAC5 siRNA-transfected HUVECs at day 7 after subcutaneous implantation (n ≥ 3 per group). (G) Representative hematoxylin and eosin–stained plug sections. Pictures were taken using an Axiovert 100M microscope, an AxioCam camera, a Plan-NEOFLUAR 10×/0.30∞/0.17 objective, and the AxioVision Rel. 4.6.3 Sp1 software (Carl Zeiss). (H) Number of invaded cells per hematoxylin and eosin–stained plug section (*P < .05). (I) Number of CD31+ structures per plug section. (J) Number of lectin+ structures per plug section. (K) Hemoglobin content of Matrigel plugs (* P < .05).
Antiangiogenic effect of HDAC5
Based on our finding that HDAC5 is a repressor of endothelial cell migration and sprouting, we focused our following study on the role of HDAC5 for angiogenesis in vitro and vivo. Transfection of endothelial cells with 2 different siRNA oligonucleotides against HDAC5 profoundly decreased HDAC5 protein expression (Figure S3). To confirm the proangiogenic effect of HDAC5 silencing, we used an additional siRNA-targeting HDAC5 and demonstrated that the second oligonucleotide sequence also effectively increased capillary sprout length (Figure S4). Moreover, HDAC5 siRNA also significantly enhanced tube formation in an additional in vitro angiogenesis assay (Figure 1F). To determine whether HDAC5 silencing also increase angiogenesis in vivo, HDAC5 siRNA-transfected HUVECs were mixed with Matrigel and subcutaneously implanted in nude mice in vivo. After 7 days, the number of invaded cells as determined by hematoxylin and eosin staining was significantly increased in HDAC5 siRNA-treated Matrigel plugs compared with scrambled controls (Figure 1G,H). To analyze vascularization, we stained the Matrigel plugs for mouse CD31 and detected fluorescent lectin, which was intravenously infused 30 minutes before the mice were killed. As shown in Figure 1I and J, the number of CD31+ structures as well as the number of lectin+ structures were significantly enhanced in response to HDAC5 siRNA. Moreover, the hemoglobin content, indicative of Matrigel plug perfusion, was significantly augmented in HDAC5-siRNA plugs (Figure 1K). Together, these data document that HDAC5 is a negative regulator of angiogenesis in vitro and in vivo.
Mechanism underlying the antiangiogenic function of HDAC5
To analyze the mechanism underlying the antiangiogenic function of HDAC5, we transfected endothelial cells with plasmids encoding wild-type HDAC5 (HDAC5 wt) or different mutants (Figure 2A). The expression of HDAC5 wt and the HDAC5 mutants is shown by RT-PCR (Figure 2B) and Western blot (Figure 2C). Overexpression of HDAC5 wt significantly inhibited sprout length (Figure 2D). Moreover, transfection of an HDAC5 S259/498A mutant, which is preferentially localized in the nucleus,36 significantly suppressed sprout length (Figures 2D, S5b), suggesting that the nuclear localization of HDAC5 is important for its repressive effect. In addition, overexpression of HDAC5 wt and the HDAC5 S259/498A mutant profoundly decreased the number of branch points and the number of sprouts per spheroid (Figure S5). PKD is known to phosphorylate HDAC5 leading to its nuclear export.37 Therefore, as a control, we cotransfected endothelial cells with constitutively active PKD and HDAC5 wt or HDAC5 S259/498A mutant (Figure S6). PKD overexpression reversed the repressive effect of HDAC5 wt on endothelial cell sprouting (Figure S6). In contrast, overexpression of the nonphosphorylatable HDAC5 mutant is refractory to PKD and still repressed sprouting (Figure S6). Because MEF2 interacts with HDAC5 in cardiac myocytes and is also crucial for the embryonic development of the vasculature38 as well as for postnatal endothelial cell signaling,39 we tested whether MEF2 is required for HDAC5 function in endothelial cells. Therefore, cells were transfected with an HDAC5 mutant construct (HDAC5 ΔMEF2), which does not bind to MEF2 as shown by immunoprecipitation (Figure 2E). Transfection of endothelial cells with HDAC5 ΔMEF2 still significantly decreased sprout length (Figure 2D), indicating that binding of HDAC5 to MEF2 is dispensable for the repressive effect of HDAC5 on endothelial cell sprouting. Having shown that the nuclear localization of HDAC5 is important and MEF2 is not required for its function, we investigated whether the enzymatic activity of HDAC5 is involved in the antiangiogenic effect. Transfection of an HDAC5 mutant lacking the deacetylase domain (HDAC5 1-767aa28 ) also significantly diminished sprout formation (Figure 2D), suggesting that the repressive effect of HDAC5 is independent of its deacetylase activity and is mediated by the N-terminal part of the protein.
Mechanism underlying the antiangiogenic function of HDAC5. (A) Scheme of different HDAC5 constructs. (B) RT-PCR analysis of HUVECs overexpressing different HDAC5 constructs after 14 hours. (C) Western blot analysis of HUVECs overexpressing different HDAC5 constructs after 14 hours. (D) Capillary-like sprout formation from spheroid cultures of HUVECs overexpressing different HDAC5 constructs (n = 3-10; *P < .05 vs mock). (E) Lysates of HUVECs overexpressing different HDAC5 constructs were immunoprecipitated with an antibody against c-myc. Western blots were performed with an anti-c-myc or an anti-MEF2 antibody in whole cell lysates or after immunoprecipitation. Ab indicates antibody control.
Mechanism underlying the antiangiogenic function of HDAC5. (A) Scheme of different HDAC5 constructs. (B) RT-PCR analysis of HUVECs overexpressing different HDAC5 constructs after 14 hours. (C) Western blot analysis of HUVECs overexpressing different HDAC5 constructs after 14 hours. (D) Capillary-like sprout formation from spheroid cultures of HUVECs overexpressing different HDAC5 constructs (n = 3-10; *P < .05 vs mock). (E) Lysates of HUVECs overexpressing different HDAC5 constructs were immunoprecipitated with an antibody against c-myc. Western blots were performed with an anti-c-myc or an anti-MEF2 antibody in whole cell lysates or after immunoprecipitation. Ab indicates antibody control.
Identification of target genes regulated by HDAC5 in endothelial cells
To identify target genes regulated by HDAC5 in endothelial cells, we profiled the transcriptome of cells transfected with HDAC5 siRNA. Analysis of the different HDAC isoenzymes revealed that the siRNA against HDAC5 used for this experiment selectively suppressed HDAC5 but none of the other class IIa HDAC isoforms (Figure S7). Bioinformatic analysis showed that 2.0% of analyzed genes were up-regulated and 1.1% were down-regulated in response to HDAC5 siRNA (> 1.5-fold vs scrambled siRNA, 3P level indicating relevant expression; Figure 3A). Among the significantly regulated genes with altered RNA levels were genes involved in angiogenesis, including angiogenic growth factors and receptors (eg, FGF2, neuropilin 2, VEGFR2, TGFBR2), guidance molecules (eg, Slit2), and homeodomain transcription factors (eg, HOXA9; Figure 3B). The expression of selected angiogenesis-related genes was further analyzed by real-time PCR. As shown in Figure 3C, FGF2, Slit2, and EphB4 were time-dependently, significantly up-regulated in HDAC5 siRNA-transfected HUVECs. Moreover, FGF2 and Slit2 mRNA expression was increased in vivo in Matrigel plugs containing HUVECs transfected with HDAC5 siRNA compared with scrambled-transfected cells (data not shown). In contrast, other genes involved in angiogenesis, such as the VEGF isoforms A to D, angiopoietins, neuropilin 1, TGFBR1, and TGFBR3, were not significantly regulated in response to HDAC5 inhibition (Figures S8, S9).
Identification of target genes regulated by HDAC5 in endothelial cells. (A) Bioinformatic analysis of the RNA profile of HUVECs transfected with HDAC5 siRNA after 24 hours (n = 3 each). Up- and down-regulated genes in response to HDAC5 siRNA versus scrambled siRNA as a proportion of all analyzed genes are shown in a gene tree analysis. The color scale is shown on the right. The brightness indicates the trust. Blue represents low expression; red, high expression. (B) Summary of selected genes relevant to angiogenesis, which were up-regulated in response to HDAC5 siRNA transfection in the oligonucleotide array. *In case of 2 different oligonucleotides representing one gene, the data were pooled. Because we focused in our following study on Slit2 and FGF2, both targets are highlighted in gray. (C) Quantitative analysis of HDAC5 and selected target gene expression in response to HDAC5 siRNA versus scrambled siRNA after 24 hours and 48 hours using real-time PCR (n = 3; *P < .05 vs scrambled).
Identification of target genes regulated by HDAC5 in endothelial cells. (A) Bioinformatic analysis of the RNA profile of HUVECs transfected with HDAC5 siRNA after 24 hours (n = 3 each). Up- and down-regulated genes in response to HDAC5 siRNA versus scrambled siRNA as a proportion of all analyzed genes are shown in a gene tree analysis. The color scale is shown on the right. The brightness indicates the trust. Blue represents low expression; red, high expression. (B) Summary of selected genes relevant to angiogenesis, which were up-regulated in response to HDAC5 siRNA transfection in the oligonucleotide array. *In case of 2 different oligonucleotides representing one gene, the data were pooled. Because we focused in our following study on Slit2 and FGF2, both targets are highlighted in gray. (C) Quantitative analysis of HDAC5 and selected target gene expression in response to HDAC5 siRNA versus scrambled siRNA after 24 hours and 48 hours using real-time PCR (n = 3; *P < .05 vs scrambled).
Contribution of FGF2 and Slit2 regulation for the repressive effect of HDAC5
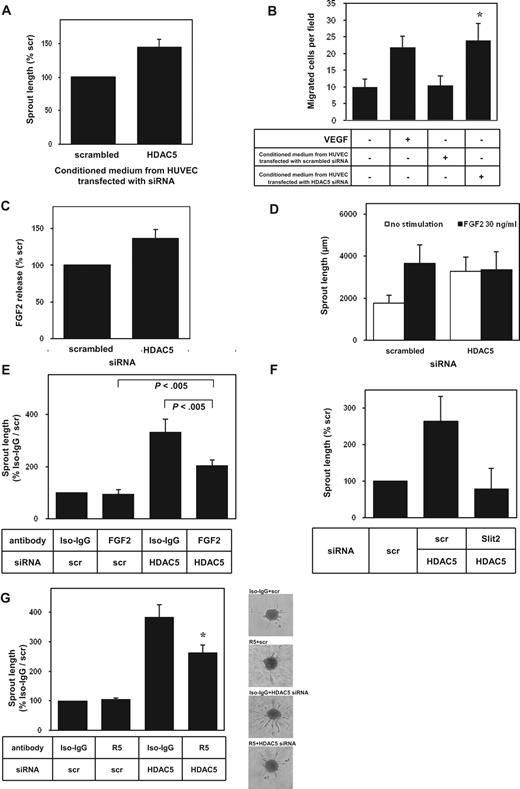
Because HDAC5 siRNA induced the expression of several secreted gene products known for their proangiogenic effects, such as FGF2 or Slit2, we investigated whether the secretion of angiogenic factors might contribute to the proangiogenic effect of HDAC5 silencing. Indeed, incubation of endothelial cells with conditioned medium from HDAC5 siRNA-transfected HUVECs increased capillary-like sprout formation (Figure 4A) and stimulated endothelial cell migration in a Boyden chamber assay (Figure 4B), indicating that HDAC5 silencing derepresses secreted proangiogenic and chemoattractive stimuli for endothelial cells.
Role of HDAC5 for sprout formation and secretion of angiogenic factors. (A) Sprout formation from endothelial cell spheroid cultures incubated with conditioned medium from HUVECs transfected with scrambled siRNA or HDAC5 siRNA for 42 hours (n = 9). (B) Boyden chamber migration of nontransfected HUVECs toward a conditioned medium. The medium was derived from HUVECs transfected with scrambled siRNA or HDAC5 siRNA for 48 hours. The number of migrated cells per field after 5 hours is given. Addition of exogenous VEGF (50 ng/mL, 5 hours) served as control. *P < .05 versus conditioned medium of scrambled siRNA-transfected HUVECs (n = 4). (C) Enzyme-linked immunosorbent assay measurement of FGF2 release into medium incubated for 2 days with HUVECs transfected with siRNA against HDAC5 (n = 4). (D) Capillary-like sprout formation from HUVEC spheroid cultures stimulated with exogenous FGF2 (30 ng/mL) after transfection of scrambled siRNA or HDAC5 siRNA for 24 hours (n = 4). (E) Capillary-like sprout formation from HUVEC spheroid cultures in the presence of a neutralizing FGF2 antibody (4 μg/mL) or IgG control after transfection of scrambled siRNA or HDAC5 siRNA for 24 hours (n = 5-7). (F) Sprout formation from spheroid cultures of scrambled versus HDAC5 siRNA-transfected HUVECs cotransfected with either of 2 independent Slit2 siRNA oligonucleotides (n = 2). (G) Capillary-like sprout formation from HUVEC spheroid cultures in the presence of R5, an IgG2b monoclonal antibody to the first immunoglobulin domain of Robo1, which neutralizes activation by Slit2, or 12CA5 IgG control antibody with no antagonistic effect at the Robo1 receptor after transfection of scrambled siRNA or HDAC5 siRNA for 24 hours. *P < .05 versus HDAC5 siRNA with 12CA5 IgG control antibody (n = 4-10). Representative images are shown on the right. Pictures were taken using an Axiovert 100M microscope, an AxioCam camera, a Plan-NEOFLUAR 10×/0.30∞/0.17 objective, and the AxioVision Rel. 4.6.3 Sp1 software (Carl Zeiss).
Role of HDAC5 for sprout formation and secretion of angiogenic factors. (A) Sprout formation from endothelial cell spheroid cultures incubated with conditioned medium from HUVECs transfected with scrambled siRNA or HDAC5 siRNA for 42 hours (n = 9). (B) Boyden chamber migration of nontransfected HUVECs toward a conditioned medium. The medium was derived from HUVECs transfected with scrambled siRNA or HDAC5 siRNA for 48 hours. The number of migrated cells per field after 5 hours is given. Addition of exogenous VEGF (50 ng/mL, 5 hours) served as control. *P < .05 versus conditioned medium of scrambled siRNA-transfected HUVECs (n = 4). (C) Enzyme-linked immunosorbent assay measurement of FGF2 release into medium incubated for 2 days with HUVECs transfected with siRNA against HDAC5 (n = 4). (D) Capillary-like sprout formation from HUVEC spheroid cultures stimulated with exogenous FGF2 (30 ng/mL) after transfection of scrambled siRNA or HDAC5 siRNA for 24 hours (n = 4). (E) Capillary-like sprout formation from HUVEC spheroid cultures in the presence of a neutralizing FGF2 antibody (4 μg/mL) or IgG control after transfection of scrambled siRNA or HDAC5 siRNA for 24 hours (n = 5-7). (F) Sprout formation from spheroid cultures of scrambled versus HDAC5 siRNA-transfected HUVECs cotransfected with either of 2 independent Slit2 siRNA oligonucleotides (n = 2). (G) Capillary-like sprout formation from HUVEC spheroid cultures in the presence of R5, an IgG2b monoclonal antibody to the first immunoglobulin domain of Robo1, which neutralizes activation by Slit2, or 12CA5 IgG control antibody with no antagonistic effect at the Robo1 receptor after transfection of scrambled siRNA or HDAC5 siRNA for 24 hours. *P < .05 versus HDAC5 siRNA with 12CA5 IgG control antibody (n = 4-10). Representative images are shown on the right. Pictures were taken using an Axiovert 100M microscope, an AxioCam camera, a Plan-NEOFLUAR 10×/0.30∞/0.17 objective, and the AxioVision Rel. 4.6.3 Sp1 software (Carl Zeiss).
Because FGF2 is a secreted gene product repressed by HDAC5, which stimulates angiogenesis in vivo,16 we assessed the functional contribution of FGF2 regulation for the repressive effect of HDAC5. Measurements of FGF2 using enzyme-linked immunosorbent assay revealed that the up-regulation of FGF2 mRNA is associated with an increased release of FGF2 protein (scrambled 24 ± 1 pg/mL vs HDAC5 siRNA 33 ± 4 pg/mL and Figure 4C). Supplementation of spheroid cultures with exogenous FGF2 stimulates sprouting under control conditions, whereas increased sprout formation in HUVECs transfected with HDAC5 siRNA was not further augmented by the addition of FGF2 (Figure 4D). To test whether an increased FGF2 secretion essentially underlies the observed increase in capillary-like sprout formation in HDAC5 siRNA-transfected HUVECs, secreted FGF2 was blocked by a neutralizing antibody.30 HDAC5 siRNA-enhanced sprouting was significantly reduced when FGF2 was neutralized (Figure 4E). Nevertheless, in the presence of the neutralizing antibody against FGF2, HDAC5 suppression still increased sprouting compared with control albeit at a lower level (Figure 4E). Thus, we conclude that FGF2 release contributes to increased sprout formation in response to HDAC5 siRNA, whereas a part of the effect of HDAC5 siRNA is mediated via FGF2-independent mechanisms, suggesting the contribution of additional effector genes. Beyond FGF2, HDAC5 siRNA modulated the expression of the axon guidance factor Slit2 (Figure 3B,C). So-called guidance molecules, known for their role in neuronal wiring, have recently been identified as essential regulators of capillary sprout formation.17 These factors control the guided and directional movement of specialized endothelial cells situated at the tips of the vascular sprouts similar to axonal growth cones.18 Guidance factors are families of surface membrane receptors and their ligands. The secreted ligand Slit2 acts as a guidance cue in cellular migration and promotes tube formation and tumor vascularization via its receptor Roundabout 1 (Robo1),24 whereas another receptor Robo4 was shown to block angiogenesis40 but stabilizes the vasculature in vivo.41 Because Slit2 is a ligand for Robo1, which is expressed in HUVECs (0.91 ± 0.05 normalized expression, Affymetrix oligonucleotide microarray), we assessed the contribution of Slit2 in HDAC5-regulated endothelial cell sprouting. siRNA oligonucleotides against Slit2 inhibited the increase in sprout formation in HUVECs cotransfected with HDAC5 siRNA (Figure 4F). To assess whether signaling via the Slit2 receptor Robo1 mediates the HDAC5 siRNA-induced increase in sprout formation, we used receptor antagonists for Robo1-mediated Slit2 signaling.24 Robo1 neutralization with R5, an IgG2b monoclonal antibody to the first immunoglobulin domain of Robo1, significantly reduced the capacity of HDAC5 siRNA to increase sprout formation (Figure 4G). Likewise, another antagonist of Robo1-mediated signaling, Robo1-Fc, also reduced the HDAC5 siRNA-induced increase in sprout formation by 57% plus or minus 25%. Of note, the inhibition of Robo1 activation with either of the 2 antagonists only partially suppressed, whereas Slit2 siRNA entirely blocked HDAC5-siRNA mediated increase in sprout formation, indicating that Slit2 exhibits effects beyond Robo1 activation.
Mechanism of the regulation of FGF2 and Slit2 by HDAC5
Having identified FGF2 and Slit2 as HDAC5 targets relevant for the suppression of angiogenic endothelial cell activity by HDAC5, we investigated the mechanism by which HDAC5 controls FGF2 and Slit2 expression. Therefore, we performed chromatin immunoprecipitation (ChIP) assays in HUVECs overexpressing HDAC5 wt and the nuclear mutant (S259/498A) to investigate the binding of HDAC5 to the promoter of FGF2 and Slit2 (Figure 5A). The immunoprecipitation of HDAC5 wt and the S259/498A mutant is shown in Figure 5B. The nuclear localized HDAC5 S259/498A mutant, and, to a minor extent, the HDAC5 wt was bound to the promoter of FGF2 and Slit2 (Figure 5C). The quantitative analysis of HDAC5 binding to the FGF2 and Slit2 promoter regions is shown by quantitative PCR in Figure S10. Consistent with the ChIP analysis, HDAC5 wt as well as the nuclear localized HDAC5 mutant decreased the expression of FGF2 and Slit2 (Figure 5D,E).
Binding of HDAC5 to the promoter of FGF2 and Slit2. (A) Overview of primer pairs amplifying the indicated promoter regions relative to the transcription start within the FGF2 and Slit2 genes. (B) Western blot analysis was performed to confirm the immunoprecipitation of myc-tagged overexpressed HDAC5 wt and HDAC5 S259/498A mutant. (C) The ChIP was performed in HUVECs overexpressing myc-tagged HDAC5 wt or HDAC5 S259/498A mutant using an antibody against c-myc. Representative PCR with primers detecting the indicated promoter regions of the FGF2 and Slit2 genes are shown (n = 3). Ab indicates antibody control. (D) Expression of FGF2 mRNA in HUVECs transfected with HDAC5 wt or HDAC5 S259/498A mutant after 48 hours (n = 4). (E) Expression of Slit2 mRNA in HUVECs transfected with HDAC5 wt or HDAC5 S259/498A mutant after 48 hours (n = 3).
Binding of HDAC5 to the promoter of FGF2 and Slit2. (A) Overview of primer pairs amplifying the indicated promoter regions relative to the transcription start within the FGF2 and Slit2 genes. (B) Western blot analysis was performed to confirm the immunoprecipitation of myc-tagged overexpressed HDAC5 wt and HDAC5 S259/498A mutant. (C) The ChIP was performed in HUVECs overexpressing myc-tagged HDAC5 wt or HDAC5 S259/498A mutant using an antibody against c-myc. Representative PCR with primers detecting the indicated promoter regions of the FGF2 and Slit2 genes are shown (n = 3). Ab indicates antibody control. (D) Expression of FGF2 mRNA in HUVECs transfected with HDAC5 wt or HDAC5 S259/498A mutant after 48 hours (n = 4). (E) Expression of Slit2 mRNA in HUVECs transfected with HDAC5 wt or HDAC5 S259/498A mutant after 48 hours (n = 3).
Discussion
Based on the cumulative evidence for an essential requirement of class I and II histone deacetylase enzymatic activity for angiogenic functions of endothelial cells, we focused our interest on the contribution of the individual mammalian HDAC isoenzymes to angiogenic signaling in endothelial cells. Our data show that, in contrast to other HDAC isoforms required for angiogenesis, HDAC5 is a negative regulator of angiogenic functions in endothelial cells. This is consistent with a study by Ha et al, which was published during the preparation of the manuscript, showing that VEGF-induced angiogenesis depends on the nuclear export of HDAC5.9 The present study, for the first time, describes the HDAC5-dependent transcriptional profile in endothelial cells and provides novel insights into the HDAC5-dependent regulation of genes, which are well known to control angiogenesis. From the list of putative targets obtained by the expression array, we here confirmed the regulation of FGF2, a potent and well-established pro-angiogenic growth factor, and the guidance factor Slit2. Blockade of FGF2 and Slit2 both significantly reduced the HDAC5 siRNA-mediated up-regulation of sprout formation. In addition, the increase of other axon guidance factors, such as the receptor EphB4, or homeobox genes, such as HOXA9, which are crucial for migration and angiogenesis,20,42,43 may contribute to the pro-migratory effect observed in HDAC5-siRNA-treated endothelial cells. In addition, we found that silencing of the other class IIa HDAC isoforms HDAC7 and HDAC9 blocked angiogenesis. The essential requirement of HDAC7 for angiogenesis shown in this study is consistent with a previous report demonstrating an embryonic lethal phenotype of HDAC7-deficient mice.6 However, the role of HDAC9 for angiogenesis has not yet been addressed. Thus, this study describes a novel proangiogenic function of HDAC9 in endothelial cells.
Having shown that HDAC5 is a repressor of angiogenic gene expression in endothelial cells, we addressed the underlying mechanism and hypothesized that HDAC5 may exert its function in the nucleus. Indeed, the repressive function of HDAC5 required its nuclear localization. A recent study demonstrated that nitric oxide stimulates nuclear shuttling of HDAC4 and HDAC5 and deacetylation in endothelial cells.44 Because nitric oxide exerts pro- or antiangiogenic activities depending on the concentration,45–48 the findings of Illi et al44 do not argue against the requirement of a nuclear localization for the antiangiogenic effect of HDAC5. Consistently, preferentially nuclear localized HDAC5 was shown to bind to the promoters of the 2 novel targets identified in the present study, namely, FGF2 and Slit2. However, it is known that class II HDACs do not directly bind DNA; therefore, the question remains how HDAC5 can bind to the FGF2 and Slit2 promoter. Using the ChIP assay, we cannot distinguish between a direct and indirect binding to the respective promoters. Thus, we hypothesize that HDAC5 is indirectly associated with the FGF2 and Slit2 promoter, eg, via binding to transcription factors. The regulation of endothelial cell functions by HDAC7 has been attributed to the repression of the transcription factor MEF2.6 HDAC5 also is well established to bind MEF2 and inhibit MEF2-dependent transcription in muscle differentiation.36,49 However, the antiangiogenic function of HDAC5 in endothelial cells is independent of MEF2 binding because a mutant unable to bind MEF2 still efficiently blocked endothelial sprout formation. Recently, Ha et al have shown that the nuclear export of HDAC5 regulates VEGF-induced MEF2 transcriptional activation.9 Based on our data, we cannot exclude an HDAC5-dependent mechanism involving MEF2 transcriptional regulation.
The repressive function of HDAC5 was also independent of the direct deacetylase activity of the enzyme. These data do not exclude a role of deacetylation for the antiangiogenic effect of HDAC5 because class II HDACs can interact with other HDAC isoenzymes (class I and II). Because broad-spectrum HDAC inhibitors block angiogenesis, one would expect a reduction of angiogenesis rather than a stimulation of angiogenesis in response to HDAC5 silencing if other HDACs might contribute to HDAC5 function. In general, class II HDACs can interact with a variety of proteins, for example, by recruiting a multiprotein complex containing the transcriptional corepressors SMRT (silencing mediator for retinoic acid receptor and thyroid-hormone receptor) and N-CoR (nuclear hormone receptor corepressor) and HDAC3 to their C-terminus as it has been shown for HDAC4 and HDAC7, respectively.50,51 In addition, the C-terminus of HDAC4 and HDAC5 interacts with Smad3, which leads to the transcriptional repression of Runx2 during osteoblast differentiation.52 However, in our study, the deacetylase-deficient HDAC5 mutant (1-767 aa), which lacks the C-terminus, still repressed endothelial cell sprouting, indicating that other mechanisms account for the repressive effects of HDAC5 on angiogenesis signaling. Indeed, the N-terminal part of class IIa HDACs might also repress transcriptional activity through the interaction with corepressors, such as HP1 or CtBP.53–55 Thus, further studies are required to address the specific underlying mechanism of HDAC5 function in endothelial cells.
The identification of a specific control of angiogenesis in vitro and in vivo by HDAC5 may have potential therapeutic implications. One may consider using HDAC5 inhibitors to improve therapeutic angiogenesis (eg, after ischemia) or to use HDAC5 activators to block pathologic angiogenesis. Interestingly, a recent publication demonstrates that inhibition of HDACs increases dendritic sprouting, learning, and memory.56 A putative regulation of axon guidance molecules by HDAC5, if confirmed in neuronal cells, may also have an impact on neuronal cell functions.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dorit Lüthje and Nicole Konecny for their expert technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (grant SFB/TR 23, subproject B5, C.U.; grant PO1306/1-1, M.P.; and grant Exc 147/1 Excellence Cluster Cardiopulmonary System), and by the Deutsche Krebshilfe (grant no. 107154; L.R.).
Authorship
Contribution: C.U. and L.R. designed research, collected data, and wrote the paper; D.K. performed research and collected and analyzed data; M.P. designed research and edited the paper; J.-N.B. performed research; A.K. and F.D. performed research and collected data; J.-G.G. provided antibodies and designed research; W.-K.H. collected and analyzed data; A.M.Z. wrote and edited the paper; and S.D. designed research and wrote and edited the paper. All authors checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefanie Dimmeler, Molecular Cardiology, Department of Internal Medicine III, University of Frankfurt, Theodor Stern-Kai 7, 60590 Frankfurt, Germany; e-mail: dimmeler@em.uni-frankfurt.de.
References
Author notes
*C.U. and L.R. contributed equally to this study.