Pretreatment aberrant DNA methylation patterns are stable at time of relapse in acute lymphocytic leukemia (ALL). We hypothesized that the detection of residual methylation alterations at the time of morphologic remission may predict for worse prognosis. We developed a real-time bisulfite polymerase chain reaction assay and analyzed the methylation levels of p73, p15, and p57KIP2 at the time of initial remission in 199 patients with Philadelphia chromosome-negative and MLL− ALL. Residual p73 methylation was detected in 18 (9.5%) patients, p15 in 33 (17.4%), and p57KIP2 in 7 (3.7%); 140 (74%) patients had methylation of 0 genes and 48 (25%) of more than or equal to 1 gene. In 123 (65%) patients, matched pretreatment samples were also studied and compared with remission ones: in 82 of those with initial aberrant methylation of at least one gene, 59 (72%) had no detectable methylation at remission and 23 (28%) had detectable residual methylation. By multivariate analysis, the presence of residual p73 methylation was associated with a significant shorter duration of first complete remission (hazard ratio = 2.68, P = .003) and overall survival (hazard ratio = 2.69, P = .002). In conclusion, detection of epigenetic alterations allows the identification of patients with ALL with standard risk but with poor prognosis.
Introduction
Acute lymphocytic leukemia (ALL) consists of a heterogeneous group of lymphoid malignancies with distinct molecular and immunophenotypic features.1 In adult ALL, current prognostic models are based on cytogenetic alterations. In particular, patients with the Philadelphia chromosome (Ph) alteration, accounting for close to 25% of adult patients, or less frequently patients with 11q23 (MLL) alterations, are known to have poor prognosis despite the use of intensive chemotherapy programs.1 The recent incorporation of imatinib mesylate to intensive chemotherapy in Ph+ ALL may be reversing this phenomenon.2 This suggests that the identification of new molecular targets may result in the development of improved risk-assessment classifications, therapies, and therefore better outcomes for selected patient populations with ALL.
Nevertheless, a large majority of patients with ALL, namely, those with Ph− or MLL− disease, are treated homogeneously despite their known molecular and phenotypic heterogeneity. Few advances in the therapy of ALL, other than the addition of tyrosine kinase inhibitors, have been made over the last decade.1,3 Most chemotherapy programs consist of polychemotherapy combinations using agents, such as vincristine, steroids, methotrexate, anthracyclines, cytarabine, and cyclophosphamide among several drugs.3,4 With this type of therapy, complete remission (CR) rates can be as high as 90%.4 Unfortunately, despite these initial responses, long-term survival occurs in only 30% to 40% of adult patients.4 In contrast to other leukemias, secondary long-lasting remissions are difficult to achieve in ALL, and most patients with relapsed ALL will die of their disease.5 Therefore, the identification of new molecular markers to predict relapse in patients with Ph− ALL is a fundamental strategy in the development of new therapies for this subset of patients.
DNA methylation refers to the addition of a methyl group to a cytosine when this is followed by a guanine in the so-called CpG pairs.6 DNA methylation of areas rich in CpG pairs in the proximity of key regulatory genetic regions, such as gene promoters, is associated with gene silencing.6 Aberrant DNA methylation of these regions is frequently observed in cancer and results in the functional inactivation of its associated genes. This is considered an alternative to the physical inactivation, via deletion or mutations, of tumor suppressor genes in cancer.7 In ALL, aberrant DNA methylation of multiple promoter-associated CpG islands is very frequent both in adult8,9 and pediatric disease10,11 and is independent of the cytogenetic characteristics of the leukemic clone. Inactivation of specific molecular pathways by aberrant DNA methylation has been shown to predict for poor prognosis, both at the DNA9,12 and protein levels.13
We had previously compared the methylation patterns before treatment and at the time of relapse in a cohort of patients with adult ALL.14 In that study, we observed that, in 70% of patients, pretreatment and relapse methylation patterns were identical. This indicated that pretreatment aberrant DNA methylation patterns are stable at the time of relapse in a majority of patients.14 This also suggested that aberrant DNA methylation patterns are an integral molecular feature of the ALL leukemic clone and could possibly serve to identify residual disease. Based on this observation, we hypothesized that the detection of residual aberrant DNA methylation at the time of remission in patients with ALL may predict for worse prognosis in patients with ALL in remission. To test this hypothesis, we have analyzed the frequency of methylation of 3 genes, p73, p15, and p57KIP2, in remission bone marrows from 199 patients with Ph−, MLL− disease treated homogeneously at one institution. These 3 genes, when methylated, are known to confer poor prognosis to patients with ALL.12 Our results indicate that residual methylation of these genes can be detected at the time of morphologic remission and may predict for poor prognosis.
Methods
Study population
Study samples corresponded to 199 patients with Ph− and MLL− adult ALL, including lymphoblastic leukemia, obtained at the time of initial documentation of remission induction, between days 14 and 28 of initial therapy that had achieved a CR. All patients have been treated with hyperfractionated cyclophosphamide, vincristine, adriamycin, dexamethasone with alternating methotrexate and cytarabine (hyper-CVAD)15 chemotherapy at M. D. Anderson Cancer Center (MDACC). Criteria for CR has been previously described.15 Median follow-up of this patient population is 8 years (range, 0.5-12.5 years). Patient characteristics are shown in Table 1. The characteristics of the study group are representative of the whole group of patients with ALL (N = 428) treated at MDACC with hyper-CVAD chemotherapy from 1992 to 2002 (data not shown). The median survival time was 209 weeks (95% confidence interval [CI], 158-514).
Patient characteristics
| Patient characteristic . | All patients (N = 199) . | Unpaired patients (n = 76) . | P* . | ||
|---|---|---|---|---|---|
| n . | % . | n . | % . | ||
| Median age, y (range) | 38 | (15-83) | 44 | (16-77) | .23 |
| Male sex | 122 | 61 | 38 | 50 | .37 |
| Median percentage of marrow blasts (range) | 86 | (1-99) | 87 | (2-97) | .44 |
| Median WBC, ×109/L (range) | 6.0 | (0.3-602.5) | 4.5 | (0.7-372.6) | .06 |
| Median Hgb, g/dL (range) | 8.8 | (2.5-16.3) | 8.85 | (4-15.8) | .82 |
| Median platelet count, ×109/L (range) | 53 | (10-529) | 53.5 | (10-348) | .95 |
| Cytogenetics | |||||
| Diploid | 64 | 32 | 21 | 28 | .07 |
| Other | 89 | 45 | 35 | 46 | |
| IM | 42 | 21 | 16 | 21 | |
| ND | 4 | 2 | 4 | 5 | |
| Relapse | 105 | 53 | 36 | 47 | .31 |
| Disease-free survival, wk, median (range) | 136 | (112-NR) | 208 | (110-NR) | .21 |
| Overall survival, wk, median (range) | 209 | (135-NR) | 234 | (179-NR) | .39 |
| Patient characteristic . | All patients (N = 199) . | Unpaired patients (n = 76) . | P* . | ||
|---|---|---|---|---|---|
| n . | % . | n . | % . | ||
| Median age, y (range) | 38 | (15-83) | 44 | (16-77) | .23 |
| Male sex | 122 | 61 | 38 | 50 | .37 |
| Median percentage of marrow blasts (range) | 86 | (1-99) | 87 | (2-97) | .44 |
| Median WBC, ×109/L (range) | 6.0 | (0.3-602.5) | 4.5 | (0.7-372.6) | .06 |
| Median Hgb, g/dL (range) | 8.8 | (2.5-16.3) | 8.85 | (4-15.8) | .82 |
| Median platelet count, ×109/L (range) | 53 | (10-529) | 53.5 | (10-348) | .95 |
| Cytogenetics | |||||
| Diploid | 64 | 32 | 21 | 28 | .07 |
| Other | 89 | 45 | 35 | 46 | |
| IM | 42 | 21 | 16 | 21 | |
| ND | 4 | 2 | 4 | 5 | |
| Relapse | 105 | 53 | 36 | 47 | .31 |
| Disease-free survival, wk, median (range) | 136 | (112-NR) | 208 | (110-NR) | .21 |
| Overall survival, wk, median (range) | 209 | (135-NR) | 234 | (179-NR) | .39 |
WBC indicates white blood cells; Hgb, hemoglobin; IM, insufficient metaphases; ND, not done; and NR, not reached.
This analysis was performed between the 76 patients without a paired diagnostic sample (n = 76) and those with a paired sample (n = 123). Thus, results are shown for the full cohort of patients (N = 199), as they are the focus of this analysis.
Samples consisted of paraffin-embedded bone marrow biopsies or bone marrow aspiration clots from adult patients with ALL obtained to document remission during days 14 through 28 of induction therapy. All patients had achieved a CR. Histologic diagnosis was confirmed for each sample by a dedicated hematopathologist (C.E.B.-R.). In a subset of patients (123), paired samples obtained before initiation of therapy were also studied. No differences were observed between patients with paired samples and those without (Table 1). Informed consent for sample collection was obtained from all patients following institutional guidelines in accordance with the Declaration of Helsinki. This study was reviewed and approved by the Institutional Review Board at MDACC.
DNA extraction and bisulfite modification
DNA extraction and bisulfite modification have been previously described.14
DNA was extracted from paraffin-embedded bone marrow biopsies and bone marrow aspiration clots. Samples were deparaffinized using xylene and ethanol followed by digestion with proteinase K. After conventional DNA extraction, DNA was modified with sodium bisulfite. Bisulfite treatment of DNA converts unmethylated CpG sites to UpG without modifying methylated sites. In summary, DNA was denatured in 0.2 N NaOH at 37°C for 10 minutes and incubated with 3 M sodium bisulfite at 50°C for 16 hours. DNA was then purified using the Wizard cleanup system (Promega, Madison, WI) and desulfonated with 0.3 N NaOH at 25°C for 5 minutes. DNA was then precipitated with ammonium acetate and ethanol, washed with 70% ethanol, dried, and resuspended in H2O.
Real-time bisulfite polymerase chain reaction
To study DNA methylation, we developed a real-time polymerase chain reaction (PCR) bisulfite assay (Q-MSP). Three genes were studied: p73, p15, and p57KIP2. These genes are known to be frequently methylated in ALL and to predict for poor prognosis when methylated.12 Primers and probes were designed to specifically amplify bisulfite-converted DNA, either of the 3 target genes or an internal reference gene interferon-γ (IFN-γ). Primers and probes for p57KIP2, p15, and p73 were designed in regions known to be inversely correlated with gene expression.12 Each Q-MSP primer set contained 9 to 11 CpG dinucleotides of the promoter sequence. To quantify methylation density, we used the IFN-γ gene as an internal control. IFN-γ has very few CpG sites, is a single copy gene, and has no homology with other known genes. Table 2 summarizes primer and probe sequences. Q-MSP assays were carried out in a reaction volume of 20 μL in 96-well plates in an Applied Biosystems Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA). PCR was done in separate wells for each primer/probe set. The final reaction mixture consisted of TaqMan Universal PCR Master mix (Applied Biosystems) according to the manufacturer's protocol, 1 μM of each primer (Invitrogen, Carlsbad, CA), 200 nmol/L probe (Applied Biosystems), and 2 μL bisulfite-converted DNA. PCR was done with the following conditions: 95°C for 10 minutes, followed by 55 cycles at 95°C for 15 seconds and 58°C for 1 minute. A standard curve was run with artificially methylated DNA treated with SssI. The methylation status of the SssI-treated DNA was confirmed using the combined restriction PCR assay (COBRA)17 (methylation was > 90%) and by pyrosequencing18 (methylation, 81.4%). The SssI-treated DNA was diluted at 1:1, 1:10, 1:100, and 1:1000. We then studied the sensitivity and specificity of methylation of the 3 genes using cell line–derived DNA treated with SssI. Methylation density was calculated using the following formula: methylation % = 2CT of EFM − CT of target gene × 100, where CT indicates the number-of-cycles threshold and EFM indicates the estimated 100% full methylation (EFM) of the target gene derived from the standard curves of the IFN-γ and target gene. Using normal controls, known to be unmethylated for all 3 genes, no false positive was detected. Using positive controls (artificially methylated DNA with SssI or cell lines known to be methylated for a particular gene), the threshold of detection was between 1% and 2%, approximately 21 to 42 copies per gene, using 1 μg DNA (data not shown).
Primers and probes for Q-MSP
| Gene . | Primer and probe sequence . | GenBank accession no.16 . |
|---|---|---|
| p57KIP2 | Forward: 5-CGCCCGACTCTACGTATACG-3 | D64137 |
| Reverse: 5-CGGACGAGATAGGCGAATTC-3 | ||
| Probe: 5-GCGACGACTACCTAACTATCCGATAATAAACTC-3 | ||
| p15 | Forward: 5-AACTCAAAACCGCTCTAACCG-3 | AC000049 |
| Reverse: 5-AGCGAGGCGGGGTAGTGAG-3 | ||
| Probe: 5-CAAAATACGAACGCGTCGCGAAATC-3 | ||
| p73 | Forward: 5-CGTCCCGACTAACCTAACGC-3 | Y11416 |
| Reverse: 5-GTATTCGTTCGGAGGTTCGC-3 | ||
| Probe: 5-CGATTTCGCTACGTCCCCTTCGC-3 | ||
| IFN-γ | Forward: 5-CCTTAACTATCACAACTAAATAAATTCCC-3 | X13274 |
| Reverse: 5-AAGATGGGTATAATATGGGTATGAAGT-3 | ||
| Probe: 5-CCACAAAATTATTAACATCATTATACTTCATACCCATAT-3 |
| Gene . | Primer and probe sequence . | GenBank accession no.16 . |
|---|---|---|
| p57KIP2 | Forward: 5-CGCCCGACTCTACGTATACG-3 | D64137 |
| Reverse: 5-CGGACGAGATAGGCGAATTC-3 | ||
| Probe: 5-GCGACGACTACCTAACTATCCGATAATAAACTC-3 | ||
| p15 | Forward: 5-AACTCAAAACCGCTCTAACCG-3 | AC000049 |
| Reverse: 5-AGCGAGGCGGGGTAGTGAG-3 | ||
| Probe: 5-CAAAATACGAACGCGTCGCGAAATC-3 | ||
| p73 | Forward: 5-CGTCCCGACTAACCTAACGC-3 | Y11416 |
| Reverse: 5-GTATTCGTTCGGAGGTTCGC-3 | ||
| Probe: 5-CGATTTCGCTACGTCCCCTTCGC-3 | ||
| IFN-γ | Forward: 5-CCTTAACTATCACAACTAAATAAATTCCC-3 | X13274 |
| Reverse: 5-AAGATGGGTATAATATGGGTATGAAGT-3 | ||
| Probe: 5-CCACAAAATTATTAACATCATTATACTTCATACCCATAT-3 |
Accessed at http://www.ncbi.nlm.nih.gov/GenBank/GenBanksearch.htm, January 1, 2007.
Statistical analysis
Fisher exact test or χ2 test was performed to determine the association of p73, p15, and p57KIP2 methylation status with categorical patient characteristics. The Wilcoxon rank sum test determined the significance of differences of continuous patient characteristics between the methylation and nonmethylation groups. All patient characteristics with P less than .2 by the Fisher exact test, χ2 test, or the Wilcoxon rank sum test were included as predictor variables in the multivariate logistic regression analysis to identify potential risk factors for methylation. The predictor variables were removed one by one until all the variables in the model were significant at a .05 significance level. Univariate Cox proportional hazards regression model characterized the association between each of the patient characteristics and methylation measures and overall survival (OS) and disease-free survival (DFS). Patient characteristics with P less than .2 in univariate analysis were included in the multivariate Cox proportional hazard model as predictor variables to predict OS and DFS. As before, they were removed one by one until all the predictor variables remain in the model were significant at a significance level of .05. The Kaplan-Meier method was used to estimate both OS and DFS probabilities. The analyses were performed in SAS 9.1 (SAS, Cary, NC) and Splus 2000 (TIBCO, Palo Alto, CA). In general, for these analyses, a sample was considered “methylated” if the methylation ratio was at least 10%.
Results
Frequency of aberrant DNA methylation of p73, p15, and p57KIP2 at the time of remission in ALL
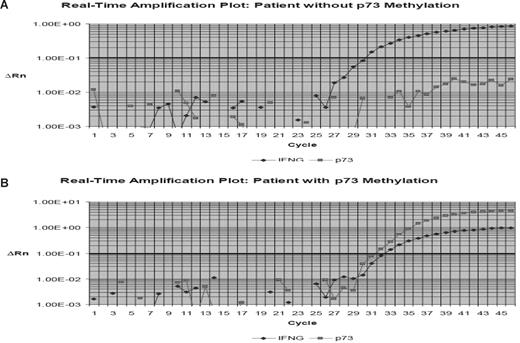
We analyzed the frequency of methylation of p73, p15, and p57KIP2 using Q-MSP in 199 patients with ALL at the time of remission. Table 1 summarizes patient characteristics. The distribution of DNA methylation is shown in Table 3. Overall, 17.4% (33 of 189 patients) had a p15 methylation density greater than 10%. For p57KIP2, 3.7% (7 of 189) had methylation density greater than 10%, and for p73, 9.5% (18 of 189) had methylation greater than 10%. Seventy-four percent (141 of 189) had methylation of 0 genes; 21% (40), 4.2% (8), and 1 had methylation of 1, 2, or 3 genes, respectively. No significant correlation was detected between methylation of any of the genes analyzed at the time of remission. Figure 1 shows representative examples of real-time PCR tracings.
Distribution of percentage of methylation at remission
| Gene . | Percent methylation, n (%) . | |||
|---|---|---|---|---|
| ≤ 5 . | 5.1-10 . | 10.1-20 . | > 20 . | |
| p57KIP2 | 182 (96) | 0 | 0 | 7 (4) |
| p15 | 152 (80) | 4 (2) | 5 (3) | 28 (15) |
| p73 | 169 (89) | 2 (1) | 2 (1) | 16 (8) |
| Gene . | Percent methylation, n (%) . | |||
|---|---|---|---|---|
| ≤ 5 . | 5.1-10 . | 10.1-20 . | > 20 . | |
| p57KIP2 | 182 (96) | 0 | 0 | 7 (4) |
| p15 | 152 (80) | 4 (2) | 5 (3) | 28 (15) |
| p73 | 169 (89) | 2 (1) | 2 (1) | 16 (8) |
Values are stated as n (%) for number (percentage) of patients.
Example of p73 real-time bisulfite PCR. Hypermethylated p73 gene promoter region DNA detected by use of the ABI Prism 7000 Sequence Detection System (TaqMan). (A) Example of a patient with hypermethylated p73. (B) Example of a case without p73 methylation. p73 methylation was estimated in relation to the detection of the interferon-γ gene (IFNG). ΔRn is defined as the cycle-to-cycle change in the reporter fluorescence signal normalized to a passive reference fluorescence signal (log scale). The horizontal line represents the threshold. CT indicates cycle threshold, the cycle number at which the Rn exceeds the baseline by the value of the threshold.
Example of p73 real-time bisulfite PCR. Hypermethylated p73 gene promoter region DNA detected by use of the ABI Prism 7000 Sequence Detection System (TaqMan). (A) Example of a patient with hypermethylated p73. (B) Example of a case without p73 methylation. p73 methylation was estimated in relation to the detection of the interferon-γ gene (IFNG). ΔRn is defined as the cycle-to-cycle change in the reporter fluorescence signal normalized to a passive reference fluorescence signal (log scale). The horizontal line represents the threshold. CT indicates cycle threshold, the cycle number at which the Rn exceeds the baseline by the value of the threshold.
Comparison of pretreatment and remission DNA methylation patterns
To evaluate the dynamics of DNA methylation changes as a result of therapy, we analyzed the methylation characteristics of paired samples from 123 (65%) patients obtained before treatment and at remission. Baseline characteristics of this subset of patients did not differ from the total group of patients studied here (Table 1). Table 4 summarizes the methylation characteristics before treatment and at the time of remission. Patients at the time of remission had significantly lower levels of methylation compared with pretreatment (Table 4).
Pretreatment and remission methylation characteristics
| Gene . | Pretreatment . | Remission . | P . | ||||
|---|---|---|---|---|---|---|---|
| Median . | Average . | Range . | Median . | Average . | Range . | ||
| p57KIP2 | 0 | 14.98 | 0-100 | 0 | 3.9 | 0-100 | .001 |
| p15 | 10.48 | 37.54 | 0-100 | 0 | 11.78 | 01-100 | .001 |
| p73 | 0 | 25.52 | 0-100 | 0 | 5.92 | 0-100 | .001 |
| Gene . | Pretreatment . | Remission . | P . | ||||
|---|---|---|---|---|---|---|---|
| Median . | Average . | Range . | Median . | Average . | Range . | ||
| p57KIP2 | 0 | 14.98 | 0-100 | 0 | 3.9 | 0-100 | .001 |
| p15 | 10.48 | 37.54 | 0-100 | 0 | 11.78 | 01-100 | .001 |
| p73 | 0 | 25.52 | 0-100 | 0 | 5.92 | 0-100 | .001 |
We analyzed patients based on whether methylation remained unchanged, decreased, or increased with therapy. For this analysis, a sample was considered methylated if the ratio of methylation was more than or equal to 10%. Methylation was considered stable (unchanged) if it was above or below that threshold at both endpoints. A decrease in methylation was considered if methylation was observed before treatment but not at remission. An increase in methylation was considered if the reverse was observed. These criteria have been previously used.14 Overall, methylation decreased in 59 (47%) patients and increased in 8 (6.5%) patients. In 33 (26.8%) patients, no significant methylation was detected before treatment or at remission. In 23 (19%) patients who had significant methylation pretreatment, this was also detected at remission. Because it is possible that the methylation changes observed are a result of the effects of chemotherapy and not necessarily the result of cellular replacement, we analyzed global methylation in 19 patients. Global methylation was assessed using the LINE assay as previously described.19 No differences in terms of global LINE methylation changes were observed between pretreatment and remission samples (Figure 2). This indicates that the methylation changes observed are not global chemical changes to methylated genes but a result of the elimination of the malignant clone.
Dynamics of global DNA methylation pretreatment and at the time of remission documentation. Global DNA methylation was studied using the LINE bisulfite pyrosequencing assay as described. No differences in LINE methylation were observed pretreatment and at the time of remission (P = .1). Lines connect individual patient LINE methylation values pretreatment and at remission. The figures indicate mean methylation levels and ranges. This indicates that gene-specific methylation changes described herein are not the result of decreased leukemia burden.
Dynamics of global DNA methylation pretreatment and at the time of remission documentation. Global DNA methylation was studied using the LINE bisulfite pyrosequencing assay as described. No differences in LINE methylation were observed pretreatment and at the time of remission (P = .1). Lines connect individual patient LINE methylation values pretreatment and at remission. The figures indicate mean methylation levels and ranges. This indicates that gene-specific methylation changes described herein are not the result of decreased leukemia burden.
Pathologic correlates
We then studied the correlation between DNA methylation of each gene, or group of genes, with the following patient characteristics: sex, performance status, presence of hepatosplenomegaly, thoracic adenopathy, immunophenotype, pretreatment cytogenetic analysis, risk of central nervous system involvement by the Kantarjian et al model,15 age, albumin, β2 microglobulin, total bilirubin, CD10 expression, creatinine, hemoglobin, lactic dehydrogenase, percentage of peripheral blood blasts, pretreatment platelet count, fraction of cells in S phase, and presenting white blood cells. By multivariate analysis, p15 was associated only with increased albumin levels as a continuous variable (P = .007). For p57KIP2, there was no characteristic associated with methylation status. p73 was associated with both increased creatinine and age (P = .046 and .03, respectively). Both were also studied as a continuous variable.
We also analyzed the relation between patient characteristics and methylation of at least one gene of the triad of p15, p73, and p57KIP2. Age (P = .02), albumin (P = .03), and β2-microglobulin (P = .04) were associated by univariate analysis. By multivariate analysis, only age (P = .01) was maintained.
Impact of residual methylation on DFS
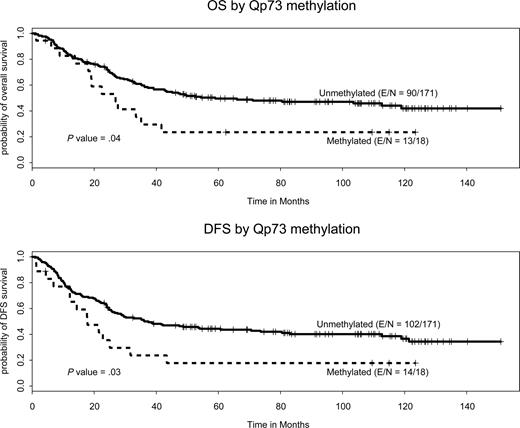
The impact of residual methylation of the 3 genes or groups on DFS was subsequently studied. By univariate analysis, age (hazard ratio [HR] = 1.02, P < .001) and residual p73 methylation (HR = 1.86, P = .03) were associated with a shorter DFS (Figure 3), whereas a higher platelet count (HR = 0.996, P = .004) was associated with a lower risk for relapse. The effect of p73 methylation on DFS was observed if a p73 cutoff of DNA methylation of 5% (P = .04), 10% (P = .03), or 20% (P = .08) was used and it was lost if a cutoff of 30% was used (P = .35). Patients with detectable p73 methylation at remission had a median DFS of 80 weeks (95% CI, 56 to not applicable) compared with a median of 213 weeks for patients with undetectable methylation (95% CI, 130 to not applicable; Figure 3). OS and DFS at 1, 2, and 5 years with CIs are shown in Table 5. No interaction between age and p73 methylation was observed. No effect on DFS was observed when methylation of p15 or p57KIP2 or the group of genes was studied or when methylation of any gene (methylation of 0, 1, 2, or 3 genes) was analyzed.
Estimated survival probabilities of OS and DFS at 1, 2, and 5 years using the Kaplan-Meier method
| Survival/p73 methylation status . | Survival probability (95% CI) . | ||
|---|---|---|---|
| 1-year . | 2-year . | 5-year . | |
| OS | |||
| Unmethylated | 0.84 (0.78-0.89) | 0.72 (0.66-0.80) | 0.496 (0.43-0.58) |
| Methylated | 0.83 (0.67-1) | 0.53 (0.34-0.83) | 0.24 (0.1-0.56) |
| DFS | |||
| Unmethylated | 0.75 (0.69-0.82) | 0.62 (0.55-0.70) | 0.44 (0.38-0.53) |
| Methylated | 0.77 (0.60-1) | 0.36 (0.19-0.68) | 0.18 (0.06-0.50) |
| Survival/p73 methylation status . | Survival probability (95% CI) . | ||
|---|---|---|---|
| 1-year . | 2-year . | 5-year . | |
| OS | |||
| Unmethylated | 0.84 (0.78-0.89) | 0.72 (0.66-0.80) | 0.496 (0.43-0.58) |
| Methylated | 0.83 (0.67-1) | 0.53 (0.34-0.83) | 0.24 (0.1-0.56) |
| DFS | |||
| Unmethylated | 0.75 (0.69-0.82) | 0.62 (0.55-0.70) | 0.44 (0.38-0.53) |
| Methylated | 0.77 (0.60-1) | 0.36 (0.19-0.68) | 0.18 (0.06-0.50) |
Impact of residual methylation on OS
By univariate analysis, age (P < .001), platelets (P = .02), and presence of residual p73 methylation (P = .03) were associated with a shorter survival (Figure 3). Patients with detectable p73 methylation at remission had a median OS of 118 weeks (95% CI, 85 to not applicable) compared with 257 weeks (95% CI, 173 to not applicable). In the final multivariate analysis (Table 6), the same characteristics were detected. No effect of p15 or p57 methylation was observed, nor of methylation of more than 1 gene of this triad (methylation of 0, 1, 2, or 3 genes) on OS. The effect of p73 methylation was observed if a p73 cutoff of DNA methylation of 5% (P = .04), 10% (P = .04), or 20% (P = .08) was used, and it was lost if a cutoff of 30% was used (P = .19).
Multivariate Cox proportional hazard model: OS (final)
| Variable . | HR (95% CI) . | P . |
|---|---|---|
| Age | 1.03 (1.02-1.04) | < .001 |
| Platelets (log 10 scale) | 0.51 (0.31-0.84) | .009 |
| Qp73 (methylation vs non) | 2.69 (1.46-4.96) | .002 |
| Variable . | HR (95% CI) . | P . |
|---|---|---|
| Age | 1.03 (1.02-1.04) | < .001 |
| Platelets (log 10 scale) | 0.51 (0.31-0.84) | .009 |
| Qp73 (methylation vs non) | 2.69 (1.46-4.96) | .002 |
Effect of the dynamics of DNA methylation on OS/DFS
Because of the potential for a prognostic value based on the dynamics of methylation changes comparing pretreatment and remission values, we analyzed this for each individual gene and group of genes. Patients were divided into 4 groups: (1) those with no methylation pretreatment and at the time of remission, (2) those with methylation pretreatment but not at the time of remission, (3) those with persistent methylation at both endpoints, and (4) those who had gained methylation at the time of remission. Methylation was defined as a cutoff positivity of 10%. No significant difference was seen between the 4 groups in terms of OS or DFS for any gene or combination of genes. The same nonsignificant results were observed if different cutoff points were used (data not shown). Overall, patients with no methylation before treatment and at remission had a nonsignificant trend toward better OS and DFS, in particular, for p73 (data not shown).
Discussion
Previously, we have demonstrated that the pretreatment DNA methylation patterns of specific genes are stable in a majority of patients at the time of relapse. These methylation patterns, therefore, could serve as a biomarker or signature of the malignant clone in the setting of minimal residual disease. In this study, we first demonstrated that residual DNA methylation of specific genes can be detected in patients with ALL at the time of morphologic remission. Because the levels of global DNA methylation did not change between the different endpoints, this residual DNA methylation is probably the result of the persistent leukemic clone. Furthermore, by multivariate analysis, the detection of residual methylation of p73 in patients with Ph−/MLL− adult ALL at the time of morphologic remission was associated with a significantly worse OS and DFS. Patients with this alteration had an estimated DFS of 80 weeks and OS of 118 weeks compared with 212 and 257 weeks, respectively, for those without these abnormalities. This alteration was observed in 9 (18%) patients. This is the first report of the use of detection of epigenetic information as a marker of minimal residual disease in ALL. A recent report has indicated that the same type of analysis may be used to detect minimal residual disease in AML.20
These results have significant implications. First, we have identified a subset of patients with poor prognosis who are currently considered as having standard risk because they are Ph- and MLL-negative. The main challenge with adult ALL is not remission induction but control of remission duration. Relapse in this patient population is almost universally associated with death. Therefore, the identification of such a subset of poor-risk patients may allow the introduction of newer therapies, such as early allogeneic stem cell transplantation, as is usually recommended for patients with Ph+ or MLL+ disease. Alternatively, if methylation of these key genes is a necessary biologic characteristic of the malignant clone, hypomethylating agents may have a role in the armamentarium of ALL therapy. Indeed, this hypothesis is currently being tested in a phase 1 study of decitabine in relapsed/refractory ALL.21
In our initial study,14 we had demonstrated that the methylation patterns at the time of relapse were almost identical in close to 70% of patients compared with the methylation patterns observed at initial presentation. In the current study, we had taken this observation further by showing that DNA methylation patterns are erased in a large proportion of patients (47%). In a subset of patients (19%), no change was observed; and in a smaller set, an observation was made of increased methylation. The significance of this last observation is not currently understood but could represent clonal evolution. Although the numbers of these patients are small in this study, this concept needs to be further investigated.
To perform these studies, we have developed an innovative bisulfite PCR technique with a reasonable turnaround time. This technique allows the analysis of DNA methylation of a particular gene(s) in less than 24 hours, with the main limiting step being the required bisulfite treatment. Because we used a conventional real-time PCR procedure, this type of assay could be then performed in any clinical laboratory. Therefore, if further validated with the right set of genes and the right methylation pattern, this could evolve into a powerful clinical tool to help guide therapy. The major advantage of this type of assay is the fact it measures a static molecular lesion of the leukemia cell and could complement other assays of minimal residual disease, such as flow cytometry, and perhaps aid in the use of specific interventions, such as the use of hypomethylating agents as a maintenance approach in ALL for a selected group of patients. This will have to be tested in clinical trials.
Despite these initial results, there are several limitations to the data presented here. The first relates to the genes selected in this study. We had based our analysis on prior data indicating that these 3 genes are frequently methylated in ALL and that their methylation confers a poor prognosis to this patient population before treatment.8,12,14 Second, we had prior evidence that the 3 genes were methylated at a high frequency in relapse samples.12,14 Despite this, however, only p73 was informative in the current analysis. It is possible that other gene(s) may prove to be as informative or more in this setting. As such, this study should be viewed as proof-of-principle endeavor that could be expanded to a broader set of genes. Indeed, these results should be expanded using a larger number of genes in a significantly higher number of patients. Furthermore, we could not define with precision the best cutoff point for methylation. Levels of 5% to 20% maintained its prognostic value, an effect lost if a cutoff of 30% was used. This was the result of the small number of patients with methylation of more than 30% at remission. The study of more genes and a larger cohort of patients may allow the analysis of more precise cutoff points. In addition, it should be noted that, in this study, we could not observe an impact on the dynamics of methylation on survival; furthermore, and in contrast with prior reports, methylation at the time of diagnosis had no prognostic impact. The reasons for this are unclear at this time but reinforce the need for validation studies using a larger cohort of patients.
Next, although the detection of p73 methylation at the time of morphologic remission identified a new subgroup of poor prognosis patients, it is notable that this was a relatively small number of patients, making up only 18% of this Ph−, MLL− study cohort. Evaluation of more genes and methylation patterns is needed to capture a larger cohort of patients at risk.
In conclusion, in this study, we have used a PCR-based real-time technique to detect residual aberrant methylation in the marrow of patients at the time of complete morphologic response. Residual p73 hypermethylation at CR in patients with Ph−, MLL− predicts for worse OS and worse DFS and may select patients for more aggressive early treatment. These results need to be confirmed in other cohorts of patients. The analysis of more genes may result in the development of new minimal residual disease detection techniques in ALL that could be used in the context of other assays, such as flow cytometry techniques or other molecular assays.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Cancer Institute (grants CA100067 and CA105771) and by the University of Texas Physician-Scientist Program funded by the Commonwealth Foundation for Cancer Research at the University of Texas M. D. Anderson Cancer Center and the Leukemia & Lymphoma Society of America (G.G.-M.).
National Institutes of Health
Authorship
Contribution: H.Y. performed all experiments; T.K. revised and helped write the manuscript; C.E.B.-R. performed all hematopathology analyses; L.X. and G.L.R. provided statistical analysis; K.H. initiated the experimental design; E.J. and S.P. contributed to data analysis; D.A.T., S.O., and H.M.K. contributed patients and helped with the write-up of the manuscript; and G.G.-M. designed, supervised, funded the studies, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guillermo Garcia-Manero, Department of Leukemia, University of Texas, M. D. Anderson Cancer Center, Box 428, 1515 Holcombe Boulevard, Houston, TX 77030; e-mail: ggarciam@mdanderson.org.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal