Abstract
High levels of aldehyde dehydrogenase (ALDH) activity have been proposed to be a common feature of stem cells. Adult hematopoietic, neural, and cancer stem cells have all been reported to have high ALDH activity, detected using Aldefluor, a fluorogenic substrate for ALDH. This activity has been attributed to Aldh1a1, an enzyme that is expressed at high levels in stem cells and that has been suggested to regulate stem cell function. Nonetheless, Aldh1a1 function in stem cells has never been tested genetically. We observed that Aldh1a1 was preferentially expressed in mouse hematopoietic stem cells (HSCs) and expression increased with age. Hematopoietic cells from Aldh1a1-deficient mice exhibited increased sensitivity to cyclophosphamide in a non–cell-autonomous manner, consistent with its role in cyclophosphamide metabolism in the liver. However, Aldh1a1 deficiency did not affect hematopoiesis, HSC function, or the capacity to reconstitute irradiated recipients in young or old adult mice. Aldh1a1 deficiency also did not affect Aldefluor staining of hematopoietic cells. Finally, Aldh1a1 deficiency did not affect the function of stem cells from the adult central or peripheral nervous systems. Aldh1a1 is not a critical regulator of adult stem cell function or Aldefluor staining in mice.
Introduction
Aldehyde dehydrogenase (ALDH) enzymes regulate retinoic acid biosynthesis, the clearance of toxic byproducts of reactive oxygen species (ROS) as well as other processes relevant to stem cell regulation. A large family of genes (20 in mice) encode ALDHs that oxidize of a variety of endogenous and exogenous aldehydes. The retinaldehyde dehydrogenase (RALDH) subfamily of ALDHs, composed of Aldh1a1, Aldh1a2, Aldh1a3, and Aldh8a1, regulate development by catalyzing retinoic acid biosynthesis.1-3 Other ALDHs can metabolize a wide range of aldehydes including the active derivative of cyclophosphamide (CY)4-7 and lipid byproducts of ROS.8,9
A variety of stem cells have been isolated based on their unusually high levels of ALDH activity. Levels of ALDH activity in cells can be inferred using Bodipy-aminoacetaldehyde (more commonly known by its commercial name Aldefluor), a fluorogenic ALDH substrate that can be taken up by live cells and measured by flow cytometry.10 Human hematopoietic stem cells (HSCs) are enriched among cells with high levels of ALDH activity.11,12 Mouse HSCs have also been isolated by the selection of cells with high levels of ALDH activity,13,14 though the utility of Aldefluor for the isolation of mouse HSCs is controversial as another study has reported mouse HSCs do not exhibit high levels of Aldefluor staining.12 Aldefluor staining has also been reported to enrich leukemic stem cells from some patients,15 human breast epithelial stem cells and breast cancer stem cells,16 and mouse neural stem cells.17 Indeed, it has been proposed that high ALDH activity might be a general marker for normal and malignant stem cells.16
The high levels of ALDH activity in stem cells have been ascribed to Aldh1a1 expression. Immunohistochemistry performed on normal human breast epithelium and breast cancer found that antibody staining for Aldh1a1 correlated with Aldefluor activity.16 Gene expression profiling of human18 and mouse HSCs19,20 has revealed that Aldh1a1 is expressed at higher levels in HSCs compared with other hematopoietic cells. Human hematopoietic progenitors expand in number and delay differentiation in culture after treatment with a chemical inhibitor of ALDHs, diethylaminobenzaldehyde (DEAB). This has been suggested to reflect a role for Aldh1a1 in the regulation of HSC differentiation.21 Over-expression of Aldh1a1 in hematopoietic cells confers cyclophosphamide resistance.6 As a result of these studies, it has been suggested that Aldh1a1 may be an important regulator of stem cell function and the main determinant of ALDH activity (Aldefluor staining) in stem cells. Nonetheless, other ALDHs are also expressed in stem cells,19,20,22 and the role of individual ALDHs in the regulation of stem cell function and ALDH activity has never been tested.
We have found that Aldh1a1 expression is elevated in mouse HSCs and that expression increases dramatically with age. To test whether Aldh1a1 is required for stem cell function, we examined Aldh1a1-deficient mice. Aldh1a1-deficient mice were constructed by targeted deletion of exon 11, which encodes the substrate binding and tetramerization domains critical for all known Aldh1a1 functions.23 This mutation eliminates detectable Aldh1a1 protein expression and dramatically reduces retinoic acid biosynthesis in the liver.23 Mating Aldh1a1-deficient mice with mice that are deficient for Aldh1a2 and Aldh1a3 completely eliminates all retinoic acid synthesis in the E10.5 dorsal retina, demonstrating that the mutant Aldh1a1 allele has a complete loss of function with respect to retinoic acid biosynthesis.24 Nonetheless, the Aldh1a1-deficient mice are born in normal numbers, are fertile, and have an apparently normal lifespan.23 We have examined hematopoiesis, HSC function, and neural stem cell function in young and old adult Aldh1a1-deficient mice. As expected, Aldh1a1 non–cell-autonomously regulated the sensitivity of hematopoietic progenitors to cyclophosphamide. However, we were unable to detect any hematopoietic or neural stem cell defects in Aldh1a1-deficient mice. Aldh1a1 deficiency also did not affect Aldefluor staining of HSCs, and mouse HSCs could not be distinguished from other bone marrow cells based on Aldefluor staining. Aldh1a1 is therefore not a major determinant of ALDH activity in mouse HSCs, is dispensable for hematopoietic and neural stem cell function, and does not affect the maintenance of these cells during aging.
Methods
Mice and reagents
All mice used in this study were housed in the Unit for Laboratory Animal Medicine at the University of Michigan, and all procedures were approved by the University Committee for the Use and Care of Animals. Aldh1a1-deficient mice23 were backcrossed to C57BL/Ka-CD45.2:Thy-1.1 mice for 4 to 8 generations before being studied. In reconstitution experiments, C57BL/Ka-CD45.2:Thy-1.2 mice aged at least 8 weeks were used as recipients. Aldefluor was purchased from StemCell Technologies (Vancouver, BC). Mobilization was induced by intraperitoneal injection of 200 mg/kg cyclophosphamide (Bristol-Myers Squibb, New York, NY) followed by 4 daily injections of 250 μg/kg human G-CSF (Amgen, Thousand Oaks, CA) as described.25 For experiments in which mice were treated weekly with 5-fluorouracil (American Pharmaceutical Partners, Schaumburg, IL), a dose of 150 mg/kg was used as described.26 Medium components were purchased from Invitrogen (Carlsbad, CA) unless otherwise noted. Statistical significance was assessed using the homoscedastic Student t test (P < .05).
Flow cytometry
HSCs were analyzed and isolated as previously described.20 Bone marrow cells were flushed from the tibia and femurs using a 25-gauge syringe (Becton Dickinson, Franklin Lakes, NJ) into Hank buffered salt solution (HBSS; without calcium or magnesium) plus 2% bovine serum. Single-cell suspensions were generated by trituration and passage through a 45-μm nylon filter (Sefar America, Kansas City, MO). CD150+CD41−CD48−Sca1+c-kit+ HSCs were isolated using phycoerythrin (PE)–conjugated anti-CD150 (TC15-12F12.2; BioLegend, San Diego, CA), fluorescein isothiocyanate (FITC)–conjugated anti-CD41 (MWReg30; BD Pharmingen, San Diego, CA), FITC-conjugated anti-CD48 (HM48-1; BioLegend), allophycocyanin (APC)–conjugated anti–Sca-1 (E13-6.7), and biotin-conjugated anti–c-kit (2B8; eBioscience, San Diego, CA). Streptavidin APC-Cy7 was used for fluorescent labeling of c-kit. c-kit+Flk-2−Lineage−Sca1+ HSCs were isolated using unconjugated antibodies against CD4 (GK1.5), CD8 (53-6.7), CD3 (KT31.1), Ter119; PE-conjugated antibodies against Gr-1 (8C5), B220 (6B2), Flk-2 (A2F10; eBioscience) Mac-1 (M1/70) anti-CD48 (HM48-1). Unconjugated antibodies were stained with a PE-conjugated anti–rat IgG secondary antibody. Antibodies against c-kit and Sca-1 were as described above. For analysis of the composition of the hematopoietic system, the same antibodies were used, in addition to APC-conjugated anti–mouse IgM (11/41; eBioscience), and PE-conjugated anti-CD43 (S7; BD Pharmingen).
All analyses excluded dead cells that stained positive for either DAPI (4′,6-diamidino-2-phenylindole; 1μg/mL) or 7-aminoactinomycin-D (Invitrogen). Cells were analyzed/sorted by fluorescence-activated cell sorting (FACS) with a FACS Vantage SE, FACS Aria, or a FACS Canto II (BD Biosciences, San Jose, CA).
Aldefluor staining
Bone marrow cells were isolated and stained as above except that the HSC stain was modified to make the FITC channel available for Aldefluor staining: antibodies used were PE/Cy5-conjugated anti-CD150 (TC15-12F12.2; BioLegend), PE-conjugated anti-CD41 (MWReg30; BD Pharmingen), and PE-conjugated anti-CD48 (HM48-1; BioLegend). HSCs were enriched using an AutoMacs with anti-biotin paramagnetic beads (Miltenyi, Auburn, CA). Enriched cells were resuspended in 1 mL Aldefluor staining medium (Aldegen) and mixed with 2 μL Aldefluor substrate (Aldegen). Half of the resuspended cells were immediately placed into a fresh vial with 5 μL DEAB (1.5mM, Aldagen) for the negative control, and both tubes were incubated at 37°C for 30 minutes. After incubation, cells were resuspended in fresh Aldefluor staining medium and analyzed.
Quantitative (real-time) PCR
A total of 1000 to 2000 HSCs or unfractionated bone marrow cells were sorted into trizol and RNA was isolated using an RNeazy Mini Kit (QIAGEN Sciences, Germantown, MD). cDNA was constructed with oligo dT primers and SuperScript II reverse transcriptase (Invitrogen). Quantitative PCR was performed with cDNA from 200 cell equivalents using a SYBR Green Kit and a LightCycler 480 (Roche Applied Science, Indianapolis, IN). Each sample was normalized to β-actin and amplification products were tested for specificity by melting curves, gel electrophoresis, and sequencing. Quantitative PCR primers are listed in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Methylcellulose culture
Totals of 500 live bone marrow cells or 1000 live spleen cells were sorted per well of a 96-well plate (Corning, Corning, NY) containing 100 μL MethoCult M3434 medium (StemCell Technologies). The medium was supplemented with 1% penicillin/streptomycin, 10 ng/mL Flt-3 (R&D Systems, Minneapolis, MN), and 10 ng/mL thrombopoietin (R&D Systems). Colonies were scored after 12 days incubation at 37°C in 6% CO2. Forty-eight wells were analyzed per sample.
Long-term hematopoietic reconstitution assay
C57BL/Ka-CD45.2:Thy-1.2 recipient mice (at least 8 weeks of age) were irradiated in 2 doses at least 2 hours apart with a total of 1140 cGy using a Cesium137 GammaCell40 Exactor Irradiator (MDS Nordia, Kanata, ON). Either 200 000 or 300 000 CD45.2+ donor bone marrow cells were transplanted with an equal number of CD45.1+ recipient bone marrow cells into the retro-orbital venous sinus. Engraftment was measured beginning at 4 weeks after transplantation by flow cytometric analysis of the frequency of donor myeloid, B cells, and T cells in blood from the tail vein of recipient mice. Red cells were lysed, and hematopoietic cells were stained with conjugated antibodies to CD45.2 (FITC, 104), B220 (PE, 6B2), and Mac-1 (APC, M1/70); or Gr-1 (PE, 8C5), and CD3 (APC, KT31.1).
Neurogenesis
Neurogenesis in the olfactory bulb was assessed as previously described.27 Animals were injected intraperitoneally with 50 mg 5-bromo-deoxyuridine (BrdU)/kg body mass (Sigma-Aldrich, St Louis, MO), and then given 1 mg/mL BrdU in their drinking water for 8 days. BrdU was chased by maintaining mice on normal drinking water for 4 weeks. Dissected brains were fixed in 4% paraformaldehyde, cryprotected in 15% sucrose, and embedded in Tissue-Tek OCT Compound (Sakura, Torrance, CA). Serial 12-μm sections were made through the olfactory bulbs. Five sections from regularly-spaced intervals thoughout each olfactory bulb were analyzed by immunohistochemistry with anti-NeuN (MAB377; Chemicon, Temecula, CA), rat anti-BrdU (Accurate Chemical, Westbury, NY), and DAPI. Five micrographs from random fields of interneurons were acquired from each slice by confocal microscopy using a Leica TSC SP5 (Leica, Wetzlar, Germany) with a 40× objective. Images were adjusted using Adobe Photoshop (Adobe Systems, San Jose, CA) and cells were quantified using ImageJ (National Institutes of Health [NIH], Bethesda, MD). Neurogenesis was determined by counting the percentage of BrdU+NeuN+DAPI+ neurons.
Isolation of central nervous system progenitor cells
Subventricular zone cells from the central nervous system (CNS) were isolated by microdissection in Opti-MEM and digested for 20 minutes at 37°C in 0.025% trypsin/0.5 mM ethylenediaminetetraacetic acid (EDTA; Calbiochem, San Diego, CA) and 200 U/mL DNase1 (Sigma-Aldrich). Enzymatic digestion was stopped by washing cells in L15 medium with 1mg/mL bovine serum albumin (BSA; Sigma-Aldrich), 10 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid] pH 7.4 (BioWhittaker, Walkersville, MD), 1% penicillin/streptomycin, and 200 U/mL DNase1. Cells were passed though a 45-μm nylon screen, counted, and plated.
CNS cell culture and self-renewal
Cells were plated at clonal density (0.67 cells/μL) in CNS “self-renewal medium”: a 5 to 3 mixture of DMEM–low glucose:neurobasal medium supplemented with 10% chick embryo extract,28 1% N2 supplement, 2% B27 supplement, 1% penicillin/ streptomycin, 50 μM β-mercaptoethanol (Sigma-Aldrich), 20 ng/mL human basic fibroblast growth factor (bFGF; R&D Systems) and 20 ng/mL epidermal growth factor (EGF; R&D Systems). Neurospheres were evaluated for size and frequency after 8 to 10 days of culture. To assess self-renewal potential, individual 8- to 9-day-old primary neurospheres were triturated to yield single-cell suspensions, then subcloned to secondary cultures, and grown for 5 to 8 days in self-renewal medium. Self-renewal was expressed as the number of secondary neurospheres generated per primary neurosphere. Multipotency was assessed by allowing neurospheres to differentiate in adherent cultures on fibronectin (Biomedical Technologies, Sloughton, MA) coated plates in “differentiation medium” that was similar to self-renewal medium except that it contained 0.5% fetal bovine serum (FBS; Invitrogen), lacked chick embryo extract, EGF, and had only 10 ng/mL bFGF. Cells were fixed after 5 to 8 days in culture and stained for oligodendrocytes (O4), neurons (TuJ1; Covance, Princeton, NJ) and astrocytes (glial fibrillary acidic protein [GFAP]; Sigma-Aldrich).
Isolation of peripheral nervous system progenitor cells
To isolate peripheral nervous system (PNS) progenitor cells, the outer muscle/plexus layers of the gut were dissected from the exterior surface of the gut wall in HBSS− without calcium or magnesium. Single-cell suspensions were generated by digestion in 1 mg/mL Collagenase type IV (Worthington, Lakewood, NJ) in HBSS− for 30 minutes at 37°C followed by a 10-minute digestion at 37°C in 10 U papain (Worthington). Enzymatic digestion was quenched by washing cells with L15 medium with 1 mg/mL BSA, 10 mM HEPES pH7.4, 1% penicillin/streptomycin, and 200 U/mL DNase1. Cells were passed though a 45-μm nylon screen, counted, and plated.
PNS cell culture and self-renewal assay
Cells were plated at clonal density (0.67 cells/μL) in PNS self-renewal medium: a 5 to 3 mixture of DMEM-low glucose:Neurobasal medium supplemented with 15% chick embryo extract, 1% N2 supplement, 2% B27 supplement, 1% penicillin/streptomycin, 50 μM β-mercaptoethanol, 35 mg/mL retinoic acid, 20 ng/mL human bFGF, and 20 ng/mL insulin-like growth factor 1 (IGF1; R&D Systems). Neurospheres were evaluated for size and frequency after 9 to 12 days of culture. To assess self-renewal potential, individual 9- to 10-day-old primary neurospheres were replated on fibronectin-coated plates and cultured adherently for 3 days, followed by trypsin/EDTA digestion and trituration to yield single-cell suspensions. The cells from individual neurospheres were then subcloned to secondary cultures and grown for 5 to 8 days in self-renewal medium. Self-renewal was expressed as the number of secondary neurospheres generated per primary neurosphere. Multipotency was assessed by allowing neurospheres to differentiate in adherent cultures on fibronectin-coated plates in PNS differentiation medium that was similar to self-renewal medium except that it contained 0.5% fetal bovine serum, and lacked chick embryo extract, IFG1, and had only 10 ng/mL bFGF. Cells were fixed after 5 to 8 days in culture and stained for smooth muscle (smooth muscle actin; Sigma-Aldrich), neurons (peripherin; Chemicon International) and astrocytes (GFAP; Sigma-Aldrich).
Results
Aldh1a1 expression increases with age in HSCs
We reanalyzed previously published gene expression profile data from Thy1.1lowSca-1+Lineage−c-kit+ HSCs and Thy1.1lowSca-1+Mac-1lowCD4lowB220− non–self-renewing multipotent hematopoietic progenitors20 to identify ALDHs that are expressed by HSCs. These microarray datasets included data on 9 of 20 ALDHs that have been identified in mice. Aldh1a1, Aldh2, Aldh1a7, Aldh3a2, and Aldh9a1 were expressed by Thy1.1lowSca-1+Lineage−c-kit+ HSCs, and all except Aldh3a2 were significantly more highly expressed in HSCs compared with multipotent progenitors (Figure 1A). Other RALDH subfamily members Aldh1a2 and Aldh1a3, as well as Aldh1b1, and Aldh3a1, were not expressed above background (data not shown). To assess whether the expression of these genes was affected by age we performed quantitative (real-time) polymerase chain reaction (qPCR) on HSCs and whole bone marrow cells obtained from young adult and old adult mice. This revealed that Aldh1a1 expression increased dramatically with age in adult HSCs but not in whole bone marrow cells (Figure 1B).
Aldh1a1 expression increases with age in HSCs, but Aldh1a1 deficiency is not compensated by increased transcription of other ALDHS. (A) Gene expression profiles for ALDHs in Thy-1.1lowSca-1+Lineage−c-kit+ HSCs, Thy-1.1lowSca-1+Mac-1lowCD4lowB220− non–self-renewing multipotent progenitors and CD45+ bone marrow cells from young adult mice. These data were extracted from genome-wide data published as supplementary material from an earlier study.20 (B) Aldh1a1 transcript levels were compared by qPCR in CD150+CD48−CD41−Sca1+c-Kit+ HSCs or whole bone marrow cells independently isolated from three 2.5-month-old mice and three 22-month-old mice. cDNA content was normalized between samples based on ß-actin expression (data not shown). Aldh1a1 transcript levels are expressed in terms of fold change relative to old bone marrow (set to 1). Aldh1a1 was expressed at significantly (P < .01) higher levels in old HSCs compared with old bone marrow or young HSCs. (C) A schematic representation of Aldh1a1 showing the position of exons (numbered black boxes), 4 sets of qPCR primer binding sites (arrows), and the neomycin resistance cassette in the targeted allele. Bone marrow cells (D) or CD150+CD48−CD41−Sca1+c-Kit+ HSCs (E) from 3 littermate pairs aged 2.5 to 6 months were tested for expression of Aldh1a1 with the above primers, and all other Aldh family members by quantitative RT-PCR. cDNA content was normalized between samples based on ß-actin expression (data not shown) and data are expressed as fold change relative to the littermate control. #P < .1; *P < .05. Error bars represent SD. The mutant Aldh1a1 transcript was present at greatly reduced levels, and we did not detect any Aldh transcript with a compensatory increase in expression.
Aldh1a1 expression increases with age in HSCs, but Aldh1a1 deficiency is not compensated by increased transcription of other ALDHS. (A) Gene expression profiles for ALDHs in Thy-1.1lowSca-1+Lineage−c-kit+ HSCs, Thy-1.1lowSca-1+Mac-1lowCD4lowB220− non–self-renewing multipotent progenitors and CD45+ bone marrow cells from young adult mice. These data were extracted from genome-wide data published as supplementary material from an earlier study.20 (B) Aldh1a1 transcript levels were compared by qPCR in CD150+CD48−CD41−Sca1+c-Kit+ HSCs or whole bone marrow cells independently isolated from three 2.5-month-old mice and three 22-month-old mice. cDNA content was normalized between samples based on ß-actin expression (data not shown). Aldh1a1 transcript levels are expressed in terms of fold change relative to old bone marrow (set to 1). Aldh1a1 was expressed at significantly (P < .01) higher levels in old HSCs compared with old bone marrow or young HSCs. (C) A schematic representation of Aldh1a1 showing the position of exons (numbered black boxes), 4 sets of qPCR primer binding sites (arrows), and the neomycin resistance cassette in the targeted allele. Bone marrow cells (D) or CD150+CD48−CD41−Sca1+c-Kit+ HSCs (E) from 3 littermate pairs aged 2.5 to 6 months were tested for expression of Aldh1a1 with the above primers, and all other Aldh family members by quantitative RT-PCR. cDNA content was normalized between samples based on ß-actin expression (data not shown) and data are expressed as fold change relative to the littermate control. #P < .1; *P < .05. Error bars represent SD. The mutant Aldh1a1 transcript was present at greatly reduced levels, and we did not detect any Aldh transcript with a compensatory increase in expression.
To test whether Aldh1a1 regulates HSC function we analyzed Aldh1a1-deficient mice.23 To confirm that these mice carried the targeted allele, we performed quantitative (real-time) RT-PCR using 4 sets of primers that spanned the Aldh1a1 transcript: primer set A was 5′ of exon 11, primer set B was in exons 11 and 12, primer set C spanned from exon 10 to 12, and primer set D was 3′ of exon 11 (see Figure 1C for a description of primer binding sites). Primer set B amplified the predicted product from heterozygous mice but nothing from Aldh1a1 deficient mice, confirming they carried the targeted allele (Figure 1D,E). Primer sets A and D amplified the predicted products, but Aldh1a1 deficient mice had template levels for these products that were 3- to 8-fold lower than in heterozygous controls. Primer set C amplified the expected products from Aldh1a1 deficient and heterozygous controls: the mutant amplicon was shortened by the lack of exon 11, carried the frameshift mutation that reflected gene targeting, and was present at 18-fold lower levels than wild type (Figure 1D,E). Mutant Aldh1a1 transcripts were thus present at significantly lower levels than wild-type transcripts, suggesting that the mutant mRNA was destabilized. When combined with the previously published inability to detect Aldh1a1 protein from this allele and the loss of retinoic acid biosynthesis,23,24 the data suggest this allele is amorphic.
To test whether the loss of Aldh1a1 expression is compensated by increases in the expression of other ALDH family members, we compared the expression levels of all 19 other ALDH family members in Aldh1a1-deficient mice and heterozygous littermate controls by qPCR. We could not detect transcripts from other RALDH subfamily members Aldh1a2, Aldh1a3, or Aldh8a1 in wild-type or in Aldh1a1-deficient bone marrow cells or HSCs. This suggests there is not redundancy among RALDH subfamily members in HSCs. We were also unable to detect the expression of Aldh1l2, Aldh3a1, or Aldh3b2 in HSCs or whole bone marrow cells. Yet all of these transcripts, including RALDH subfamily members, were readily amplified from E13.5 embryo cDNA. We detected the expression of all other ALDHs in bone marrow cells (Figure 1D) and HSCs (Figure 1E) of Aldh1a1-deficient mice and littermate controls. Aldh1a7 was the only family member that showed significantly altered expression within Aldh1a1-deficient mice, and its expression was 2- to 3-fold reduced in HSCs and bone marrow cells from in Aldh1a1-deficient mice (Figure 1D,E). Because Aldh1a1 and Aldh1a7 are adjacent to each other on chromosome 19 and the mutant allele retains the neomycin resistance gene, is it possible the neomycin cassette interfered with Aldh1a7 expression.29 The mutant mice therefore had partially reduced Aldh1a7 expression in addition to a loss of Aldh1a1 expression and function, but we could find no evidence of a compensatory increase in the expression of any ALDH in the absence of Aldh1a1.
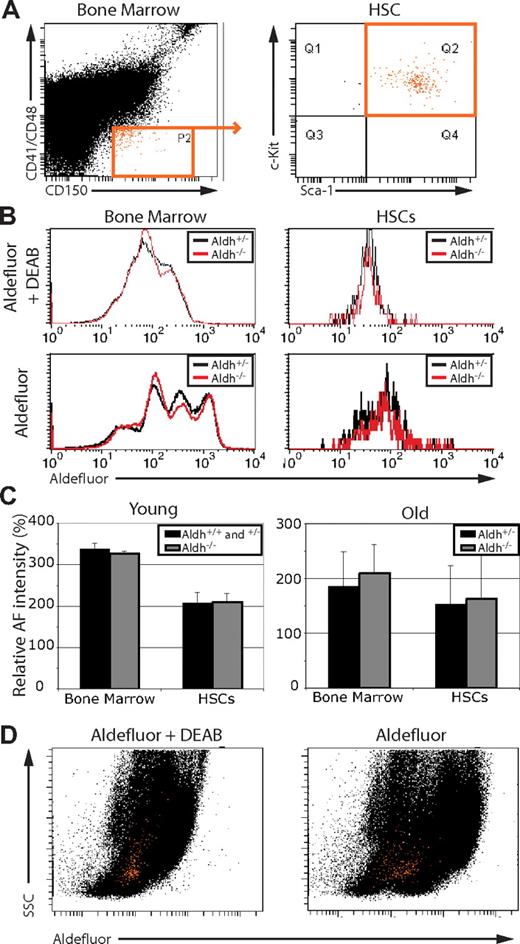
Aldefluor staining does not distinguish mouse HSCs and is not determined by Aldh1a1
To test whether Aldefluor staining reflects Aldh1a1 activity in hematopoietic cells, we compared the Aldefluor staining of bone marrow and HSCs from wild-type and Aldh1a1-deficient mice. The fluorescent Aldefluor substrate becomes membrane impermeable as a result of ALDH activity, leading to its accumulation and fluorescence within cells in proportion to the amount of ALDH activity within those cells. We found that Aldefluor fluorescence within unfractionated mouse bone marrow cells and CD150+CD41−CD48−Sca1+c-kit+ HSCs (Figure 2A,B) was unaffected by Aldh1a1 deficiency in both young adult and old adult mice (Figure 2B,C). CD150+CD41−CD48−Sca1+c-kit+ cells include at least 95% of all long-term multilineage reconstituting cells in the bone marrow, and more than 40% of single cells within this population give long-term multilineage reconstitution after transplantation into irradiated mice.20,30,31 This demonstrates that Aldefluor fluorescence does not reflect Aldh1a1 activity within HSCs.
Aldefluor staining is not determined by Aldh1a1 function, and HSCs cannot be distinguished from bone marrow cells based on Aldefluor staining. (A) whole bone marrow (left) and CD150+CD48−CD41−Sca1+c-kit+ HSCs (right; cells highlighted in orange) were (B) analyzed for Aldefluor fluorescence in the presence and absence of the ALDH inhibitor DEAB. Aldh1a1 deficiency had no effect on Aldefluor staining. (C) Mean Aldefluor fluorescence intensity minus background fluorescence in the presence of DEAB (at least 2 independent experiments per age involving a total of 3 to 5 mice per treatment). Error bars represent standard deviation. (D) Representative plots of whole bone marrow cells (black dots) and HSCs (orange dots) from (A) show SSC versus Aldefluor staining. Note that mouse HSCs did not exhibit high levels of Aldefluor staining relative to whole bone marrow. Similar results were obtained using c-kit+Flk-2−Lineage−Sca1+ HSCs (Figure S1).
Aldefluor staining is not determined by Aldh1a1 function, and HSCs cannot be distinguished from bone marrow cells based on Aldefluor staining. (A) whole bone marrow (left) and CD150+CD48−CD41−Sca1+c-kit+ HSCs (right; cells highlighted in orange) were (B) analyzed for Aldefluor fluorescence in the presence and absence of the ALDH inhibitor DEAB. Aldh1a1 deficiency had no effect on Aldefluor staining. (C) Mean Aldefluor fluorescence intensity minus background fluorescence in the presence of DEAB (at least 2 independent experiments per age involving a total of 3 to 5 mice per treatment). Error bars represent standard deviation. (D) Representative plots of whole bone marrow cells (black dots) and HSCs (orange dots) from (A) show SSC versus Aldefluor staining. Note that mouse HSCs did not exhibit high levels of Aldefluor staining relative to whole bone marrow. Similar results were obtained using c-kit+Flk-2−Lineage−Sca1+ HSCs (Figure S1).
CD150+CD41−CD48−Sca1+c-kit+ HSCs did not exhibit high levels of Aldefluor staining relative to unfractionated bone marrow. The bone marrow cells with the highest levels of Aldefluor staining did not include CD150+CD41−CD48−Sca1+c-kit+ HSCs, which had moderate levels of Aldefluor staining similar to many other bone marrow cells (Figure 2D). Similar results were obtained by gating on c-kit+Flk-2−Lineage−Sca1+ HSCs, which also exhibited moderate levels of Aldefluor staining similar to most other bone marrow cells (Figure S1). These results are consistent with recently published results in demonstrating that mouse HSCs cannot be distinguished from other hematopoietic cells on the basis of high levels of Aldefluor staining.12 Nonetheless, they do not rule out the possibility that Aldefluor staining may be used to enrich HSCs when combined with lineage depletion and additional fractionation steps.13,14
Aldh1a1 is dispensable for normal hematopoiesis
Because Aldh1a1 was strongly expressed by HSCs (Figure 1) and has been suggested to regulate HSC function, we tested whether Aldh1a1 was required for normal hematopoiesis. Wild-type and Aldh1a1+/− mice were indistinguishable and were, therefore, used interchangeably as littermate controls. We did not detect any difference in the bone marrow or spleen of Aldh1a1-deficient compared with littermate control mice in terms of cellularity (Figure 3A), CD150+CD41−CD48−Sca1+c-kit+ HSC frequency (Figure 3A), colony-forming progenitor frequency (Figure 3B), or composition with respect to the frequency of B, T, erythroid, and myeloid lineage cells (Figure 3C). These data suggest Aldh1a1 is not required for steady-state hematopoiesis in young adult mice.
Aldh1a1 is not required for normal hematopoiesis. (A) Cellularity (left) and HSC frequency (right) were determined in young (2-3 months) and old (21-27 months) adult Aldh1a1+/− and Aldh1a1−/− mice (n = 6-14 mice per treatment in at least 6 independent experiments). (B) The frequency of colony-forming progenitors in bone marrow (top row) and spleen (second row) of young (left column) and old adult (right column) Aldh1a1+/− and Aldh1a1−/− mice (n = 6-7 mice per treatment in 5 independent experiments). (C) The frequency of various subsets of B (B220+), T (CD3+), erythroid (Ter119+), and myeloid (Mac-1+Gr-1+) lineage cells in the bone marrow (top row) and spleen (bottom row) from young (left column) and old (right column) Aldh1a1+/− and Aldh1a1−/− mice (n = 5 mice per treatment in 5 independent experiments). No significant differences were observed between Aldh1a1−/− and control mice. Error bars represent SD.
Aldh1a1 is not required for normal hematopoiesis. (A) Cellularity (left) and HSC frequency (right) were determined in young (2-3 months) and old (21-27 months) adult Aldh1a1+/− and Aldh1a1−/− mice (n = 6-14 mice per treatment in at least 6 independent experiments). (B) The frequency of colony-forming progenitors in bone marrow (top row) and spleen (second row) of young (left column) and old adult (right column) Aldh1a1+/− and Aldh1a1−/− mice (n = 6-7 mice per treatment in 5 independent experiments). (C) The frequency of various subsets of B (B220+), T (CD3+), erythroid (Ter119+), and myeloid (Mac-1+Gr-1+) lineage cells in the bone marrow (top row) and spleen (bottom row) from young (left column) and old (right column) Aldh1a1+/− and Aldh1a1−/− mice (n = 5 mice per treatment in 5 independent experiments). No significant differences were observed between Aldh1a1−/− and control mice. Error bars represent SD.
Because Aldh1a1 expression increases with age in HSCs (Figure 1B), we examined whether Aldh1a1 becomes a critical regulator of hematopoiesis in old mice. To test this, we aged Aldh1a1-deficient and littermate control mice and analyzed them between 21 and 27 months of age. However, we still observed no effect of Aldh1a1 deficiency on cellularity (Figure 3A), CD150+CD41−CD48−Sca1+c-kit+ HSC frequency (Figure 3A), colony-forming progenitor frequency (Figure 3B), or lineage composition (Figure 3C) of the bone marrow or spleen. These data indicate that although Aldh1a1 expression increases dramatically with age in HSCs, Aldh1a1 remains dispensable for normal hematopoiesis in old mice.
Aldh1a1 is dispensable for normal HSC function
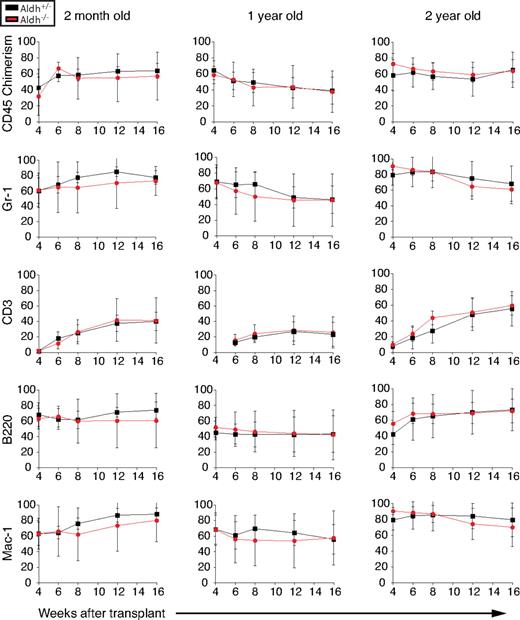
To test whether Aldh1a1 is required for normal HSC function, we performed competitive reconstitution assays using whole bone marrow cells from 2-month-old, 1-year-old, and 2-year-old Aldh1a1-deficient and littermate control mice. In each case, 300 000 donor (CD45.2+) bone marrow cells from Aldh1a1−/− or littermate control mice were transplanted into irradiated wild-type recipient (CD45.1+) mice along with an equal number of recipient bone marrow cells. Aldh1a1−/− bone marrow cells consistently gave long-term multilineage reconstitution of the myeloid, B-cell, and T-cell lineages, and the levels of donor-cell reconstitution did not significantly differ between Aldh1a1−/− and control cells (Figure 4). Serial transplantation of bone marrow cells into secondary recipients demonstrated that Aldh1a1-deficient cells could also give long-term multilineage reconstitution of secondary recipients as effectively as control cells (data not shown). These data demonstrate that Aldh1a1 is not required for the self-renewal or differentiation of HSCs from young, middle-aged, or old adult mice.
Aldh1a1 is not required for HSC maintenance or function. Irradiated CD45.1+ recipient mice were competitively reconstituted with 3 × 105 whole bone marrow cells from 2-month-old, 13-month-old, or 26-month-old Aldh1a1+/− (black lines) or Aldh1a1−/− (red lines) CD45.2 donor cells along with a radioprotective dose of 3 × 105 CD45.1+ bone marrow cells. Similar levels of long-term multilineage reconstitution were observed at all ages from Aldh1a1+/− and Aldh1a1−/− cells. At least 3 independent experiments were performed at each age, from which one representative experiment is shown. Error bars represent SD.
Aldh1a1 is not required for HSC maintenance or function. Irradiated CD45.1+ recipient mice were competitively reconstituted with 3 × 105 whole bone marrow cells from 2-month-old, 13-month-old, or 26-month-old Aldh1a1+/− (black lines) or Aldh1a1−/− (red lines) CD45.2 donor cells along with a radioprotective dose of 3 × 105 CD45.1+ bone marrow cells. Similar levels of long-term multilineage reconstitution were observed at all ages from Aldh1a1+/− and Aldh1a1−/− cells. At least 3 independent experiments were performed at each age, from which one representative experiment is shown. Error bars represent SD.
Aldh1a1 non–cell-autonomously regulates the cyclophosphamide sensitivity of HSCs
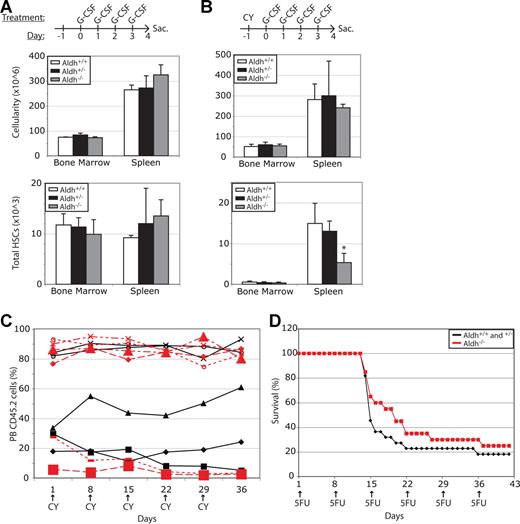
HSC mobilization in response to G-CSF treatment involves an expansion in HSCs followed by migration from the bone marrow to the spleen.25,32 To test whether these processes are regulated by Aldh1a1 we administered G-CSF on 4 consecutive days to Aldh1a1-deficient mice and littermate controls. Aldh1a1 deficiency did not significantly affect bone marrow cellularity, spleen cellularity, or total CD150+CD41−CD48−Sca1+c-kit+ HSC numbers in the bone marrow or spleen (Figure 5A). These data demonstrate that HSCs do not require Aldh1a1 to expand in number or to mobilize in response to G-CSF.
Aldh1a1 is non–cell-autonomously required to reduce the toxicity of cyclophosphamide. (A) Aldh1a1−/− and littermate control mice were treated with 250 μg G-CSF/kg body mass/day for 4 days to induce HSC mobilization. Aldh1a1 deficiency did not significantly affect bone marrow or spleen cellularity or HSC content (n = 2-5 mice per treatment in 2 independent experiments). (B) Littermates were treated with 200 mg/kg cyclophosphamide (CY) followed by 4 days of 250 μg/kg G-CSF. Aldh1a1−/− mice had significantly (*P < .05) fewer HSCs in their spleen compared with littermate controls but did not exhibit a significant difference in overall bone marrow or spleen cellularity (n = 3-6 mice per treatment in 2 independent experiments). (C) Donor-cell chimerism in wild-type mice reconstituted with Aldh1a1−/− (red lines) or littermate control (black lines) bone marrow cells that were treated weekly with 200 mg/kg cyclophosphamide. Because cyclophosphamide did not generally reduce the levels of Aldh1a1−/− donor cells compared with control donor cells these results indicate that the toxic effect of cyclophosphamide on HSCs is primarily non–cell-autonomously attenuated by Aldh1a1. (D) Survival of Aldh1a1−/− and littermate control mice treated weekly with 150 mg/kg 5-fluorouracil. Data represent the combined results from 2 independent experiments (n = 20-22 mice per treatment). Error bars represent SD.
Aldh1a1 is non–cell-autonomously required to reduce the toxicity of cyclophosphamide. (A) Aldh1a1−/− and littermate control mice were treated with 250 μg G-CSF/kg body mass/day for 4 days to induce HSC mobilization. Aldh1a1 deficiency did not significantly affect bone marrow or spleen cellularity or HSC content (n = 2-5 mice per treatment in 2 independent experiments). (B) Littermates were treated with 200 mg/kg cyclophosphamide (CY) followed by 4 days of 250 μg/kg G-CSF. Aldh1a1−/− mice had significantly (*P < .05) fewer HSCs in their spleen compared with littermate controls but did not exhibit a significant difference in overall bone marrow or spleen cellularity (n = 3-6 mice per treatment in 2 independent experiments). (C) Donor-cell chimerism in wild-type mice reconstituted with Aldh1a1−/− (red lines) or littermate control (black lines) bone marrow cells that were treated weekly with 200 mg/kg cyclophosphamide. Because cyclophosphamide did not generally reduce the levels of Aldh1a1−/− donor cells compared with control donor cells these results indicate that the toxic effect of cyclophosphamide on HSCs is primarily non–cell-autonomously attenuated by Aldh1a1. (D) Survival of Aldh1a1−/− and littermate control mice treated weekly with 150 mg/kg 5-fluorouracil. Data represent the combined results from 2 independent experiments (n = 20-22 mice per treatment). Error bars represent SD.
An alternative method of mobilization involves cyclophosphamide treatment, followed by daily injections of G-CSF.25 Aldh1a1 is known to detoxify cyclophosphamide by metabolizing the biologically active derivative, a process that at least partially occurs in the liver.5,6,33 This suggests that HSC numbers should be reduced in Aldh1a1-deficient mice treated with cyclophosphamide/G-CSF compared with littermate controls, as a result of the slower metabolism of cyclophosphamide. To test this, we administered cyclophosphamide/G-CSF to Aldh1a1-deficient mice and littermate controls. Although overall bone marrow cellularity and spleen cellularity were not significantly affected by Aldh1a1 deficiency, total numbers of CD150+CD41−CD48−Sca1+c-kit+ HSCs in the spleen were significantly reduced in Aldh1a1-deficient mice compared with littermate controls (Figure 5B).
To test whether this reflects a cell-autonomous or non–cell-autonomous requirement of HSCs for Aldh1a1, we transplanted CD45.2+Aldh1a1−/− or Aldh1a1+/− bone marrow cells into irradiated wild-type CD45.1+ recipients in competitive reconstitution assays. Twenty weeks later, after the donor cells had achieved stable chimerism, we treated with weekly doses of cyclophosphamide (Figure 5C). If HSCs cell-autonomously require Aldh1a1 to reduce cyclophosphamide toxicity, we would expect to observe a reduction in donor-cell chimerism in the recipients of Aldh1a1-deficient cells. In contrast, if HSCs non–cell-autonomously require Aldh1a1 to reduce cyclophosphamide toxicity, we would not expect to observe a reduction in donor-cell chimerism because the recipient mice were wild type in all cases. Overall, we did not observe a clear or consistent reduction in donor chimerism within recipients of Aldh1a1-deficient cells, demonstrating that under physiological conditions Aldh1a1 acts primarily non–cell-autonomously with respect to HSCs to detoxify cyclophosphamide.
To assess whether the increased sensitivity of Aldh1a1-deficient HSCs to cyclophosphamide reflects a specific role for Aldh1a1 in the metabolism of cyclophosphamide, or a more general role in the response to myeloablative compounds, we also tested the sensitivity of Aldh1a1-deficient mice to weekly injections of 5-fluorouracil. 5-fluorouracil is not thought to be metabolized by ALDHs. The survival of Aldh1a1-deficient mice was not significantly different compared with littermate controls in response to weekly 5-fluoruracil administration (Figure 5D). This suggests that Aldh1a1 is required for the metabolism of cyclophosphamide, not for a general protective effect against myeloablative or proliferation-inducing compounds.
Aldh1a1 is dispensable for CNS stem cell function
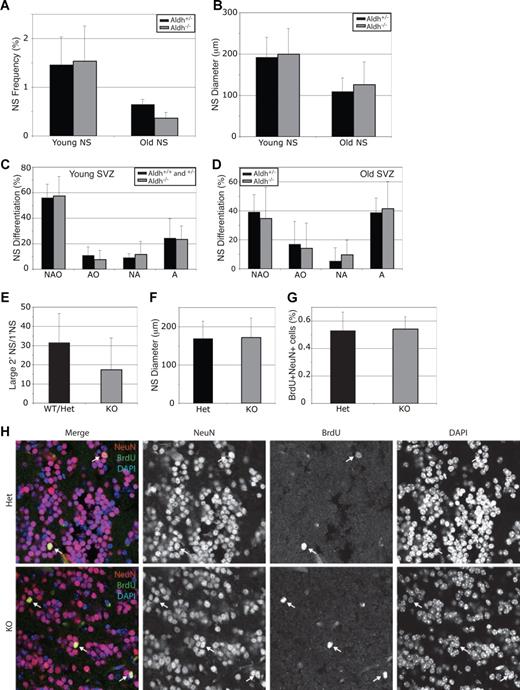
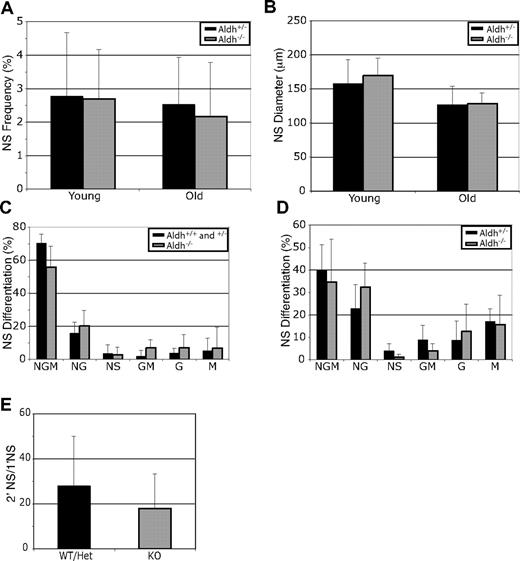
Aldh1a1 is expressed by CNS stem cells,22 and Aldefluor has been used to isolate these stem cells.17 To test whether Aldh1a1 regulates CNS stem cell function, we dissociated lateral ventricle subventricular zone (SVZ) cells from young and old adult mice and cultured them at clonal density (1 cell/1.5 μL) for 8 to 9 days.34 Aldh1a1 deficiency did not significantly affect the percentage of SVZ cells that formed neurospheres (Figure 6A), the diameter of these neurospheres (Figure 6B), or their ability to undergo multilineage differentiation (Figure 6C,D). Aldh1a1 deficiency also did not significantly affect the ability of primary neurospheres to form secondary neurospheres upon subcloning (Figure 6E) or the size of these secondary neurospheres (Figure 6F). These data suggest that Aldh1a1 deficiency does not affect CNS stem cell frequency or function in young or old adult mice.
Aldh1a1 is not required for central nervous system stem cell maintenance or function. (A) Lateral ventricle SVZ cells from young (2-3 months) and old (24-27 months) Aldh1a1+/− ( ) and Aldh1a1−/− (
) and Aldh1a1−/− ( ) mice were cultured. Aldh1a1 deficiency did not affect the frequency of neurospheres (A), neurosphere diameter after 8 to 10 days in culture (B), neurosphere differentiation (C,D), or the capacity of primary neurospheres to form large (> 100 μm; these are almost always multipotent) secondary neurospheres upon subcloning (E). Colonies were assessed for the presence of neurons (N), astrocytes (A) and oligodendrocytes (O). The size of secondary neurospheres was also not affected (F). All data represent a total of 3 to 7 mice per treatment, in at least 3 independent experiments except for F which represents 2 independent experiments with 1 mouse per experiment. (G, H) Aldh1a1 deficiency also did not affect the rate of neurogenesis (frequency of BrdU + NeuN + neurons) in the olfactory bulb of 18-month-old adult mice in vivo (n = 2 mice with 25 sections per mouse). Error bars represent SD.
) mice were cultured. Aldh1a1 deficiency did not affect the frequency of neurospheres (A), neurosphere diameter after 8 to 10 days in culture (B), neurosphere differentiation (C,D), or the capacity of primary neurospheres to form large (> 100 μm; these are almost always multipotent) secondary neurospheres upon subcloning (E). Colonies were assessed for the presence of neurons (N), astrocytes (A) and oligodendrocytes (O). The size of secondary neurospheres was also not affected (F). All data represent a total of 3 to 7 mice per treatment, in at least 3 independent experiments except for F which represents 2 independent experiments with 1 mouse per experiment. (G, H) Aldh1a1 deficiency also did not affect the rate of neurogenesis (frequency of BrdU + NeuN + neurons) in the olfactory bulb of 18-month-old adult mice in vivo (n = 2 mice with 25 sections per mouse). Error bars represent SD.
Aldh1a1 is not required for central nervous system stem cell maintenance or function. (A) Lateral ventricle SVZ cells from young (2-3 months) and old (24-27 months) Aldh1a1+/− ( ) and Aldh1a1−/− (
) and Aldh1a1−/− ( ) mice were cultured. Aldh1a1 deficiency did not affect the frequency of neurospheres (A), neurosphere diameter after 8 to 10 days in culture (B), neurosphere differentiation (C,D), or the capacity of primary neurospheres to form large (> 100 μm; these are almost always multipotent) secondary neurospheres upon subcloning (E). Colonies were assessed for the presence of neurons (N), astrocytes (A) and oligodendrocytes (O). The size of secondary neurospheres was also not affected (F). All data represent a total of 3 to 7 mice per treatment, in at least 3 independent experiments except for F which represents 2 independent experiments with 1 mouse per experiment. (G, H) Aldh1a1 deficiency also did not affect the rate of neurogenesis (frequency of BrdU + NeuN + neurons) in the olfactory bulb of 18-month-old adult mice in vivo (n = 2 mice with 25 sections per mouse). Error bars represent SD.
) mice were cultured. Aldh1a1 deficiency did not affect the frequency of neurospheres (A), neurosphere diameter after 8 to 10 days in culture (B), neurosphere differentiation (C,D), or the capacity of primary neurospheres to form large (> 100 μm; these are almost always multipotent) secondary neurospheres upon subcloning (E). Colonies were assessed for the presence of neurons (N), astrocytes (A) and oligodendrocytes (O). The size of secondary neurospheres was also not affected (F). All data represent a total of 3 to 7 mice per treatment, in at least 3 independent experiments except for F which represents 2 independent experiments with 1 mouse per experiment. (G, H) Aldh1a1 deficiency also did not affect the rate of neurogenesis (frequency of BrdU + NeuN + neurons) in the olfactory bulb of 18-month-old adult mice in vivo (n = 2 mice with 25 sections per mouse). Error bars represent SD.
SVZ stem cells undergo neurogenesis throughout adult life, forming interneurons that migrate into the olfactory bulb.27 To test whether Aldh1a1 deficiency affects the rate of neurogenesis in vivo, we administered BrdU to 18-month-old Aldh1a1-deficient and littermate control mice for 8 days to mark dividing SVZ progenitors, followed by 1 month without BrdU to allow these cells to migrate into the olfactory bulb and differentiate into neurons.27 Then we killed the mice, cut sagittal sections through the olfactory bulb, and stained with antibodies against BrdU and NeuN (a mature neuronal marker). Aldh1a1 deficiency did not affect the frequency of BrdU+NeuN+ neurons observed in sections (Figure 6G, H). This indicates that Aldh1a1 is not required for normal CNS stem cell function or neurogenesis in vivo.
Aldh1a1 is dispensable for PNS stem cell function
Aldh1a1 is also expressed by the neural crest stem cells (NCSCs) that give rise to the enteric nervous system during fetal development35 and that persist throughout adult life in the gut wall.36 qPCR analysis of Aldh1a1 expression levels of flow-cytometrically isolated p75+ gut NCSCs36-38 revealed that Aldh1a1 expression levels increase with age in NCSCs in a manner similar to that observed in HSCs: Aldh1a1 expression levels increased 2.4-fold from embryonic day 14 to postnatal day 15 and 144-fold from postnatal day 15 to 2 years of age (data not shown). To test whether Aldh1a1 regulates NCSC function, we cultured dissociated cells from the outer plexus/muscle layers of the gut from young and old adult Aldh1a1-deficient and littermate control mice according to protocols that we developed previously to study adult NCSC function.34,39 Aldh1a1 deficiency had no effect on the percentage of gut cells that formed neurospheres in culture (Figure 7A), neurosphere diameter (Figure 7B), the differentiation of these neurospheres (Figure 7C,D), or the ability of neurospheres to give rise to secondary neurospheres upon subcloning (Figure 7E). These data suggest that Aldh1a1 deficiency does not affect gut NCSC frequency or function in young or old adult mice.
Aldh1a1 is not required for peripheral nervous system stem cell maintenance or function. Outer muscle/plexus layer cells from the guts of young (2-3 months) and old (24-27 months) Aldh1a1+/− ( ) and Aldh1a1−/− (
) and Aldh1a1−/− ( ) mice were cultured. Aldh1a1 deficiency did not affect the frequency of neurospheres (A), neurosphere diameter after 9 to 11 days in culture (B), neurosphere differentiation (C,D), or the capacity of primary neurospheres to form multipotent secondary neurospheres upon subcloning (E). Colonies were assessed for the presence of neurons (N), glia (G), and myofibroblasts (M). All data represent a total of 3 to 8 mice per treatment, in at least 3 independent experiments. Error bars represent SD.
) mice were cultured. Aldh1a1 deficiency did not affect the frequency of neurospheres (A), neurosphere diameter after 9 to 11 days in culture (B), neurosphere differentiation (C,D), or the capacity of primary neurospheres to form multipotent secondary neurospheres upon subcloning (E). Colonies were assessed for the presence of neurons (N), glia (G), and myofibroblasts (M). All data represent a total of 3 to 8 mice per treatment, in at least 3 independent experiments. Error bars represent SD.
Aldh1a1 is not required for peripheral nervous system stem cell maintenance or function. Outer muscle/plexus layer cells from the guts of young (2-3 months) and old (24-27 months) Aldh1a1+/− ( ) and Aldh1a1−/− (
) and Aldh1a1−/− ( ) mice were cultured. Aldh1a1 deficiency did not affect the frequency of neurospheres (A), neurosphere diameter after 9 to 11 days in culture (B), neurosphere differentiation (C,D), or the capacity of primary neurospheres to form multipotent secondary neurospheres upon subcloning (E). Colonies were assessed for the presence of neurons (N), glia (G), and myofibroblasts (M). All data represent a total of 3 to 8 mice per treatment, in at least 3 independent experiments. Error bars represent SD.
) mice were cultured. Aldh1a1 deficiency did not affect the frequency of neurospheres (A), neurosphere diameter after 9 to 11 days in culture (B), neurosphere differentiation (C,D), or the capacity of primary neurospheres to form multipotent secondary neurospheres upon subcloning (E). Colonies were assessed for the presence of neurons (N), glia (G), and myofibroblasts (M). All data represent a total of 3 to 8 mice per treatment, in at least 3 independent experiments. Error bars represent SD.
Discussion
In agreement with prior studies,18,19,22 we found that Aldh1a1 is preferentially expressed by HSCs compared with other hematopoietic cells, particularly in old mice (Figure 1). We also observed increasing Aldh1a1 expression in neural stem cells with age (data not shown). However, our data demonstrate that Aldh1a1 is not an important regulator of stem cell function in the hematopoietic and nervous systems. Aldh1a1 deficiency did not affect hematopoiesis, HSC maintenance, or HSC function in either young or old adult mice (Figure 3 and Figure 4). Aldh1a1 deficiency also did not affect CNS stem cell frequency, function, or olfactory bulb neurogenesis (Figure 6). Finally, Aldh1a1 deficiency did not affect the frequency or function of gut NCSCs (Figure 7). Moreover, the fact that Aldh1a1-deficient mice are developmentally normal, fertile, and have a normal lifespan23 suggests that there are also no gross defects in stem cell function in other tissues. Although Aldh1a1 tends to be expressed at higher levels in stem cells compared with other cells in the hematopoietic and nervous systems (Figure 1),18,19,22 it is not a critical regulator of adult stem cell function in mice.
There is good reason to believe that Aldh1a1-deficient mice have a complete loss of Aldh1a1 function. The targeted exon contains a domain required for substrate binding and tetramerization, both of which are necessary for all known Aldh1a1 functions.23 Splicing around the targeted exon results in a frameshift. These mice lack the expression of all detectable Aldh1a1 protein by western blot using a polyclonal antiserum and exhibit reduced retinoic acid synthesis.23,24 Consistent with this, our data suggest that the mutant mRNA is destabilized (Figure 1D,E). Our data further confirm the loss of Aldh1a1 function by demonstrating the increased sensitivity of hematopoietic cells to cyclophosphamide. Overall, the data from this and prior studies of Aldh1a1-deficient mice indicate that gene targeting has deleted a domain critical for Aldh1a1 function, destabilized mRNA expression, eliminated all detectable protein expression, and eliminated all known functions for Aldh1a1. It remains formally possible that an undetected fragment of Aldh1a1 with unknown function is still expressed, but such a possibility exists for most knockout mice and usually cannot be formally excluded.
We did not detect elevated expression of other ALDH family members in the absence of Aldh1a1, suggesting that Aldh1a1 function in HSCs is not compensated by increased expression of other family members. Our results also cannot easily be explained by redundancy as other RALDH subfamily members (Aldh1a2, Aldh1a3, and Aldh8a1) that share overlapping functions with Aldh1a1 were not detected in HSCs. Nonetheless, it is possible that more distantly related ALDHS could exhibit functional redundancy with Aldh1a1 in stem cells or that other ALDHS could compensate for the loss of Aldh1a1 by posttranscriptional mechanisms. Aldh1a1 is therefore not required for stem cell function, either because it does not regulate stem cells or because its functions are redundantly performed by multiple ALDHS. It remains possible that other Aldhs do regulate critical stem cell functions.
We were unable to distinguish mouse HSCs from other hematopoietic cells based on Aldefluor staining. HSCs were similar to most other bone marrow cells in the degree to which they stained with Aldefluor (Figure 2). This observation agrees with the results from another recent study.12 It remains possible that Aldefluor staining can help to enrich HSCs when combined with other criteria, such as lineage depletion,13,14 but Aldefluor staining on its own is not an effective marker of mouse HSCs. Thus, while high levels of Aldefluor staining enrich human hematopoietic and breast epithelial stem cells,11,15,16 strong Aldefluor staining is not a general stem cell marker.
Although Aldefluor staining has often been assumed to reflect Aldh1a1 activity, Aldh1a1 deficiency did not affect Aldefluor staining of mouse HSCs or bone marrow cells (Figure 2B). This suggests that most ALDH activity in mouse hematopoietic cells derives from other ALDHs, despite the prominent expression of Aldh1a1. We also observed clear expression of Aldh2, Aldh3a2, and Aldh9a1 in HSCs and other hematopoietic cells (Figure 1), raising the possibility that one or more of these ALDHs could be responsible for Aldefluor staining. ALDH activity may also be conferred by different enzymes in mouse and human stem cells.
Although Aldh1a1 did not regulate stem cell function in any discernible way under physiological conditions, it was non–cell-autonomously required to detoxify cyclophosphamide. Aldh1a1-deficient mice exhibited significantly lower numbers of HSCs in the spleen after cyclophosphamide/G-CSF treatment, but Aldh1a1-deficient HSCs were not at a compe-titive disadvantage to control HSCs when transplanted into wild-type mice and treated with cyclophosphamide (Figure 5). These results are consistent with a known role for Aldh1a1 in the liver in the metabolism of the active derivative of cyclophosphamide.4-7
Our study represents the first direct test of Aldh1a1 function in stem cells. We detected no defects in the physiological function of neural or hematopoietic stem cells in young or old adult mice. Critical ALDH functions in stem cells are likely provided by other ALDH family members.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Shenghui He, Mark J. Kiel, Jae Lee, Daisuke Nakada, and S. Guy Slutsky for technical advice and assistance, and Martin White and David Adams for flow cytometry assistance.
This work was supported by the Howard Hughes Medical Institute and NIH (1 R01 AG024945-01 to S.J.M. and 2 R01 EY013969-05 to G.D). B.P.L. was supported by an American Heart Association Postdoctoral Fellowship (0725726Z) and a Cancer Research Institute Postdoctoral Fellowship. O.H.Y. was supported by a predoctoral fellowship from the University of Michigan (UM) Institute of Gerontology. Flow cytometry was supported in part by the UM–Comprehensive Cancer, NIH CA46592. Thanks to Elizabeth Smith (Hybridoma Core Facility) for antibody production, supported in part through the Rheumatic Core Disease Center (P30 AR48310).
National Institutes of Health
Authorship
Contribution: B.P.L. and O.H.Y. performed all of the experiments; G.D. made the Aldh1a1-deficient mice; and B.P.L., O.H.Y., and S.J.M. designed, executed, and interpreted experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sean J. Morrison, Howard Hughes Medical Institute, Department of Internal Medicine and Center for Stem Cell Biology, Life Sciences Institute, 5435 Life Sciences Institute, 210 Washtenaw Avenue, Ann Arbor, MI, 48109-2216; e-mail: seanjm@umich.edu.