In this issue of Blood, Grubor and colleagues investigate genomic changes in a large series of patients with CLL. They report novel genetic lesions, ascertain the frequency of losses and gains of genomic areas with greater accuracy, track down candidate genes, and analyze intraclonal heterogeneity.
Despite its phenotypic homogeneity, chronic lymphocytic leukemia (CLL) is clinically heterogeneous. Some patients have an aggressive disease and a poor prognosis, others an indolent course and a virtually normal life expectancy.1 The possibility of identifying CLL patients at diagnosis who harbor lesions known to be associated with poor prognosis would offer an opportunity for selective and more effective treatment for some while sparing others from toxic effects of therapy. This has led to a quest for biologically defined prognostic factors, the most widely used being scoring of the mutational status of immunoglobulin (IG)VH genes, expression of surface CD38 or cytoplasmic ZAP70, and fluorescent in situ hybridization (FISH) analysis for specific chromosomal lesions.2 FISH and comparative genomic hybridization (CGH) have clearly documented the clinical importance of the genetic subtyping of CLL.3 With FISH or CGH as compared to conventional cytogenetics, a much higher frequency of CLL patients have been found to have recurrent abnormalities, especially deletions (eg 13q14.2, 11q22-q23, 17p13) while DNA segment gains (such as trisomy 12) are less frequent.1 Still, with both FISH and conventional CGH platforms 15% to 20% of samples do not have detectable abnormalities.
Here begins the relevance of the findings described by Grubor et al, who use Representational Oligonucleotide Microarray Analysis (ROMA),4 a form of CGH that allows the examination of genomic changes in tumor samples at an unprecedented resolution. The power of ROMA technology is in the numbers, as only 1.7% (1/58) samples did not have detectable lesions. The implication is that ROMA brings to light lesions too fine to be identified with cytogenetics/FISH or lower- resolution CGH technologies. For example, a lesion as small as 20 kb could be identified. While an increased resolution is expected to yield a more accurate definition of genetic lesions, it also has an inherent risk—which is irrelevant at lower resolution—that is, to confuse the normal copy number variation of genome that frequently occurs in the human gene pool with leukemia-associated abnormalities. To safeguard against this problem, the authors have obtained DNA from both CLL cells and neutrophils from each patient, providing an internal normal genome control for comparison with the tumor cells.
By increasing the sensitivity to an unprecedented magnitude, Grubor et al have observed a changing landscape and documented not only far more cases with genetic lesions than hitherto described but also far more lesions per individual case. At least 1 genomic lesion was observed in virtually every CLL sample examined, the boundaries of known events were observed with greater clarity, and previously undescribed lesions were reported, with some being recurrent. All previously reported major cytogenetic abnormalities except one (6q21) were validated. Further, it was confirmed that the majority of CLL genetic lesions are deletions and not amplifications Newly identified abnormal regions are also reported, and span genes of considerable interest because of their potential influence on the disease history. Finally, the authors have taken advantage of the surface expression of CD38 by a proportion of circulating CLL cells5 to separate CD38+and CD38− fractions from individual patients and to ROMA profile the 2 different cell fractions. The demonstration that in 3 of 4 cases intraclonal diversification had indeed occurred indicates the existence of distinct subclones that may have implication for future investigations.
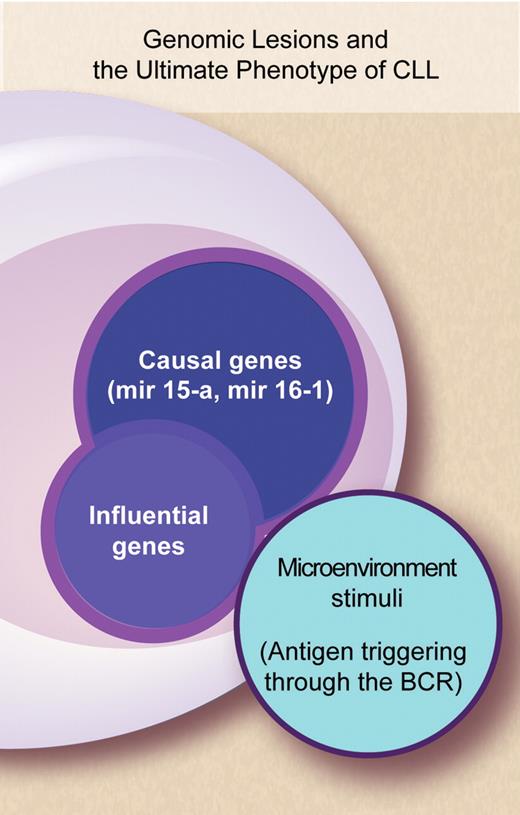
Cytogenetic studies paved the way to investigating which genes might be CLL-specific and led to the discovery that in CLL microRNA (miRNA) genes reside in chromosomal hot spots.6 The emerging view is that mir15a and mir161—which are located at 13q14.3, the most frequent deletion in CLL—are among the likely “causal genes” of CLL.7,8 Still the use of ROMA confirms that our knowledge of the genomic abnormalities that occur in CLL is far from satisfactory. The ultimate phenotype of individual patients—their clinical presentation and natural history—conceivably results from the interplay of a number of genes and of microenvironmental factors whose complexity we are just starting to unravel. It is plausible to consider that besides “causal genes” a number of other “influential genes” may work and that the products of all these genes may interact with the microenvironment and concur to shape the natural history of CLL (see figure). New technologies such as ROMA are leading toward a proper dissection of which gene lesions are involved in the natural history of CLL, to what extent they are implicated, what is their clinical relevance, and whether the proteins they encode may be candidates for new treatment approaches.
Complex interactions between genes and microenvironment concur to shape the phenotype of CLL patients—their clinical presentation and natural history. Professional illustration by Debra T. Dartez.
Complex interactions between genes and microenvironment concur to shape the phenotype of CLL patients—their clinical presentation and natural history. Professional illustration by Debra T. Dartez.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal