Abstract
TNFRSF13B encodes transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI), a B cell– specific tumor necrosis factor (TNF) receptor superfamily member. Both biallelic and monoallelic TNFRSF13B mutations were identified in patients with common variable immunodeficiency disorders. The genetic complexity and variable clinical presentation of TACI deficiency prompted us to evaluate the genetic, immunologic, and clinical condition in 50 individuals with TNFRSF13B alterations, following screening of 564 unrelated patients with hypogammaglobulinemia. We identified 13 new sequence variants. The most frequent TNFRSF13B variants (C104R and A181E; n = 39; 6.9%) were also present in a heterozygous state in 2% of 675 controls. All patients with biallelic mutations had hypogammaglobulinemia and nearly all showed impaired binding to a proliferation-inducing ligand (APRIL). However, the majority (n = 41; 82%) of the pa-tients carried monoallelic changes in TNFRSF13B. Presence of a heterozygous mutation was associated with antibody deficiency (P <.001, relative risk 3.6). Heterozygosity for the most common mutation, C104R, was associated with disease (P < .001, relative risk 4.2). Furthermore, heterozygosity for C104R was associated with low numbers of IgD−CD27+ B cells (P = .019), benign lymphoproliferation (P < .001), and autoimmune complications (P = .001). These associations indicate that C104R heterozygosity increases the risk for common variable immunodeficiency disorders and influences clinical presentation.
Introduction
Common variable immunodeficiency disorders (CVIDs) are the most prevalent primary immunodeficiencies in adults, with a frequency of about 1:25 000. They are characterized by hypogammaglobulinemia and a failure to mount protective antibody responses upon vaccination or pathogen exposure.1 Individuals affected with CVIDs are susceptible to recurrent infections by encapsulated bacteria, emphasizing the role of antibodies in the defense against these pathogens. Other clinical problems seen at increased frequency in CVID patients include: autoimmunity,2,3 nonmalignant lymphoproliferation (eg, splenomegaly, follicular nodular hyperplasia of the gastrointestinal tract, and lymphadenopathy),1 and malignancies, especially lymphoma and gastric cancer.1,4,5
The immunologic and cellular phenotypes of CVID patients are manifold and the majority of patients show a perturbed antigen-dependent terminal B-cell differentiation. Analysis of peripheral B-cell subsets identified a reduction of switched CD27+ memory B cells as a consistent feature of the immunological phenotype in CVID patients, which has provided a basis for disease classification models in CVID.3,6,7
Most cases of CVID occur sporadically, but in approximately 10% of CVID cases, at least one additional family member has either CVID or selective IgA deficiency (sIgAD).8 In the majority of multiplex CVID families, the mode of inheritance is autosomal dominant, but in about 20%, the mode is autosomal recessive. Biallelic mutations of any of the 4 genes: ICOS,9 TNFRSF13B,10,11 CD19,12 and TNFRSF13C13 have previously been shown to cause recessive CVID. In addition, heterozygous mutations in TNFRSF13B have been associated with CVID.10,11 The purpose of this study was to clarify and quantify aspects of that association by evaluating a large cohort of CVID patients.
The tumor necrosis factor receptor (TNFR) superfamily member transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI) is encoded by TNFRSF13B. TACI is preferentially expressed on marginal zone B cells, CD27+ memory B-cell subsets, and plasma cells.14–17 It binds the 2 ligands a proliferation inducing ligand (APRIL) and B-cell activating factor (BAFF). These interactions induce class switch recombination (CSR) in human and murine B cells.18–21 TACI−/− mice show impaired T cell–independent type II (TI-2) responses against polysaccharide antigens.22 In addition, these mice spontaneously develop lymphoproliferation and a fatal autoimmune syndrome.23
Previous studies in CVID patients have identified a variety of sequence variants in TNFRSF13B of which the most common are A181E and C104R.10,11,24,25 Both these variants are also seen in a heterozygous state in healthy individuals, albeit at lower frequencies.24,25 We undertook a large study to estimate the risk of CVID associated with these monoallelic variants. We thus recruited a cohort of 564 unrelated patients, including 433 newly described patients, and analyzed them for TNFRSF13B mutations. We evaluated their immunological and clinical status and assessed whether there is any genotype/phenotype association between the more common mutations and the different pathologies of CVIDs. Our evaluation of the relevance for the rare mutations included: absence of the mutation in a large collection of controls, presence of the mutation along with a more common mutation in a compound heterozygous state, and/or bioinformatic sequence analysis indicating that the mutation is likely to affect TACI function.
Methods
Patient and control cohorts
The study initially included 579 patients with hypogammaglobulinemia from 564 unrelated families. Of the 579 patients, 533 patients were diagnosed with CVID according to the European Society for Immunodeficiencies (ESID) criteria (see Supplemental Methods in Document S1 for diagnostic criteria; available on the Blood website; see the Supplemental Materials link at the top of the online article),26 41 patients suffered from IgG subclass deficiencies, and 5 patients were diagnosed with Good syndrome. Among the 564 index patients, 520 cases were sporadic and 44 came from multiplex families. Among the patients, 204 originated from the United Kingdom and Ireland, 105 from Germany, 101 from the United States, 60 from Scandinavia (49 from Sweden and 11 from Norway), 46 from Italy, and 26 from Brazil and Colombia. Most of the remaining 37 patients were from scattered locations in Europe and were white or had incomplete country/ethnicity information. One hundred thirty-one patients, including 72 from Germany and 48 from Sweden were included in a cohort described in a previously published report.25 Informed written consent was obtained from each patient or parental guardian prior to participation, in accordance with the Declaration of Helsinki. The research was conducted under the internal ethics review board–approved research protocol no. 239/99 (ZERM, University Hospital Freiburg, Freiburg, Germany) and no. 186/06 (ZERM, University Hospital Freiburg and REC Great Ormond Street Hospital and Royal Free Hospital, London, United Kingdom).
A total of 675 healthy controls were included in this study. This control cohort comprised 283 anonymous healthy individuals from the United Kingdom, 100 anonymous healthy blood donors from Germany, 50 controls from Italy, and 25 from South America. An additional 217 healthy donors were collected from personnel and students working at or attending the University of Freiburg. These last 217 controls were also included in a cohort described in a previously published report.25 These participants provided consent under a separate ethics protocol for healthy donors, no. 186/05 (ZERM, University Hospital Freiburg and REC Great Ormond Street Hospital and Royal Free Hospital).
We analyzed all 5 coding exons of TNFRSF13B in 579 patients. In 183 patients all exons were analyzed by direct sequencing; in 396 patients exons 1, 2, 3, and 4 were screened for mutations by heteroduplex analysis and subsequent sequencing, and by direct sequencing of exon 5.
In 107 control samples, all 5 coding exons of TNFRSF13B were analyzed by direct sequencing, and TNFRSF13B exons 3 and 4 were screened for mutations by heteroduplex analysis and subsequent sequencing in additional 568 control samples.
Heteroduplex analysis
Heteroduplex analysis was performed using fluorescence-labeled primers and conditions as previously described.27 Heteroduplex formation was induced by mixing equal amounts of patient polymerase chain reaction (PCR) product with a control DNA PCR product followed by denaturation at 95°C for 5 minutes with 5°C decrements every 2 minutes until the base holding temperature (25°C) was reached. Heteroduplex analysis was carried out on an ABI Prism 377 DNA Sequencer (PE Applied Biosystems, Foster City, CA) and on a MegaBACE 1000 DNA Analyser (GE Healthcare, Chalfont St Giles, United Kingdom). The results were analyzed using the Genetic Profiler v.2.0 software (GE Healthcare) or GENOTYPER software (PE Applied Biosystems). Peaks were compared with control DNA homoduplexes. Any with an abnormal morphology (eg, split or extra peaks) were reamplified using unlabeled primers and then sequenced.
Sequence analysis of TNFRSF13B
PCR amplification of TNFRSF13B and sequencing were performed with primers and conditions as previously described.10 Sequence analysis was performed with DNA Sequencing Analysis software, version 3.4 (PE Applied Biosystems), Sequencer version 3.4.1 (Gene Codes Corporation, Ann Arbor, MI), and BioEdit (Ibis Biosciences, Carlsbad, CA).
TACI expression
We stained 106 Epstein-Barr virus–transformed B cells28 with either a monoclonal rat anti–human TACI antibody (1A1; Abcam, Cambridge, MA or Alexis Biochemicals, San Diego, CA) or a biotinylated polyclonal goat anti–human TACI antibody (Peprotech EC, London, United Kingdom) followed by phycoerythrin (PE)–labeled goat anti–rat IgG or streptavidin-PE or the appropriate PE-labeled isotype controls together with CD19-PC7 (J4.119; Beckman Coulter, Paris, France). At least 104 cells, gated according to their forward and sideward scatters, were collected on a FACSCalibur (Becton Dickinson, Franklin Lakes, NJ) and analyzed using FlowJo software (TreeStar, Ashland, OR).
Binding of Flag-APRIL
Staining of EBV-transformed cell lines was performed as described previously.10 Briefly, 106 EBV-transformed cells were stained with 10-50 ng Flag-ACRP-hAPRIL29 in the presence of 0.1 μL heparin (Liquemin; Roche Pharma, Grenzach-Wyhlen, Germany) and detected with monoclonal mouse anti-Flag antibody M2 (Sigma, Seelze, Germany) and PE-labeled goat anti–mouse antibodies (Caltag, Hamburg, Germany). The specificity of Flag-ACRP-hAPRIL binding to EBV-transformed B cells was assessed by simultaneous staining of an EBV-transformed B-cell line known to carry a TNFRSF13B null mutation (S144X).10
Results
Sequence analysis of TNFRSF13B in 564 index patients with antibody deficiency
We analyzed a cohort of 564 patients with antibody deficiency and found that 50 (8.9%) carried at least one altered TNFRSF13B allele (Figure 1; Table 1). Of those 50 unrelated patients, 2 (4%) carried homozygous mutations, 7 (14%) carried compound heterozygous mutations, and 41 (82%) carried heterozygous mutations in TNFRSF13B. One of these patients carried a double mutation (D41H;c.298insT) on one TNFRSF13B allele. The polymorphisms R72H (rs55916807), V220A (rs56063729), and P251L (rs34562254) were observed in the present cohort of antibody-deficient patients and controls at frequencies comparable to previously published frequencies in CVID patient cohorts and healthy controls, respectively.10,24,25
Mutations of TNFRSF13B in 50 of 564 index patients with antibody deficiency. Distribution of the observed TNFRSF13B mutations across the TNFRSF13B/TACI protein. Each symbol represents one patient. ● indicates patients with homozygous TNFRSF13B mutations. ○ represents patients with heterozygous TNFRSF13B mutations.  designates a patient with a double mutation on one allele of TNFRSF13B. Compound heterozygous mutated TACI-deficient patients are labeled as indicated in the figure. CRD indicates cysteine-rich domain; stalk, stalk region; and TM, transmembrane domain.
designates a patient with a double mutation on one allele of TNFRSF13B. Compound heterozygous mutated TACI-deficient patients are labeled as indicated in the figure. CRD indicates cysteine-rich domain; stalk, stalk region; and TM, transmembrane domain.
Mutations of TNFRSF13B in 50 of 564 index patients with antibody deficiency. Distribution of the observed TNFRSF13B mutations across the TNFRSF13B/TACI protein. Each symbol represents one patient. ● indicates patients with homozygous TNFRSF13B mutations. ○ represents patients with heterozygous TNFRSF13B mutations.  designates a patient with a double mutation on one allele of TNFRSF13B. Compound heterozygous mutated TACI-deficient patients are labeled as indicated in the figure. CRD indicates cysteine-rich domain; stalk, stalk region; and TM, transmembrane domain.
designates a patient with a double mutation on one allele of TNFRSF13B. Compound heterozygous mutated TACI-deficient patients are labeled as indicated in the figure. CRD indicates cysteine-rich domain; stalk, stalk region; and TM, transmembrane domain.
Mutations in TNFRSF13B
| Exon . | NCBI_SS no. . | cDNA* . | Protein† . | Domain . |
|---|---|---|---|---|
| 2 | 10516792 | c.118T>C | p.W40R | CRD1, extracellular |
| 10516793 | c.121delG | p.D41Ifs*43 | CRD1, extracellular | |
| 10516794 | c.121G>C | p.D41H | CRD1, extracellular | |
| 3 | 10516795 | c.204insA | p.L69Tfs*12 | CRD2, extracellular |
| 10516796 | c.236A>G | p.Y79C | CRD2, extracellular | |
| 10516797 | c.260T>A | p.I87N | CRD2, extracellular | |
| 10516798 | c.298insT | p.C100Lfs*6 | CRD2, extracellular | |
| 48424922 | c.310T>C | p.C104R | CRD2, extracellular | |
| 10516799 | c.311G>A | p.C104Y | CRD2, extracellular | |
| 10516800 | c.445G>A | p.A149T | Stalk region | |
| 4 | 10516801 | c.455G>A | p.G152E | Stalk region |
| 10516802 | c.492C>A | p.Y164X | Stalk region | |
| 10516803 | c.542C>A | p.A181E | Transmembrane | |
| 10516804 | c.571insG | p.D191Gfs*46 | Intracellular | |
| 10516805 | c.579C>A | p.C193X | Intracellular | |
| 5 | 10516806 | c.736G>T | p.V246F | Intracellular |
| Exon . | NCBI_SS no. . | cDNA* . | Protein† . | Domain . |
|---|---|---|---|---|
| 2 | 10516792 | c.118T>C | p.W40R | CRD1, extracellular |
| 10516793 | c.121delG | p.D41Ifs*43 | CRD1, extracellular | |
| 10516794 | c.121G>C | p.D41H | CRD1, extracellular | |
| 3 | 10516795 | c.204insA | p.L69Tfs*12 | CRD2, extracellular |
| 10516796 | c.236A>G | p.Y79C | CRD2, extracellular | |
| 10516797 | c.260T>A | p.I87N | CRD2, extracellular | |
| 10516798 | c.298insT | p.C100Lfs*6 | CRD2, extracellular | |
| 48424922 | c.310T>C | p.C104R | CRD2, extracellular | |
| 10516799 | c.311G>A | p.C104Y | CRD2, extracellular | |
| 10516800 | c.445G>A | p.A149T | Stalk region | |
| 4 | 10516801 | c.455G>A | p.G152E | Stalk region |
| 10516802 | c.492C>A | p.Y164X | Stalk region | |
| 10516803 | c.542C>A | p.A181E | Transmembrane | |
| 10516804 | c.571insG | p.D191Gfs*46 | Intracellular | |
| 10516805 | c.579C>A | p.C193X | Intracellular | |
| 5 | 10516806 | c.736G>T | p.V246F | Intracellular |
SS indicates submitted SNP.
NCBI nucleotide entry NM_012452.
NCBI protein entry NP_036584.
Sixteen different genetic alterations were observed, 13 of which have not been previously described. Two new nonsense alleles and 4 frameshift mutations (3 single nucleotide insertions and 1 single nucleotide deletion) were found. Among the 564 patients, the most frequently mutated allele was C104R, which was present in 26 patients (4.6%), followed by the A181E allele, which was present in 13 patients (2.3%). Only these 2 alleles were observed in a homozygous state, each in one individual. In addition, C104R was also present in 5 of 7 compound heterozygous mutations.
Sequence variants of TNFRSF13B in healthy subjects
A total of 675 controls were analyzed for sequence variants in TNFRSF13B. Seven (1%) controls were heterozygous for the A181E variant, 6 (0.9%) healthy controls were heterozygous for the C104R allele, and 1 control was heterozygous for the I87N allele. The missense mutations W40R, D41H, Y79C, C104Y, A149T, G152E, and V246F were not found in the healthy control cohort. In addition, no biallelic mutation was found in the control cohort.
Table 2 gives estimates of the statistical significance and relative risk of developing an antibody deficiency associated with different types of mutations in different patient subsets relative to the corresponding set of controls. Having a monoallelic or biallelic mutation in TACI was strongly associated with hypogammaglobulinemia (P < .001; relative risk, 4.3; 95% CI 2.4-7.6). The association of a monoallelic C104R mutation with antibody deficiency was found to be highly significant (P < .001; relative risk 4.2; 95% CI 1.7-10.1). In contrast, the association with the A181E allele was not found to be statistically significant. The association of the A181E variant with CVID has, however, previously been described as significant in an even larger cohort of CVID patients and controls.25
Frequencies, significance, and relative risk for any mutation, any monoallelic mutation, or heterozygous C104R in different subsets of patients
| Patient subset (n) . | Genotype . | n (%) . | P* . | Relative risk (95% CI) . |
|---|---|---|---|---|
| All patients (564) | Any mutation | 50 (8.9) | < .001 | 4.3 (2.4-7.6) |
| Sporadic patients, nonfamilial (520) | Any mutation | 42 (8.1) | < .001 | 3.9 (2.2-6.9) |
| All patients, no biallelic mutation (555) | Monoallelic mutation | 41 (7.4) | < .001 | 3.6 (2.0-6.4) |
| All patients, no mutation other than het. C104R (535) | Heterozygous C104R | 20 (3.7) | .008 | 4.2 (1.7-10.1) |
| Patient subset (n) . | Genotype . | n (%) . | P* . | Relative risk (95% CI) . |
|---|---|---|---|---|
| All patients (564) | Any mutation | 50 (8.9) | < .001 | 4.3 (2.4-7.6) |
| Sporadic patients, nonfamilial (520) | Any mutation | 42 (8.1) | < .001 | 3.9 (2.2-6.9) |
| All patients, no biallelic mutation (555) | Monoallelic mutation | 41 (7.4) | < .001 | 3.6 (2.0-6.4) |
| All patients, no mutation other than het. C104R (535) | Heterozygous C104R | 20 (3.7) | .008 | 4.2 (1.7-10.1) |
CI indicates confidence interval.
P values are one-sided and calculated by a χ2 test.
Bioinformatic analysis of TACI amino acid substitutions
We carried out bioinformatic analysis of 17 amino acid substitutions in TACI including those reported here and some detected only in earlier studies (Table 3). The general aim was to predict which changes are likely to be deleterious and, if possible, determine which aspects of protein function (eg, the folding energy) are affected. We used the same approach and most of the software packages used by Thusberg and Vihinen30 to carry out a similar analysis of substitutions in the neutropenia-associated protein ELA2.
Summary of results from 18 bioinformatics methods applied to 17 amino acid substitutions observed in TACI in either this study or earlier studies
| Substitution . | Seq conserv . | Stability center . | Disorder . | Energy change . | Steric clashes . | β-Aggregation . | Reference . |
|---|---|---|---|---|---|---|---|
| W40R | XX | X | NA | NA | This study | ||
| D41H | XX | NA | NA | This study | |||
| R72H | XX | This study, 10,24 | |||||
| Y79C | XX | X | This study | ||||
| I87N | XX | X | XX | This study | |||
| C104R | XX | X | XX | X | This study, 10,11,24,25,29 | ||
| C104Y | XX | XX | X | This study | |||
| R122W | XX | X | NA | NA | 25 | ||
| A149T | NA | NA | This study | ||||
| G152E | XX | NA | NA | This study | |||
| L171R | X | NA | NA | 24,29 | |||
| C172Y | XX | X | NA | NA | X | 29 | |
| A181E | X | X | NA | NA | This study, 10,11,24,25,29 | ||
| R202H | X | NA | NA | 10,11,25 | |||
| V220A | X | NA | NA | This study, 10,11,24,25 | |||
| V246F | X | NA | NA | This study | |||
| P251L | XX | NA | NA | This study, 10,11,24,25 |
| Substitution . | Seq conserv . | Stability center . | Disorder . | Energy change . | Steric clashes . | β-Aggregation . | Reference . |
|---|---|---|---|---|---|---|---|
| W40R | XX | X | NA | NA | This study | ||
| D41H | XX | NA | NA | This study | |||
| R72H | XX | This study, 10,24 | |||||
| Y79C | XX | X | This study | ||||
| I87N | XX | X | XX | This study | |||
| C104R | XX | X | XX | X | This study, 10,11,24,25,29 | ||
| C104Y | XX | XX | X | This study | |||
| R122W | XX | X | NA | NA | 25 | ||
| A149T | NA | NA | This study | ||||
| G152E | XX | NA | NA | This study | |||
| L171R | X | NA | NA | 24,29 | |||
| C172Y | XX | X | NA | NA | X | 29 | |
| A181E | X | X | NA | NA | This study, 10,11,24,25,29 | ||
| R202H | X | NA | NA | 10,11,25 | |||
| V220A | X | NA | NA | This study, 10,11,24,25 | |||
| V246F | X | NA | NA | This study | |||
| P251L | XX | NA | NA | This study, 10,11,24,25 |
For categories in which there were numerous methods, we distinguish between XX (consensus of the methods predicts that this change is deleterious), X (some methods predict that the change is deleterious and some do not), and no mark (most if not all methods predict that the change is benign). This table is a compilation of summaries of results that can be found in Tables S1 through S16.
Seq conserv indicates sequence conservation; and NA, not available.
We categorized the methods based on which aspect of the protein they evaluate: sequence conservation, stability, disordered regions, folding energy and contacts, steric conflicts, and aggregation of β-sheets. Predictions that depend on having a protein structure in Protein Data Bank (PDB) format31 are necessarily limited to 5 substitutions at positions in the structure for the CRD2 domain of TACI bound to APRIL.32 The majority (11/17) of the mutations were predicted to be deleterious by at least 2 sequence conservation methods. Only 3 mutations, A149T, V220A, and V246A, were predicted to be benign by all sequence conservation methods, but V220A and V246A may disturb stability centers of the protein. The most frequent mutation, C104R, was predicted to lead to energy changes, an increase in disorder, and steric clashes within the protein. Only one sequence conservation method predicted the A181E mutation to be deleterious, but the mutation is located at a predicted stability center of TACI. The I87N mutation was predicted to cause energy changes and found at a stability center of the protein. The bioinformatic analysis for TACI is less definitive than for ELA2 because heterozygous mutations in ELA2 are generally sufficient to cause at least a cyclic form of neutropenia, while heterozygous mutations in TACI may increase the risk of developing hypogammaglobulinemia but not lead to CVID in some carriers. Additional details and tabular results for the bioinformatic methods can be found in Supplemental Methods in Document S1 and Tables S1 through S16.
Familial segregation
Among the 564 unrelated patients, 520 cases were sporadic and 44 were from multiplex families. In 28 families the pattern of inheritance was autosomal dominant, in 13 the pattern was autosomal recessive, and in 3 the pattern was undetermined.
Of the 50 index patients with mutations in TNFRSF13B, 8 had first-degree relatives with a history of humoral immunodeficiency. One of these families (A181E heterozygous) has been previously reported (family 5 in Figure S1).25 The pedigrees of the remaining 7 familial cases are shown as families A through G in Figure 2 (arrows indicate index patients) and a summary of clinical, immunological, and genetic data for these families is shown in Table 4.
Pedigrees of familial TACI deficiency. Pedigrees of 7 families with TACI deficiency are shown. Symbols: circles indicate female; squares, male; black filled symbols, CVID; gray filled symbols, IgG subclass deficiency or dysgammaglobulinemia; hatched symbols, selective IgA deficiency; slash, deceased; and arrow, index patient. Mutations shown in brackets are inferred. §IDs refer to Voerechovsky et al.33
Pedigrees of familial TACI deficiency. Pedigrees of 7 families with TACI deficiency are shown. Symbols: circles indicate female; squares, male; black filled symbols, CVID; gray filled symbols, IgG subclass deficiency or dysgammaglobulinemia; hatched symbols, selective IgA deficiency; slash, deceased; and arrow, index patient. Mutations shown in brackets are inferred. §IDs refer to Voerechovsky et al.33
Families with mutations in TNFRSF13B
| Family . | Individual . | Age, y . | Affection status . | TNFRSF13B mutation . | IgG, g/L . | IgA, g/L . | IgM, g/L . |
|---|---|---|---|---|---|---|---|
| A | I.1 | 42 | Healthy | C104R | 12.9 | 0.83 | 1.24 |
| I.2 | 40 | Healthy | C104Y | 12.04 | 1.6 | 0.82 | |
| II.1 | 14 | CVID | C104R/C104Y | 2.87 | 0.06 | 0.14 | |
| II.2 | 4 | CVID | C104R/C104Y | 3.59 | 0.12 | 0.19 | |
| B | I.1 | 55 | Dysgamma | I87N* | 5.5 | 0.36 | 0.99 |
| I.2 | 54 | Healthy | Y79C | 7.6 | 1.4 | 1.3 | |
| II.1 | 33 | IgG subclass deficiency | Y79C/I87N | 5.9 | 1.1 | 1.3 | |
| II.2 | 32 | IgG subclass deficiency | Y79C/I87N | 5.6 | 0.9 | 0.5 | |
| C | II.1 | 53 | CVID | c.204insA/C104R | 2.30 | 0.23 | 0.31 |
| III.1 | 37 | IgG1 subclass deficiency | c.204insA | 5.94 | 0.69 | 1.31 | |
| D | I.2 | 66 | CVID | C104R | 3.31 | 0.07 | 1.18 |
| II.1 | 36 | Dysgamma | C104R | 7.14 | 0.34 | 0.64 | |
| II.2 | 32 | CVID | C104R | 3.4 | 0 | 0.23 | |
| III.1 | 6 | Healthy | wt | 7.43 | 0.58 | 0.36 | |
| III.2 | 4 | Dysgamma | wt | 5.53 | 0.24 | 0.53 | |
| E(cv78) | I.1 | 87 | SIgAD | unknown | 16.0 | 0 | 3.6 |
| I.2 | 77 | Healthy | wt | 12.0 | 1.05 | 0.7 | |
| II.1 | 52 | CVID | wt | 8.8† | 0.1 | 0.1 | |
| II.2 | 57 | CVID | C104R | 7.4† | 0.1 | 0.1 | |
| II.3 | 56 | SIgAD | wt | 16.0 | 0 | 1.6 | |
| II.4 | 54 | SIgAD | wt | 15.0 | 0 | 1.6 | |
| F(cv132) | I.1 | 61 | Healthy | A181E | NA | NA | NA |
| I.2 | 56 | CVID | wt | 9.2† | 0.1 | < 0.1 | |
| II.1 | 19 | CVID | A181E | 2.73 | 0.17 | 0.25 | |
| II.2 | 26 | Healthy | A181E | 7.8 | 2.0 | 0.9 | |
| G | I.1 | 34 | Healthy | wt | 9.78 | 4.86 | 0.81 |
| I.2 | 32 | Healthy | D41H;c.298insT | 14.92 | 3.23 | 2.11 | |
| II.1 | 9 | CVID | D41H;c.298insT | 0.19 | 0.02 | 3.51 | |
| II.2 | 1 | Healthy | D41H;c.298insT | 8.98 | 0.62 | 0.71 |
| Family . | Individual . | Age, y . | Affection status . | TNFRSF13B mutation . | IgG, g/L . | IgA, g/L . | IgM, g/L . |
|---|---|---|---|---|---|---|---|
| A | I.1 | 42 | Healthy | C104R | 12.9 | 0.83 | 1.24 |
| I.2 | 40 | Healthy | C104Y | 12.04 | 1.6 | 0.82 | |
| II.1 | 14 | CVID | C104R/C104Y | 2.87 | 0.06 | 0.14 | |
| II.2 | 4 | CVID | C104R/C104Y | 3.59 | 0.12 | 0.19 | |
| B | I.1 | 55 | Dysgamma | I87N* | 5.5 | 0.36 | 0.99 |
| I.2 | 54 | Healthy | Y79C | 7.6 | 1.4 | 1.3 | |
| II.1 | 33 | IgG subclass deficiency | Y79C/I87N | 5.9 | 1.1 | 1.3 | |
| II.2 | 32 | IgG subclass deficiency | Y79C/I87N | 5.6 | 0.9 | 0.5 | |
| C | II.1 | 53 | CVID | c.204insA/C104R | 2.30 | 0.23 | 0.31 |
| III.1 | 37 | IgG1 subclass deficiency | c.204insA | 5.94 | 0.69 | 1.31 | |
| D | I.2 | 66 | CVID | C104R | 3.31 | 0.07 | 1.18 |
| II.1 | 36 | Dysgamma | C104R | 7.14 | 0.34 | 0.64 | |
| II.2 | 32 | CVID | C104R | 3.4 | 0 | 0.23 | |
| III.1 | 6 | Healthy | wt | 7.43 | 0.58 | 0.36 | |
| III.2 | 4 | Dysgamma | wt | 5.53 | 0.24 | 0.53 | |
| E(cv78) | I.1 | 87 | SIgAD | unknown | 16.0 | 0 | 3.6 |
| I.2 | 77 | Healthy | wt | 12.0 | 1.05 | 0.7 | |
| II.1 | 52 | CVID | wt | 8.8† | 0.1 | 0.1 | |
| II.2 | 57 | CVID | C104R | 7.4† | 0.1 | 0.1 | |
| II.3 | 56 | SIgAD | wt | 16.0 | 0 | 1.6 | |
| II.4 | 54 | SIgAD | wt | 15.0 | 0 | 1.6 | |
| F(cv132) | I.1 | 61 | Healthy | A181E | NA | NA | NA |
| I.2 | 56 | CVID | wt | 9.2† | 0.1 | < 0.1 | |
| II.1 | 19 | CVID | A181E | 2.73 | 0.17 | 0.25 | |
| II.2 | 26 | Healthy | A181E | 7.8 | 2.0 | 0.9 | |
| G | I.1 | 34 | Healthy | wt | 9.78 | 4.86 | 0.81 |
| I.2 | 32 | Healthy | D41H;c.298insT | 14.92 | 3.23 | 2.11 | |
| II.1 | 9 | CVID | D41H;c.298insT | 0.19 | 0.02 | 3.51 | |
| II.2 | 1 | Healthy | D41H;c.298insT | 8.98 | 0.62 | 0.71 |
Patients and relatives were assigned to their respective affection status as being healthy, or having sIgAD, dysgamma, IgG subclass deficiency, or CVID.
Values below normal range for age are printed in italics. Dysgamma indicates dysgammaglobulinemia; wt, wild type; and NA, not available.
This mutation was inferred.
IgG level while the patient received intravenous immunoglobulin substitution therapy.
In these 7 families, 2 important observations are notable: first, there are healthy or only mildly affected family members who have monoallelic or biallelic mutations, therefore mutations in TNFRSF13B are not 100% penetrant; and second, CVID patients E.II.1 and F.I.2 have 2 wild-type TNFRSF13B alleles, hence these patients teach us that mutations in TACI are a contributing factor to develop CVID but are not solely causative. More clinical information on the families can be found in Table 4 and Supplemental Results in Document S1.
TACI expression and APRIL binding
EBV-transformed B-cell lines were established from 25 patients with mutations in TNFRSF13B, 5 CVID patients with wild-type alleles of TNFRSF13B, and 5 healthy controls. These cell lines were analyzed for TACI expression and for binding of APRIL. An additional EBV-transformed cell line, homozygous for a stop codon mutation, S144X, and previously shown to lack TACI expression,10 served as a negative control. EBV-transformed cells were stained with (1) a polyclonal Ab directed against the extracellular domain of TACI, (2) a monoclonal Ab (mAb; clone 1A1), which requires Cys 104 for TACI recognition, and (3) the fluorescence-tagged APRIL.
Surface expression of TACI in EBV-transformed cell lines from controls was readily detected (Figure 3A) but was variable in controls and in patients. In all cases it was much higher than that of the negative homozygous S144X control. Cell lines from CVID patients without a mutation in TACI and healthy donors were used as controls (Figure 3A).
TACI expression and APRIL binding on EBV-transformed cell lines. (A) The histograms show the mean fluorescence intensity (MFI) of stainings of EBV-transformed B-cell lines with a polyclonal antibody against TACI (left), a monoclonal antibody recognizing the TACI extracellular domain (clone 1A1; middle), and a Flag-tagged recombinant APRIL construct. ♦ indicates C104R homozygous (n = 2); ◇, C104R heterozygous (n = 12);  , healthy donors (n = 5); and
, healthy donors (n = 5); and  , CVID patients without TNFRSF13B mutation (n = 5). The thick gray line in each graph indicates the background staining of a previously described homozygous S144X cell line.10 Mean values and SDs are shown at the right side of the scatter diagram of each group. Testing for significance was performed with the Student t test. (B) TACI expression and APRIL binding on EBV-transformed cell lines with homozygous or compound heterozygous TNFRSF13B mutations, stained with a polyclonal antibody directed against the TACI extracellular domain (top panels), stained with a monoclonal antibody recognizing the TACI extracellular domain (middle panels), and stained with a Flag-tagged APRIL construct (bottom panels). Thick black lines indicate patients; thick dashed black lines, healthy donors; and gray filled histograms, patients with TNFRSF13B null mutation (S144X homozygous, negative control).
, CVID patients without TNFRSF13B mutation (n = 5). The thick gray line in each graph indicates the background staining of a previously described homozygous S144X cell line.10 Mean values and SDs are shown at the right side of the scatter diagram of each group. Testing for significance was performed with the Student t test. (B) TACI expression and APRIL binding on EBV-transformed cell lines with homozygous or compound heterozygous TNFRSF13B mutations, stained with a polyclonal antibody directed against the TACI extracellular domain (top panels), stained with a monoclonal antibody recognizing the TACI extracellular domain (middle panels), and stained with a Flag-tagged APRIL construct (bottom panels). Thick black lines indicate patients; thick dashed black lines, healthy donors; and gray filled histograms, patients with TNFRSF13B null mutation (S144X homozygous, negative control).
TACI expression and APRIL binding on EBV-transformed cell lines. (A) The histograms show the mean fluorescence intensity (MFI) of stainings of EBV-transformed B-cell lines with a polyclonal antibody against TACI (left), a monoclonal antibody recognizing the TACI extracellular domain (clone 1A1; middle), and a Flag-tagged recombinant APRIL construct. ♦ indicates C104R homozygous (n = 2); ◇, C104R heterozygous (n = 12);  , healthy donors (n = 5); and
, healthy donors (n = 5); and  , CVID patients without TNFRSF13B mutation (n = 5). The thick gray line in each graph indicates the background staining of a previously described homozygous S144X cell line.10 Mean values and SDs are shown at the right side of the scatter diagram of each group. Testing for significance was performed with the Student t test. (B) TACI expression and APRIL binding on EBV-transformed cell lines with homozygous or compound heterozygous TNFRSF13B mutations, stained with a polyclonal antibody directed against the TACI extracellular domain (top panels), stained with a monoclonal antibody recognizing the TACI extracellular domain (middle panels), and stained with a Flag-tagged APRIL construct (bottom panels). Thick black lines indicate patients; thick dashed black lines, healthy donors; and gray filled histograms, patients with TNFRSF13B null mutation (S144X homozygous, negative control).
, CVID patients without TNFRSF13B mutation (n = 5). The thick gray line in each graph indicates the background staining of a previously described homozygous S144X cell line.10 Mean values and SDs are shown at the right side of the scatter diagram of each group. Testing for significance was performed with the Student t test. (B) TACI expression and APRIL binding on EBV-transformed cell lines with homozygous or compound heterozygous TNFRSF13B mutations, stained with a polyclonal antibody directed against the TACI extracellular domain (top panels), stained with a monoclonal antibody recognizing the TACI extracellular domain (middle panels), and stained with a Flag-tagged APRIL construct (bottom panels). Thick black lines indicate patients; thick dashed black lines, healthy donors; and gray filled histograms, patients with TNFRSF13B null mutation (S144X homozygous, negative control).
In 2 EBV-transformed cell lines with homozygous C104R mutations (from a newly identified patient and one from a previous report10 ), the binding of APRIL and of the mAb 1A1 were completely abolished (Figure 3A). This was expected as TACI C104R has previously been shown to be defective in APRIL binding, and because Cys 104 is part of the epitope recognized by mAb 1A1.10 Staining of C104R homozygous B-cell lines with the polyclonal antibody was reduced 2- to 3-fold on average when compared with wild type, suggesting that this mutation in a homozygous state also impairs surface expression of TACI (Figure 3A), or influences the stability of the protein. It is unlikely that this reduced staining results from decreased binding of the polyclonal antibody to TACI C104R, but indeed reflects reduced surface expression of the TACI C104R mutant. This was demonstrated by the reduced staining of the polyclonal antibody in 293T cells transfected with TACI C104R full-length constructs and the unimpaired staining of cells expressing a TACI-C104R-TRAIL-R3 fusion protein (Figure S3). More details on this experiment can be found in Supplemental Methods and Results in Document S1.
The analysis of 12 cell lines with the heterozygous C104R mutation revealed TACI surface expression by all 3 staining techniques but with a lower average staining intensity compared with TACI wild-type controls. These results are consistent with the surface expression of both wild-type TACI and C104R TACI. The results are also consistent with wild-type TACI staining with mAb anti-TACI (1A1) and with Flag-APRIL, while the mutated C104R allele was stained with neither. This also indicates that the TACI C104R allele does not interfere with surface expression and APRIL binding of the coexpressed wild-type TACI.
Single EBV-transformed cell lines carrying heterozygous frameshift/nonsense mutations (c.121delG; c.204insA; C193X) or the heterozygous missense mutation A181E displayed no significant differences in TACI expression and ligand-binding capacity compared with controls (Figure S1). A slight but specific decrease of APRIL binding was observed in the 2 I87N heterozygotes, suggesting that these mutant cells have a decreased binding of APRIL (Figure S1).
There were 5 EBV-transformed cell lines from CVID patients with compound heterozygous TNFRSF13B mutations available for testing; all showed severely impaired APRIL binding, despite surface expression of TACI (Figure 3B). It is noteworthy that the Y79C and I87N mutations are located in the ligand-binding domain of TACI (CRD2; Figures 1 and S2), and although these residues do not mediate direct contacts with APRIL,32 they may disturb the protein architecture, preventing APRIL binding.
Interestingly, the compound heterozygous mutations involving C104R in combination with a frameshift or nonsense mutation resemble closely the staining pattern that is seen in EBV-transformed cell lines carrying a C104R homozygous mutation (Figure 3B). This suggests that the frameshift or nonsense alleles are not expressed at the cell surface.
B-cell phenotype and immunoglobulin levels in TACI-deficient individuals
The CD19+ B-cell percentage of total blood lymphocytes was analyzed in 45 of the 50 index patients with TNFRSF13B mutations (Figure 4A left panel). B cells were present at normal percentages (6%-19% of lymphocytes) in the majority of patients (25 of 45). Severe B-cell lymphopenia (< 1%) was only identified in 1 of the 45 patients (c.121delG). This patient also showed a relative expansion of transitional IgD++IgM++CD24++CD38++ B cells (26.1% of B cells).
Immunologic phenotype of TACI-deficient patients. (A) CD19+ B-cell percentages of lymphocytes and IgD+IgM+CD27−, IgD−IgM+CD27+, and IgD−IgM−CD27+ B-cell subsets in patients with TACI deficiency. (B) Immunoglobulin levels at time of diagnosis in patients with TACI deficiency. Each circle represents a single value from one patient. ● indicates homozygous or compound heterozygous TNFRSF13B mutations;  , heterozygous TNFRSF13B mutations other than C104R; ○, C104R heterozygous mutations; and
, heterozygous TNFRSF13B mutations other than C104R; ○, C104R heterozygous mutations; and  , mean values for each patient group. Testing for significant differences between patient groups was performed with the Student t test.
, mean values for each patient group. Testing for significant differences between patient groups was performed with the Student t test.
Immunologic phenotype of TACI-deficient patients. (A) CD19+ B-cell percentages of lymphocytes and IgD+IgM+CD27−, IgD−IgM+CD27+, and IgD−IgM−CD27+ B-cell subsets in patients with TACI deficiency. (B) Immunoglobulin levels at time of diagnosis in patients with TACI deficiency. Each circle represents a single value from one patient. ● indicates homozygous or compound heterozygous TNFRSF13B mutations;  , heterozygous TNFRSF13B mutations other than C104R; ○, C104R heterozygous mutations; and
, heterozygous TNFRSF13B mutations other than C104R; ○, C104R heterozygous mutations; and  , mean values for each patient group. Testing for significant differences between patient groups was performed with the Student t test.
, mean values for each patient group. Testing for significant differences between patient groups was performed with the Student t test.
B-cell subset analysis was available for 37 of the 50 patients (Figure 4A right panel). IgD−IgM−CD27+ switched memory B cells were reduced in 28 of the 37 patients (76%) and below 2% of total B cells in 18 of the 37 analyzed patient samples (49%). This is comparable to the B-cell phenotypes of CVID patients in general.3,6 The proportion of IgD−IgM+CD27+ memory B cells and IgD+IgM+CD27− naive B cells varied considerably among patients. In particular, 13 of 17 (76%) C104R heterozygous patients had switched memory B cells below 2% of total B cells, whereas only 2 of 9 patients with homozygous or compound heterozygous mutations in TNFRSF13B were found to have switched memory B cells below 2% of total B cells. The difference in percentages of switched memory B cells was found to be significant, when the C104R heterozygous patient group was compared with the patients carrying 2 mutated TNFRSF13B alleles (P = .019; Student t test).
Serum IgG levels were available at diagnosis and/or prior to the onset of IgG substitution therapy for 40 of 50 patients (Figure 4B); these IgG levels showed major variation between patients. In 11 patients the total IgG level was below 1 g/L at the time of diagnosis, whereas 4 patients had IgG levels of about 5 g/L at the time of diagnosis. IgA and IgM levels were available for 46 patients (Figure 4B). IgA was deficient (< 0.07 g/L) in 25 patients, low (0.07-0.7 g/L) in 18 patients, and normal in 3 patients. Of the patients with normal IgA levels (0.07-4.0 g/L), one carried a compound heterozygous Y79C/I87N mutation, one had a heterozygous I87N mutation, and the third had a C104R heterozygous mutation. IgM was low in 31 patients (< 0.4 g/L), normal in 12 patients (0.4-2.3 g/L), and elevated (> 2.3 g/L) in 3 patients. Of the 3 patients with elevated IgM levels, all had CVID and one had a C104R heterozygous mutation, the second had a homozygous A181E mutation, and the third carried the double mutation (D41H;c.298insT).
Clinical phenotypes observed in TACI-deficient individuals
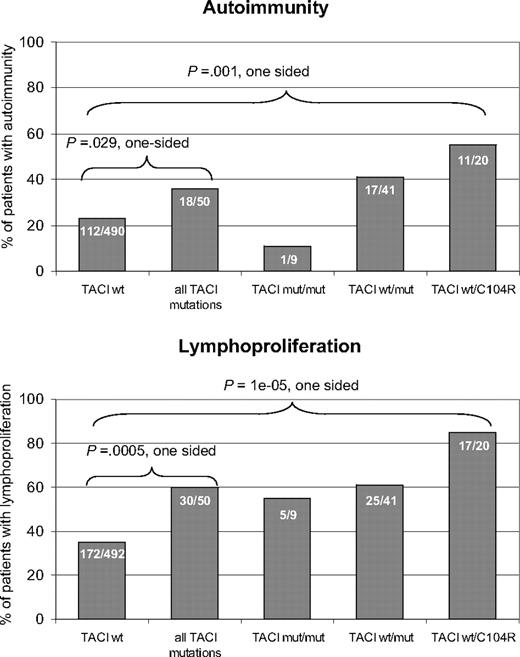
Among all index patients, 540 were evaluated for the presence of autoimmunity. Of these, 130 (24%) had at least one autoimmune manifestation. A total of 542 patients were evaluated for the presence of lymphoproliferation, which was found in 202 of these patients (37%). All patients suffered from recurrent infections as a consequence of the hypogammaglobulinemia. In the index patients with a wild-type TACI sequence, autoimmunity was present in 112 of 490 (23%), and lymphoproliferation was found in 172 of 492 (35%). In the TACI-mutated patient cohort, autoimmunity was present in 18 of 50 patients (36%; P = .029) and signs of lymphoproliferation were present in 30 of 50 patients (60%; P < .001) (Figure 5). Clinical details on all 50 TACI-deficient patients can be found in Table S17. Splenomegaly was the predominant sign of lymphoproliferation (22 patients). Other manifestations of lymphoproliferation were persistent lymphadenopathy (13 patients) and nodular lymphatic hyperplasia in the gut (2 patients). Eleven patients showed more than one feature of lymphoproliferation. Malignant lymphoproliferation was noted in 4 CVID patients with mutated TACI. One patient developed a cerebral lymphoma, a second had a low- grade malignant B-cell lymphoma, a third had T-cell lymphoma, and a fourth suffered from B-cell non-Hodgkin lymphoma and T-cell lymphoma. Three of the 4 patients carried heterozygous A181E mutations; the fourth carried a heterozygous C104R mutation. Notably, one A181E heterozygous patient reported by Castigli et al11 also suffered from a B-cell non-Hodgkin lymphoma.
Autoimmunity and lymphoproliferation in TACI deficiency. Histograms show the prevalence of autoimmune diseases (top panel) and lymphoproliferation (bottom panel) as percentages in CVID patients without TACI mutation, TACI-deficient patients, and subgroups of TACI-deficient patients with homozygous or compound heterozygous, C104R heterozygous, or other heterozygous mutations. Absolute numbers of patients are printed on the bars.
Autoimmunity and lymphoproliferation in TACI deficiency. Histograms show the prevalence of autoimmune diseases (top panel) and lymphoproliferation (bottom panel) as percentages in CVID patients without TACI mutation, TACI-deficient patients, and subgroups of TACI-deficient patients with homozygous or compound heterozygous, C104R heterozygous, or other heterozygous mutations. Absolute numbers of patients are printed on the bars.
The most prevalent autoimmune disease was autoimmune thrombocytopenia (ITP), seen in 7 patients. Other autoimmune manifestations included autoimmune hemolytic anemia (3 patients), the presence of anti-IgA antibodies (2 patients), vitiligo, diabetes mellitus, celiac disease, arthritis, autoimmune urticaria, autoimmune hepatitis, and recurrent parotitis (one patient each). These findings are comparable to previously reported surveys of CVID patients in general.1,3,34
However, most patients with autoimmunity and a TACI mutation (12 of 18) and with lymphoproliferation and a TACI mutation (21 of 30) carried at least one C104R allele (Figure 5). The prevalence of autoimmunity was significantly higher (P = .001) in the TNFRSF13B C104R heterozygous group than the group of patients with a wild-type TNFRSF13B sequence (Figure 5 top histogram). The prevalence of lymphoproliferation was also significantly higher (P < .001) in the C104R heterozygous group than in the patient groups with no mutation in TNFRSF13B (Figure 5 bottom histogram).
Discussion
In previous studies, the proportion of CVID patients carrying at least one TNFRSF13B mutation was estimated at 10 to 20%.10,11 Of a cohort of 564 antibody deficiency patients in the present study, 50 patients had at least one TACI allele mutated, yielding a more precise estimate of 9%. We confirmed the previous hypothesis that biallelic TNFRSF13B mutations are not found in healthy individuals; however, it should be noted that individuals with some biallelic mutations such as homozygous S144X10 and Y79C/I87N do not meet all current criteria for a diagnosis of CVID and may be clinically well. We identified 13 new mutations, including 5 nonsense and frameshift mutations. One limitation of this study is that we sequenced only exons of TNFRSF13B and the intron-exon borders; patients may have had mutations in the promoter or in regulatory sequences in introns that went undetected.
In our large CVID cohort, the only significant association of a specific TACI variant with CVID was that of TACI C104R (P < .001). Other sequence variants, including frameshift and nonsense mutations, were often observed only in single individuals and thus were too rare to reach statistical significance. Functional studies have demonstrated that the C104R variant impairs the function of TACI both in homozygous and heterozygous states.10,35 Heterozygous TNFRSF13B C104R is associated with CVID, but neither this mutation nor any other heterozygous mutation in TNFRSF13B can explain a sufficient percentage of the multiplex CVID families with dominant inheritance.
Although TACI expression on human peripheral blood B cells correlates well with the expression of CD27,14,17 we observed that TACI does not seem to be required for the development of memory B cells in humans. Several TACI-deficient patients accumulate normal numbers of switched and nonswitched memory B cells, including one hypogammaglobulinemic individual with a previously described TNFRSF13B null mutation (S144X)10 and a CVID patient with a compound heterozygous mutation presented here (C104R/c.204insA).
The IgG levels at diagnosis or prior to intravenous immunoglobulin therapy were low in the majority of TACI-deficient patients. Agammaglobulinemia (total Ig ≤ 1 g/L) was observed in 7 patients. IgA was the most severely affected isotype, which is typical for CVID and also consistent with our previous findings in patients with TACI deficiency.10 However, 3 patients with TNFRSF13B mutations had normal IgA levels at the time of diagnosis, which was based partly on low IgG levels. This suggests that in humans, switching to IgA can still occur when the APRIL-TACI interaction is impaired, in contrast to the findings in APRIL−/− mice.21 The BAFF receptor (BAFFR), another member of the TNFR superfamily, is also unlikely to be the major factor in class switching to IgA because BAFF-R–deficient mice and humans are capable of switching to IgA and have normal IgA levels in serum.13,36 In contrast to our initial study,10 the IgM levels in this cohort of TACI-deficient CVID patients were less affected, and we found 3 patients with elevated IgM serum levels at the time of diagnosis.
Because TACI-knockout mouse models show a prominent autoimmune phenotype and massive signs of lymphoproliferation,23 it has been speculated that human TACI deficiency in CVID patients is primarily associated with these features. In our initial study, 5 of 13 patients (38%) showed signs of autoimmunity.10 However, most of the autoimmune phenomena observed were mild (vitiligo, anti-IgA antibodies, autoimmune thyroiditis). Castigli et al11 reported no autoimmune phenomena in their 4 CVID patients with TNFRSF13B mutations. Zhang et al37 observed a higher prevalence of ITP in 6 of 13 TACI-deficient CVID patients (46%; 5/10 heterozygotes and 1/3 compound heterozygotes) versus 20 of 163 CVID patients without TNFRSF13B mutations (12%), but they did not compare the incidence of types of autoimmunity other than ITP.
Our current results suggest a more complex association of TNFRSF13B mutations and autoimmunity. Many types of autoimmunity other than ITP are seen in patients with TACI mutations, although ITP may be particularly common in C104R heterozygotes. The prevalence of autoimmunity is significantly lower (1/9, 11%) in patients with biallelic mutations in TNFRSF13B than in patients with the specific monoallelic mutation C104R (11/20, 55%; 2-sided P = .04; Fisher exact test). We speculate that the higher rate of autoimmunity in TACI heterozygotes arises because the wild-type TACI allele in heterozygotes may be able to promote the survival of autoreactive B-cell clones better than any of the 2 mutated TACI alleles available in individuals with biallelic mutations.
We have previously shown that individuals with homozygous TNFRSF13B mutations have a selective impairment of APRIL binding, APRIL induced B-cell proliferation, and CSR.10 Every cell line that we have studied from patients with compound heterozygous mutations failed to bind APRIL. Hence, it is assumed that primary B cells from patients with biallelic mutations do not respond to APRIL-mediated stimuli either.
The effects of heterozygous sequence variants are less clear. The recruitment of a TACI mutant in the APRIL signaling complex may exert a dominant-negative effect by disturbing the 3-fold symmetry required for proper signaling.29,31 However, as TACI C104R does not bind APRIL, a dominant negative effect would require preassociation of TACI C104R with wild-type TACI in a ligand-independent manner, as suggested by the recent report of Garibyan et al.35 In support of this model, we demonstrate surface expression of TACI in cell lines derived from patients with homozygous TACI C104R mutations. Thus, in heterozygous patients, TACI C104R will most likely be coexpressed at the surface with wild-type TACI. TACI was recently shown to respond to oligomeric BAFF or APRIL only, implying that at least 6 wild-type receptors are likely to be required in an active signaling complex,38 rendering the system particularly sensitive to dominant- negative effects of mutated receptors.
In summary, we found that about 9% of patients with antibody deficiency carry at least one mutated TNFRSF13B allele, and nearly 2% carry biallelic mutations. These estimates are consistent with previous studies, but are now based on a larger sample. Homozygous or compound heterozygous mutations in TNFRSF13B were exclusively found in individuals with clinical or immunological features of antibody deficiency. Incomplete penetrance and familial segregation were, however, demonstrated for heterozygous TNFRSF13B sequence variants and imply that heterozygous mutations increase the risk, but are neither necessary nor sufficient to cause CVID. Further studies are needed to unravel the additional genetic and environmental factors that act in concert with simple heterozygous genetic alterations in TNFRSF13B to give rise to antibody deficiencies in humans.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are indebted to Viviane Knerr, Sarita Workman, and Mara-Lynn Metzger for excellent technical assistance and to Dr Daniel Myrtek for critical reading and correction of the manuscript.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Bonn, Germany) GR1617/3 (B.G.), SFB620/C2 (B.G., U.S.), SFB620/Z1 (H.-H.P.), and SFB620/C1 (H.-H.P.); from the European Commission (Brussels, Belgium) SP23-CT-2005-006411 (H.C., B.G.), MEXT-CT-2006-042316 (B.G.), and HEALTH-F2-2008-201549 (U.S.); from the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID; Bethesda, MD)/USIDnet no. NO1-A1-30070 (B.G.); from the Primary Immunodeficiency Association (PIA; London, United Kingdom), Medical Research Council (MRC; London, United Kingdom), and The Wellcome Trust (London, United Kingdom; C.B., S.B., H.B.G., A.J.T., H.C.); from the Swedish Research Council (Stockholm, Sweden; Q.P.-H., L.H.); from the Fondazione C. Golgi (Brescia, Italy), and PRIN2006 (Rome, Italy; A.P.); from the Swiss National Science Foundation (Bern, Switzerland; P.S.); from the Grant Colciencias (Bogota, Colombia) 1115-05-16784 (J.L.F.); from the NIH (grant HD 37091 to H.D.O., H.C.), and from the Jeffrey Modell Foundation (New York, NY; H.D.O., H.C.). This research was supported in part by the Intramural Research Program of the National Institutes of Health, National Library of Medicine (NLM; E.M.G., A.A.S.).
National Institutes of Health
Authorship
Contribution: U.S. and C.B. wrote the manuscript and performed and evaluated the experiments; S.B., Q.P.-H., S.J., A.B., T.H., S.A.-S., and J.B. performed sequencing, TACI expression, and APRIL binding studies; V.L., A.P., A.D.B.W., H.-H.P., D.S., H.C., A.M.-T., G.P.S., H.D.O., S. Urschel, B.H.B., S. Ugrinovic, D.S.K., T.C.L., A.M.H., J.L.F., I.S., L.H., and A.J.T. provided patient samples, ascertained patients' clinical and immunological data, and edited the manuscript; P.S. provided Flag-APRIL constructs and participated in writing the manuscript; E.M.G. performed sequence analysis and edited the manu-script; A.A.S. performed statistical analyses and edited the manuscript; and H.B.G. and B.G. designed the research and wrote the manuscript.
Conflict of interest disclosure: The authors declare no competing financial interests.
Correspondence: Bodo Grimbacher, MD, Department of Immunology and Molecular Pathology, Royal Free Hospital & University College London, Pond Street, London NW3-2QG, United Kingdom; e-mail: bgrimbac@medsch.ucl.ac.uk.
References
Author notes
*U.S. and C.B. contributed equally to this work.