Abstract
Down syndrome (DS) children have a unique genetic susceptibility to develop leukemia, in particular, acute megakaryocytic leukemia (AMkL) associated with somatic GATA1 mutations. The study of this genetic susceptibility with the use of DS as a model of leukemogenesis has broad applicability to the understanding of leukemia in children overall. On the basis of the role of GATA1 mutations in DS AMkL, we analyzed the mutational spectrum of GATA1 mutations to begin elucidating possible mechanisms by which these sequence alterations arise. Mutational analysis revealed a predominance of small insertion/deletion, duplication, and base substitution mutations, including G:C>T:A, G:C>A:T, and A:T>G:C. This mutational spectrum points to potential oxidative stress and aberrant folate metabolism secondary to genes on chromosome 21 (eg, cystathionine-β-synthase, superoxide dismutase) as potential causes of GATA1 mutations. Furthermore, DNA repair capacity evaluated in DS and non-DS patient samples provided evidence that the base excision repair pathway is compromised in DS tissues, suggesting that inability to repair DNA damage also may play a critical role in the unique susceptibility of DS children to develop leukemia. A model of leukemogenesis in DS is proposed in which mutagenesis is driven by cystathionine-β-synthase overexpression and altered folate homeostasis that becomes fixed as the ability to repair DNA damage is compromised.
Introduction
Down syndrome (DS) children have an estimated 10- to 20-fold increased risk of developing acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) compared with non-DS children1 and overall represent approximately 2% and 15% of childhood ALL and AML cases, respectively. The majority of DS AML cases (> 90%) are the acute megakaryocytic phenotype (AMkL). It is estimated that DS children have a 500-fold increased risk of developing AMkL compared with non-DS children, highlighting a unique predisposition to develop a specific leukemia phenotype.2 A variant of AMkL, the transient myeloproliferative disorder (TMD), is diagnosed in DS neonates with blast cells that display the same morphologic and immunophenotypic characteristics of AMkL.3 The majority of TMD cases resolve spontaneously, although approximately 20% of infants will subsequently develop AMkL and require chemotherapy treatment.
A seminal finding from the laboratory of Dr John Crispino4 and subsequently confirmed by other groups was the occurrence of acquired somatic mutations at exon 2 of the transcription factor gene GATA1 (localized to Xp11.23), with nearly 100% penetrance in DS TMD and AMkL cases.4-10 The reported sequence alterations in the region encoding the N-terminal activation domain of GATA1 included insertion, deletion, missense, nonsense, and splice-site mutations at the exon 2/intron boundary that invariably result in the synthesis of a short-form GATA1 protein (GATA1s, 40-kDa protein compared with the 50-kDa wild-type protein) with altered transactivation capacity.4 These mutations are believed to result in accumulation of poorly differentiated megakaryocytic precursors and represent initiating ”genetic hits“ in a multistep process of leukemogenesis. The observation that these mutations occur exclusively in DS children with TMD/AMkL suggests the possibility that trisomy 21 induces a “mutator phenotype.” The prenatal presence of the GATA1 mutations establishes that genomic instability precedes leukemia and further supports the hypothesis that DS may induce a mutator phenotype.10,11 This finding is supported by additional observations, including reports that up to 18% of DS children with ALL have somatic mutations in a highly conserved region of the JAK2 gene12 and that persons with DS have atypical background somatic mutations at the hypoxanthine phosphoribosyl transferase locus.13
Analyzing the mutational spectrum of DS could provide critical insight into mechanisms of mutagenesis that may account for the significantly increased risk of this group to develop leukemia. To date, no studies have analyzed the mutation spectrum of sequence alterations at GATA1/exon2 in DS TMD/AMkL cases. In this report, our analysis of the mutational spectrum at GATA1 in DS suggests that uracil accumulation, oxidative stress, and reduced DNA base excision repair (BER) capacity may account for the generation of mutations as early leukemogenic events leading to the development of leukemia in DS children.
Methods
Clinical TMD and AMkL samples
Diagnostic blast cells from DS children with AMkL (n = 14) and TMD (n = 5) and from non-DS children with AMkL (n = 13) were obtained from the Children's Hospital of Michigan leukemia cell bank and from the Pediatric Oncology Group 9421 study as previously described.14,15 The diagnosis of AMkL was confirmed by flow cytometric detection of the megakaryocytic antigens CD41 and CD61. Mononuclear cells were isolated on Ficoll-Hypaque gradients to obtain highly purified mononuclear cell fractions consisting mostly of leukemic blasts (median, 66% and 81.5% in the DS and non-DS samples, respectively). DNA was isolated as described previously.14 Blast cells were screened for the presence of GATA1 exon 2 mutations in genomic DNAs by the use of forward (5′-GAGGGGGAAAGGAGGGAAGAGGAGCAGGTG-3′) and reverse (5′-CACTCAGCCAATGCCAAGACAGCCACTCAATG-3′) primers. Amplicons were sequenced directly and after subcloning in pGEM-T-Easy vector (Promega). Sequence comparisons and translation analyses were performed by the use of programs available from the National Center for Biotechnology Information. Total RNAs were extracted from the samples by the use of TRIzol reagent (Invitrogen Life Technologies) per manufacturer protocol.14 cDNAs were synthesized from 2 μg of RNA by the use of random hexamer primers and an reverse-transcription polymerase chain reaction (RT-PCR) kit (Perkin Elmer) and purified with the QIAquick PCR Purification Kit (QIAGEN). The research protocol was approved by the Human Investigation Committee of Wayne State University School of Medicine. Informed consent was provided according to the Declaration of Helsinki.
Sequence analysis of GATA1 exon 2 mutations in the literature
GATA1 exon 2 mutations reported in the literature4-9 were compiled and analyzed along with the mutations identified in our patient samples to expand the number of mutations being analyzed. Following the initial description of GATA1 exon 2 mutations (Wechsler et al4 ), we numbered the mutations from the initiating ATG in exon 2. The numbering systems used in other reports were modified to be consistent for easier identification of specific sites within exon 2. In Rainis et al,7 Groet et al,8 and Hitzler et al,6 the adenine of the ATG which was identified as base 1 was modified to be base 113, whereas in Xu et al,9 the numbering begins at the 5′ end of exon 2, 19 basepairs from our start site.
Fetal liver tissue
Fetal liver samples (17-23 weeks' gestation) from 5 DS and 7 non-DS fetuses were obtained from therapeutically terminated pregnancies. Total RNAs were isolated from the fetal liver by use of the VersaGene RNA isolation system (Gentra) per the manufacturer's protocol. The use of fetal tissue was approved by the Human Investigation Committee of Wayne State University School of Medicine.
Gene expression by real-time PCR
DNA polymerase beta (β-pol) and uracil DNA glycosylase (UDG) transcripts were quantitated by real-time RT-PCR (Roche). PCRs contained 2 μL of purified cDNA or standard plasmid, 4 mmol/L MgCl2, 0.5 μmol/L sense and antisense primers, and 2 μL of FastStart DNA Master SYBR Green I enzyme-SYBR reaction mix (Roche). Sense (5′-ggtgggccaagcaaggtgttc-3′) and antisense (5′-ggggagggatgagccgtctg-3′) primers to UDG were designed to detect the nuclear isoform of UDG. PCRs were carried out under the following conditions: initial denaturation at 95°C for 10 minutes, then samples were denatured by heating to 95°C, annealing at 58°C for 10 seconds, elongation at 72°C for 5 seconds for 50 cycles, followed by melting curve analysis from 40°C to 95°C, and a final cooling step to 40°C. Proprietary primers for detection of the low abundance β-pol transcript were purchased from SuperArray Biosciences. Transcript levels for β-pol and UDG were quantitated against standard curves derived from serial dilutions of the respective amplified regions and were normalized to either 18S RNA or glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Real-time RT-PCR results were presented as mean values from 3 independent experiments using the same cDNA preparation.
Uracil detection
DNA was isolated from fetal tissue by the use of QIAGEN gravity tip columns per manufacturer's protocol and assayed for uracil incorporation as described in Cabelof et al.16 In brief, 4 μg of DNA was blocked for 2 hours at 37°C in a 2X Tris/methoxyamine buffer (final concentration: 100 mmol/L methoxyamine; Sigma–Aldrich); 50 mmol Tris–HCl, pH 7.4). DNA was precipitated with 7.5% volume of 4 mol/L NaCl and 4 volumes of ice-cold 100% ethanol and resuspended in TE buffer, pH 7.6. DNA was then treated with 0.4 units of UDG (New England Biolabs) for 15 minutes at 37°C, immediately precipitated, and resuspended in TE buffer, pH 7.6. DNA was then probed with 2 mmol/L aldehydic reactive probe (Dojindo Molecular Technology) for 15 minutes at 37°C followed by ethanol precipitation and resuspension in TE buffer, pH 7.6. DNA was then heat-denatured, immobilized onto a nitrocellulose membrane (Schleicher and Schuell), and baked under vacuum. The dried membrane was washed in 5× saline sodium citrate for 15 minutes at 37°C, then incubated in a prehybridization buffer (20 mmol/L Tris, pH 7.5; 0.1 mol/L NaCl; 1 mmol/L EDTA; 0.5% casein wt/vol; 0.25% bovine serum albumin wt/vol; 0.1% Tween-20 vol/vol) for 30 minutes at room temperature. Streptavidin–peroxidase conjugate (Roche) was added at a 1:2000 dilution for 45 minutes at room temperature. Membranes were washed in TBS-Tween 3 times at 37°C, incubated in ECL solution (Pierce) for 5 minutes at room temperature, then visualized and quantified by the use of a ChemiImager system (Alpha Innotech). Data were expressed as the integrated density value of the band per microgram of DNA loaded on the membrane.
Base excision repair activity
Nuclear protein extracts were isolated from DS and non-DS fetal tissues by use of the CelLytic Nuclear Extraction kit from Sigma-Aldrich. Purified radio-end-labeled 30-basepair (bp) oligonucleotides (upper strand, 5′-ATATACCGCGGUCGGCCGATCAAGCTTATTdd-3′; lower strand, 3′-ddTATATGGCGCCGGCCGGCTAGTTCGAATAA-5′) containing a G:U mismatch and a HpaII restriction site (CCGG) were incubated in a BER reaction mixture containing 50 μg of nuclear protein as previously described.17 Repair of the G:U mismatch to a correct G:C base pair was determined by treatment with 20 U of HpaII (Promega) for 1 hour at 37°C and analysis by electrophoresis on a 20% denaturing 19:1 acrylamide:bis-acrylamide gel (SequaGel sequencing system; National Diagnostics). DNA oligonucleotides were visualized and quantified by the use of a Molecular Imager system (Bio-Rad), and repair was calculated by taking the ratio of repaired substrate (16-mer) to unrepaired substrate (30-mer; product/substrate). Data were expressed as machine counts per microgram of protein.
F2 isoprostane detection
F2 isoprostanes were measured in fetal liver tissue by the use of gas chromatography-mass spectrometry methods as previously described.18 In brief, tissue was homogenized in chloroform:methanol containing butylated hydroxytoluene (0.005%) to prevent auto-oxidation, dried under a stream of nitrogen, and resuspended in methanol containing butylated hydroxytoluene. Esterified F2 isoprostanes in phospholipids were saponified with potassium hydroxide. Samples were acidified, diluted with water, deuterated, and an internal standard was added to the mixture. The mixture was then run on a silica column to separate isoprostanes from bulk fatty acids. The eluate was converted to pentafluorobenzyl esters by treatment with pentafluorobenzyl bromide to improve separation by gas chromatography. The mixture was than subjected to thin-layer chromatography to remove excess pentafluorobenzyl bromide and unreacted fatty acids. The F2 isoprostane fraction was extracted with ethyl acetate and analyzed by injection into a Thermo Finnigan TRACE DSQ single quadrupole mass spectrometer. Peak height was used for quantification and data were corrected with the internal standard expressed as nanogram of F2 isoprostane per gram of tissue.
Statistical analysis
Differences in transcript levels and oxidative stress markers between different AML patient groups and in fetal tissues were compared with the use of the nonparametric Mann–Whitney 2-sample test. Statistical analyses were performed with GraphPad Prism 4.0.
Results and discussion
Mutational specificity of DS
Because GATA1 mutations are specifically linked to both the TMD and AMkL phenotypes in persons with DS, we evaluated the mutational spectrum at this locus in diagnostic blast cells from DS children with AMkL (n = 14) and TMD (n = 5) to provide insights into the sources of genomic instability in DS and potential mechanisms linked to increased risk of developing leukemia. Samples were screened for GATA1 mutations by PCR amplification of exon 2, followed by direct sequencing and/or subcloning and sequencing at least 10 clones per samples. As reported previously,15 mutations at GATA1/exon 2 were detected in DS TMD and AMkL samples (Table 1) but not in non-DS AML samples, non-AMkL DS AML samples, or DS ALL samples (data not shown). Table 1 summarizes the spectrum of GATA1 mutations and the sequence characteristics of the exon 2. The net effect of the mutations is the introduction of early stop codons and the synthesis of a shorter GATA1 (designed GATA1s, 40 kDa), initiated from a downstream translation start site and distinguishable from the wild-type GATA1 (50 kDa). Both GATA1s and the wild-type GATA1 show similar DNA binding abilities and interact with a partner protein named “friend of GATA1” (FOG1), although GATA1s protein exhibits altered transactivation capacity because of the loss of the N-terminal activation domain.4
Summary of mutations at GATA1/exon 2 in DS TMD and AMkL patients
| Patient . | Sex . | FAB . | Gata1 exon 2 mutations . | Analysis of sequence characteristics . | ||
|---|---|---|---|---|---|---|
| Mutation . | Consequence of mutation . | Sequence context . | Notable sequence elements . | |||
| 1 | F | TMD | 114-159del46 | Stop codon after Met84 | ctgggcc[tga…gcc]acc | gcc direct repeat abuts CTGG |
| 2 | M | TMD | 172dup166-171delG | Stop codon after Met84 | –gca(g)ct– | Shares endpoint with 182Dup; direct repeat 175-179 |
| 3 | M | TMD | 221 G>A | Splice mutant | nonCpG | |
| 4 | M | TMD | 174dup159-173insGC | Stop codon after Met84 | (gc)—caccgctgcagctgc– | gctgc direct repeat within duplication |
| 5 | F | TMD | 192dup187-191 | Stop codon after Med84 | tac–tacag– | Direct repeat 205-209; endpoint disrupts TACT |
| 6 | F | M7 | 182dup172-181insGGG | Stop codon before Met84 | ctgg–gcggcactgg–(ggg)cctact | CTGG 5′ end of duplication; shares endpoint with 172Dup; within 2bp of TACT |
| 7 | F | M7 | 114-159del46 | Stop codon after Met84 | ctgggcc[tga…gcc]acc | gcc direct repeat abuts CTGG |
| 8 | M | M7 | 115 G>T | Stop codon before Met84 | nonCpG | |
| 9 | F | M7 | Indel del4-37ins134 | Stop codon before Met84 | atg[gag…cag]agcccct | 3′ endpoint 2 bp from mononculeotide repeats; ins/del disrupts ctgggg/ccccag palindrome |
| 10 | M | M7 | 49 C>T | Stop codon before Met84 | nonCpG; base substitution disrupts palindrome ctgggg/ccccag; disrupted base in bold | |
| 11 | M | M7 | 161-162ins5 | Stop codon before Met84 | cagccac(agtgg)cgctgcagc | Direct repeat |
| 160A>G | G>T is a CpG site | |||||
| 163G>T | Base substitutions surrounding insertion in bold | |||||
| 12 | M | M7 | 149-150insG | Stop codon after Met84 | gcccc(g) agcac | 3′ to GC4 hotspot |
| 13 | M | M7 | 162delCinsTGT | Stop codon after Met84 | cagccacc(tgt)(c)gctgcagc | Direct repeat |
| 14 | M | M7 | 99-100ins4 | Stop codon before Met84 | ggggttttc(nnaa)ttcccct | 3′ to C4T hotspot; within polypyrimidine run; unable to unequivocally determine sequence of 2 bases within insertion |
| 15 | F | M7 | 106-107insT 5A>G | Stop codon before Met84 | ttttcttcccct(t)ttcccctct | Within polypyrimidine run |
| 16 | F | M7 | 184dup84-183 | Stop codon before Met84 | acc—aga…gcc–tact | 100-bp duplication contains CTGG and 6 mononucleotide repeats; TACT endpoint |
| 17 | M | M7 | 2-3insT | No translation from Met1 | at(t)ggag | |
| 18 | M | M7 | 31-32insA | Stop codon before Met84 | ggggtccctgggga(a)cctc | 3′ to 2 TG4 IG heavy-chain hotspots; between palindrome ctgggg/ccccag |
| 143-168del26 | [ctgc…gca]gctgc | Direct repeat | ||||
| 19 | M | M7 | 45dup41-44 | Stop codon before Met84 | agc–ccct–ccccc | Endpoint in mononucleotide run; duplication endpoint at palindrome ctgggg/ccccag |
| Patient . | Sex . | FAB . | Gata1 exon 2 mutations . | Analysis of sequence characteristics . | ||
|---|---|---|---|---|---|---|
| Mutation . | Consequence of mutation . | Sequence context . | Notable sequence elements . | |||
| 1 | F | TMD | 114-159del46 | Stop codon after Met84 | ctgggcc[tga…gcc]acc | gcc direct repeat abuts CTGG |
| 2 | M | TMD | 172dup166-171delG | Stop codon after Met84 | –gca(g)ct– | Shares endpoint with 182Dup; direct repeat 175-179 |
| 3 | M | TMD | 221 G>A | Splice mutant | nonCpG | |
| 4 | M | TMD | 174dup159-173insGC | Stop codon after Met84 | (gc)—caccgctgcagctgc– | gctgc direct repeat within duplication |
| 5 | F | TMD | 192dup187-191 | Stop codon after Med84 | tac–tacag– | Direct repeat 205-209; endpoint disrupts TACT |
| 6 | F | M7 | 182dup172-181insGGG | Stop codon before Met84 | ctgg–gcggcactgg–(ggg)cctact | CTGG 5′ end of duplication; shares endpoint with 172Dup; within 2bp of TACT |
| 7 | F | M7 | 114-159del46 | Stop codon after Met84 | ctgggcc[tga…gcc]acc | gcc direct repeat abuts CTGG |
| 8 | M | M7 | 115 G>T | Stop codon before Met84 | nonCpG | |
| 9 | F | M7 | Indel del4-37ins134 | Stop codon before Met84 | atg[gag…cag]agcccct | 3′ endpoint 2 bp from mononculeotide repeats; ins/del disrupts ctgggg/ccccag palindrome |
| 10 | M | M7 | 49 C>T | Stop codon before Met84 | nonCpG; base substitution disrupts palindrome ctgggg/ccccag; disrupted base in bold | |
| 11 | M | M7 | 161-162ins5 | Stop codon before Met84 | cagccac(agtgg)cgctgcagc | Direct repeat |
| 160A>G | G>T is a CpG site | |||||
| 163G>T | Base substitutions surrounding insertion in bold | |||||
| 12 | M | M7 | 149-150insG | Stop codon after Met84 | gcccc(g) agcac | 3′ to GC4 hotspot |
| 13 | M | M7 | 162delCinsTGT | Stop codon after Met84 | cagccacc(tgt)(c)gctgcagc | Direct repeat |
| 14 | M | M7 | 99-100ins4 | Stop codon before Met84 | ggggttttc(nnaa)ttcccct | 3′ to C4T hotspot; within polypyrimidine run; unable to unequivocally determine sequence of 2 bases within insertion |
| 15 | F | M7 | 106-107insT 5A>G | Stop codon before Met84 | ttttcttcccct(t)ttcccctct | Within polypyrimidine run |
| 16 | F | M7 | 184dup84-183 | Stop codon before Met84 | acc—aga…gcc–tact | 100-bp duplication contains CTGG and 6 mononucleotide repeats; TACT endpoint |
| 17 | M | M7 | 2-3insT | No translation from Met1 | at(t)ggag | |
| 18 | M | M7 | 31-32insA | Stop codon before Met84 | ggggtccctgggga(a)cctc | 3′ to 2 TG4 IG heavy-chain hotspots; between palindrome ctgggg/ccccag |
| 143-168del26 | [ctgc…gca]gctgc | Direct repeat | ||||
| 19 | M | M7 | 45dup41-44 | Stop codon before Met84 | agc–ccct–ccccc | Endpoint in mononucleotide run; duplication endpoint at palindrome ctgggg/ccccag |
Mutations from patient samples were detected as described in ″Methods.″
FAB indicates French-American-British classification; M7, AMkL leukemia; and TMD, transient myeloproliferative disorder. Hyphens indicate duplicated sequence; square brackets indicate deletion endpoints; parentheses enclose inserted or deleted bases; and direct repeats are underlined.
Patients 1 and 7 represent the same person, first presenting with TMD and subsequently presenting with AMkL.
On the basis of the presence of multiple mutations in several patient samples, 23 distinct mutations from 19 patients were identified and analyzed. One DS patient developed AMkL (#7) after the spontaneous remission of TMD (#1), and blast samples from both conditions were analyzed and found to harbor the same GATA1 mutation. Of the 23 GATA1 mutations identified in the TMD and AMkL samples, 17 (74%) were insertion/deletion/duplication mutations, and 6 (26%) were base substitutions. Both single-base changes and small insertions/deletions in exon 2 of GATA1 were described in previous studies.4-9 Figure 1 provides a schematic of the mutations and the sequence context of the mutations.
Schematic of GATA1/exon 2 mutations in DS AMkL and TMD patients. Sites of GATA1/exon 2 mutations in DS AMkL and TMD patients are shown. DNA sequence for exon 2 is shown in 3 lines of nucleotide sequence, with nucleotide 1 as the first translated nucleotide of GATA1. Ins/del/dup sites are indicated below the line of the target sequence and identified by patient number.  indicating sites of deletion are inserted immediately 5′ to the deleted base.
indicating sites of deletion are inserted immediately 5′ to the deleted base.  above the line of sequence indicate sites of base substitution and are identified by patient number. More complete information is provided in Table 1. *Identical mutation observed in one person presenting first with TMD (#1) and subsequently with AMkL (#7).
above the line of sequence indicate sites of base substitution and are identified by patient number. More complete information is provided in Table 1. *Identical mutation observed in one person presenting first with TMD (#1) and subsequently with AMkL (#7).
Schematic of GATA1/exon 2 mutations in DS AMkL and TMD patients. Sites of GATA1/exon 2 mutations in DS AMkL and TMD patients are shown. DNA sequence for exon 2 is shown in 3 lines of nucleotide sequence, with nucleotide 1 as the first translated nucleotide of GATA1. Ins/del/dup sites are indicated below the line of the target sequence and identified by patient number.  indicating sites of deletion are inserted immediately 5′ to the deleted base.
indicating sites of deletion are inserted immediately 5′ to the deleted base.  above the line of sequence indicate sites of base substitution and are identified by patient number. More complete information is provided in Table 1. *Identical mutation observed in one person presenting first with TMD (#1) and subsequently with AMkL (#7).
above the line of sequence indicate sites of base substitution and are identified by patient number. More complete information is provided in Table 1. *Identical mutation observed in one person presenting first with TMD (#1) and subsequently with AMkL (#7).
Insertions/deletions/duplications
Insertion/deletion/duplication (ins/del/dup) mutations represent the predominant mutational signature of DS. This finding is consistent with the reported frequencies of ins/del/dup mutations in GATA1/exon 2 from other laboratories (including 100%,4 83%,6 76%,7 and 64%9 ). Ins/del/dup mutations frequently arise as a result of polymerase slippage and in response to single- and double-strand breaks in DNA,19,20 both of which are impacted by local sequence context. Table 1 summarizes the ins/del/dup events in GATA1/exon 2 and indicates sequence elements that may contribute to the mutagenesis. A compilation of these mutations (including those reported in this work) can be found in Table 2. The list of published mutations is in order, starting from the ATG as described previously, for a total of 85 ins/del/dup mutations across studies. Table 3 lists a summary of the sequence elements surrounding these mutations. Twenty-seven (32%) of all 85 mutations fell within the mononucleotide repeat sequence. One factor that had an impact on mutagenesis at mononucleotide repeat sites was the loss of DNA BER capacity,21 suggesting a role for reduced DNA repair capacity in the mutagenesis of DS.
GATA1 exon 2 mutations across studies
| Number . | Mutation . |
|---|---|
| 1 | 2-3 insT (Cabelof et al) |
| 2 | 4-37del ins134 (Cabelof et al) |
| 3 | 5-6 delAG (Xu et al9 ) |
| 4 | 9 insC (Xu et al9 ) |
| 5 | 12-13 insT (Hitzler et al6 ) |
| 6 | 31-32 insA (Cabelof et al) |
| 7 | 33 dup18-33 (Rainis et al7 ) |
| 8 | 36-37 delAG (Xu et al9 ) |
| 9 | 38-39 del2 (Hitzler et al6 ) |
| 10 | 40 insC (Xu et al9 ) |
| 11 | 45 insC (Xu et al9 ) |
| 12 | 45 dup 41-44 (Cabelof et al) |
| 13 | 49-50 del2bp (Groet et al8 ) |
| 14 | 84-85 insT (Weschler et al4 ) |
| 15 | 90 del AG (Rainis et al7 ) |
| 16 | 90 del AG (Rainis et al7 ) |
| 17 | 90-91 delAG (Xu et al9 ) |
| 18 | 90-91 delAG (Xu et al9 ) |
| 19 | 90 insAGGG (Xu et al) (dup9 ) |
| 20 | 90-91 delAG (Xu et al9 ) |
| 21 | 99-100 ins4 (Cabelof et al) |
| 22 | 105 insC (Rainis et al7 ) |
| 23 | 105-106 insC (Hitzler et al6 ) |
| 24 | 106-107 insT (Cabelof et al) |
| 25 | 114-115 del47 (Hitzler et al6 ) |
| 26 | 114-159 del46 (Cabelof et al) |
| 27 | 114-159 del46 (Cabelof et al) |
| 28 | 116 delA (Xu et al9 ) |
| 29 | 121-131 del11 (Hitzler et al6 ) |
| 30 | 127-128 ins1 (Mundschau et al5 ) |
| 31 | 133-154 del22bp (Groet et al8 ) |
| 32 | 135-158 del24 (Hitzler et al6 ) |
| 33 | 139 delT (Groet et al8 ) |
| 34 | 140 del35bp (Rainis et al7 ) |
| 35 | 143-168 del26 (Hitzler et al6 ) |
| 36 | 143-168 del26 (Cabelof et al) |
| 37 | 146-180 del (Mundschau et al) |
| 38 | 147 (dup 147-181) (Groet et al8 ) |
| 39 | 149-150 insG (Cabelof et al) |
| 40 | 150 delG (Rainis et al7 ) |
| 41 | 150 insT (Xu et al9 ) |
| 42 | 151 del43 (Rainis et al7 ) |
| 43 | 151 delG (Groet et al8 ) |
| 44 | 152-210 del (Weschler et al4 ) |
| 45 | 154-155 insCAGC (Hitzler et al6 ) (dup) |
| 46 | 158-159 insACAG (Hitzler et al6 ) (dup) |
| 47 | 158-159 ins7bp (Groet et al8 ) |
| 48 | 158-159 ins7bp (Groet et al8 ) |
| 49 | 159-160 ins20 (Mundschau et al5 ) |
| 50 | 160 ins10bp (Rainis et al7 ) |
| 51 | 161-162 insAGCACAGCCAC (Hitzler et al6 ) (dup) |
| 52 | 161-162 insAGTG (Hitzler et al6 ) |
| 53 | 161-162 ins5 (Cabelof et al) |
| 54 | 162 del1 (Hitzler et al6 ) |
| 55 | 162 delCinsTGT (Cabelof et al) |
| 56 | 166 insT (Hitzler et al6 ) |
| 57 | 166-167 del (Weschler et al4 ) |
| 58 | 169 ins22bp (Xu et al9 ) |
| 59 | 170-171 ins1 (Mundschau et al5 ) |
| 60 | 172 dup166-171delG (Cabelof et al) |
| 61 | 173-174 ins 15bp (Weschler et al4 ) |
| 62 | 173-174 ins16 (Mundschau et al5 ) |
| 63 | 174 dup159-173insGC (Cabelof et al) |
| 64 | 174-175 ins14 (Mundschau et al5 ) |
| 65 | 174 ins19bp (Xu et al9 ) |
| 66 | 180 ins11bp (Rainis et al7 ) |
| 67 | 180 insT (Rainis et al7 ) |
| 68 | 182 dup172-181insGGG (Cabelof et al) |
| 69 | 184 ins19 (Rainis et al7 ) |
| 70 | 184 dup84-183 (Cabelof et al) |
| 71 | 185 insA (Rainis et al7 ) |
| 72 | 186 dup177-186 (Rainis et al7 ) |
| 73 | 186-187 insT (Hitzler6 ) |
| 74 | 187 insT (Xu et al9 ) |
| 75 | 187 dup166-187 (Rainis et al7 ) |
| 76 | 187-188 ins TACT (Weschler et al4 ) (dup) |
| 77 | 187-188 ins TACT (Weschler et al4 ) (dup) |
| 78 | 189 delC (Xu et al9 ) |
| 79 | 189 Del11bpIns22bp (Rainis et al7 ) |
| 80 | 192 dup187-191 (Cabelof et al) |
| 81 | 192 delGins3bp (Rainis et al7 ) |
| 82 | 193 ins5bp (Rainis et al7 ) |
| 83 | 205-218 del (Mundschau et al5 ) |
| 84 | 209 delGA (Rainis et al7 ) |
| 85 | 220 del10bp (Rainis et al7 ) |
| Number . | Mutation . |
|---|---|
| 1 | 2-3 insT (Cabelof et al) |
| 2 | 4-37del ins134 (Cabelof et al) |
| 3 | 5-6 delAG (Xu et al9 ) |
| 4 | 9 insC (Xu et al9 ) |
| 5 | 12-13 insT (Hitzler et al6 ) |
| 6 | 31-32 insA (Cabelof et al) |
| 7 | 33 dup18-33 (Rainis et al7 ) |
| 8 | 36-37 delAG (Xu et al9 ) |
| 9 | 38-39 del2 (Hitzler et al6 ) |
| 10 | 40 insC (Xu et al9 ) |
| 11 | 45 insC (Xu et al9 ) |
| 12 | 45 dup 41-44 (Cabelof et al) |
| 13 | 49-50 del2bp (Groet et al8 ) |
| 14 | 84-85 insT (Weschler et al4 ) |
| 15 | 90 del AG (Rainis et al7 ) |
| 16 | 90 del AG (Rainis et al7 ) |
| 17 | 90-91 delAG (Xu et al9 ) |
| 18 | 90-91 delAG (Xu et al9 ) |
| 19 | 90 insAGGG (Xu et al) (dup9 ) |
| 20 | 90-91 delAG (Xu et al9 ) |
| 21 | 99-100 ins4 (Cabelof et al) |
| 22 | 105 insC (Rainis et al7 ) |
| 23 | 105-106 insC (Hitzler et al6 ) |
| 24 | 106-107 insT (Cabelof et al) |
| 25 | 114-115 del47 (Hitzler et al6 ) |
| 26 | 114-159 del46 (Cabelof et al) |
| 27 | 114-159 del46 (Cabelof et al) |
| 28 | 116 delA (Xu et al9 ) |
| 29 | 121-131 del11 (Hitzler et al6 ) |
| 30 | 127-128 ins1 (Mundschau et al5 ) |
| 31 | 133-154 del22bp (Groet et al8 ) |
| 32 | 135-158 del24 (Hitzler et al6 ) |
| 33 | 139 delT (Groet et al8 ) |
| 34 | 140 del35bp (Rainis et al7 ) |
| 35 | 143-168 del26 (Hitzler et al6 ) |
| 36 | 143-168 del26 (Cabelof et al) |
| 37 | 146-180 del (Mundschau et al) |
| 38 | 147 (dup 147-181) (Groet et al8 ) |
| 39 | 149-150 insG (Cabelof et al) |
| 40 | 150 delG (Rainis et al7 ) |
| 41 | 150 insT (Xu et al9 ) |
| 42 | 151 del43 (Rainis et al7 ) |
| 43 | 151 delG (Groet et al8 ) |
| 44 | 152-210 del (Weschler et al4 ) |
| 45 | 154-155 insCAGC (Hitzler et al6 ) (dup) |
| 46 | 158-159 insACAG (Hitzler et al6 ) (dup) |
| 47 | 158-159 ins7bp (Groet et al8 ) |
| 48 | 158-159 ins7bp (Groet et al8 ) |
| 49 | 159-160 ins20 (Mundschau et al5 ) |
| 50 | 160 ins10bp (Rainis et al7 ) |
| 51 | 161-162 insAGCACAGCCAC (Hitzler et al6 ) (dup) |
| 52 | 161-162 insAGTG (Hitzler et al6 ) |
| 53 | 161-162 ins5 (Cabelof et al) |
| 54 | 162 del1 (Hitzler et al6 ) |
| 55 | 162 delCinsTGT (Cabelof et al) |
| 56 | 166 insT (Hitzler et al6 ) |
| 57 | 166-167 del (Weschler et al4 ) |
| 58 | 169 ins22bp (Xu et al9 ) |
| 59 | 170-171 ins1 (Mundschau et al5 ) |
| 60 | 172 dup166-171delG (Cabelof et al) |
| 61 | 173-174 ins 15bp (Weschler et al4 ) |
| 62 | 173-174 ins16 (Mundschau et al5 ) |
| 63 | 174 dup159-173insGC (Cabelof et al) |
| 64 | 174-175 ins14 (Mundschau et al5 ) |
| 65 | 174 ins19bp (Xu et al9 ) |
| 66 | 180 ins11bp (Rainis et al7 ) |
| 67 | 180 insT (Rainis et al7 ) |
| 68 | 182 dup172-181insGGG (Cabelof et al) |
| 69 | 184 ins19 (Rainis et al7 ) |
| 70 | 184 dup84-183 (Cabelof et al) |
| 71 | 185 insA (Rainis et al7 ) |
| 72 | 186 dup177-186 (Rainis et al7 ) |
| 73 | 186-187 insT (Hitzler6 ) |
| 74 | 187 insT (Xu et al9 ) |
| 75 | 187 dup166-187 (Rainis et al7 ) |
| 76 | 187-188 ins TACT (Weschler et al4 ) (dup) |
| 77 | 187-188 ins TACT (Weschler et al4 ) (dup) |
| 78 | 189 delC (Xu et al9 ) |
| 79 | 189 Del11bpIns22bp (Rainis et al7 ) |
| 80 | 192 dup187-191 (Cabelof et al) |
| 81 | 192 delGins3bp (Rainis et al7 ) |
| 82 | 193 ins5bp (Rainis et al7 ) |
| 83 | 205-218 del (Mundschau et al5 ) |
| 84 | 209 delGA (Rainis et al7 ) |
| 85 | 220 del10bp (Rainis et al7 ) |
Mutations are written as reported by the authors in the original reports, except that the numbering of the mutations was adapted to reflect a uniform numbering system as described in “Methods.” In addition, if a mutation was found to be a duplication that was not indicated in the original report(s), the designation (dup) follows the mutation. Several mutations from other reports have not been included because they occurred in regions outside exon 2.
Summary of sequence elements identified across studies
| Element . | Mutations in this region . |
|---|---|
| Short direct repeats | |
| 111GCCtgagggc ttggatgcag cagcttcctc cactGCCccg… | 24, 25, 26, 27 |
| 141cactgccCCG agcacagcca CCGctgcagc… | 25, 26, 27, 31, 32, 35, 36, 37, 38, 39*, 40*, 41*, 42*, 43, 44*, 45, 46, 47, 48, 49, 50, 51*, 52*, 53*, 54*, 55* |
| 151agcaCAGCca ccgctgCAGC… | 25, 26, 27, 32*, 34, 35*, 36*, 37, 38, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57*, 58*, 59 |
| 171tgcggcactG GCCTACtaca gggacgctga GGCCTACaga… | 38*, 44, 66*, 67*, 68*, 69*, 70*, 71*, 72*, 73*, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83* |
| Indel hotspots | |
| Ig Switch sites: TGGGG, CCCCA, GGGGT | 6, 11, 12, 13, 15, 16, 17, 18, 19, 20 |
| Pol α Hotspot: 41cccTCCCCCa… | 11, 12, 13 |
| Fragile X Hotspot: 141cactgccCCG…agcacagcca CCGctgcagc… (note: this hotspot is also a short direct repeat). | 25, 26, 27, 31, 32, 35, 36, 37, 38, 39*, 40*, 41*, 42*, 43, 44*, 45, 46, 47, 48, 49, 50, 51*, 52*, 53*, 54*, 55* |
| Mononucleotide repeats | |
| 9CCC | 4 |
| 18GGGgtCCCtGGGG | 2, 7 |
| 40CCCCtCCCCC | 10, 11, 12, 13 |
| 91GGGG | 15, 16, 17, 18, 19, 20 |
| 95TTTTcTT | 21 |
| 102CCCC | 22, 23 |
| 117GGG | 28 |
| 137CCtCC | 33 |
| 146CCCC | 37, 38, 39, 40, 41 |
| 191GGG | 80, 81, 82 |
| 215CCCC | 83 |
| Element . | Mutations in this region . |
|---|---|
| Short direct repeats | |
| 111GCCtgagggc ttggatgcag cagcttcctc cactGCCccg… | 24, 25, 26, 27 |
| 141cactgccCCG agcacagcca CCGctgcagc… | 25, 26, 27, 31, 32, 35, 36, 37, 38, 39*, 40*, 41*, 42*, 43, 44*, 45, 46, 47, 48, 49, 50, 51*, 52*, 53*, 54*, 55* |
| 151agcaCAGCca ccgctgCAGC… | 25, 26, 27, 32*, 34, 35*, 36*, 37, 38, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57*, 58*, 59 |
| 171tgcggcactG GCCTACtaca gggacgctga GGCCTACaga… | 38*, 44, 66*, 67*, 68*, 69*, 70*, 71*, 72*, 73*, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83* |
| Indel hotspots | |
| Ig Switch sites: TGGGG, CCCCA, GGGGT | 6, 11, 12, 13, 15, 16, 17, 18, 19, 20 |
| Pol α Hotspot: 41cccTCCCCCa… | 11, 12, 13 |
| Fragile X Hotspot: 141cactgccCCG…agcacagcca CCGctgcagc… (note: this hotspot is also a short direct repeat). | 25, 26, 27, 31, 32, 35, 36, 37, 38, 39*, 40*, 41*, 42*, 43, 44*, 45, 46, 47, 48, 49, 50, 51*, 52*, 53*, 54*, 55* |
| Mononucleotide repeats | |
| 9CCC | 4 |
| 18GGGgtCCCtGGGG | 2, 7 |
| 40CCCCtCCCCC | 10, 11, 12, 13 |
| 91GGGG | 15, 16, 17, 18, 19, 20 |
| 95TTTTcTT | 21 |
| 102CCCC | 22, 23 |
| 117GGG | 28 |
| 137CCtCC | 33 |
| 146CCCC | 37, 38, 39, 40, 41 |
| 191GGG | 80, 81, 82 |
| 215CCCC | 83 |
Mutations are depicted in columns to the left, with critical sequences indicated in bold. For ease of comparison, sequences are represented in the same 10-base blocks as shown in Figure 2, with a superscript indicating the location within exon 2. In the right column, the mutations presented and enumerated in Table 3 are indicated.
Mutations that interrupt at least one of the direct repeats.
Other sequence elements that drive ins/del/dup mutations include repeat or symmetrical elements. Sixteen of the 23 mutations in our patient samples involved these types of sequences: 8 direct repeats, 4 palindromes, and 4 mononucleotide runs (Table 1 “Notable sequence elements”). Examining the symmetrical elements of all 85 mutations (Table 2), we found that regions between 4 short direct repeats were highly mutable across studies (Table 3). Forty-nine mutations occurred in regions involving direct repeats (58% of all ins/del/dup mutations), and 25 of these mutations directly interrupted one of the direct repeat elements, pointing to DNA strand breaks as a source of mutagenesis. In a meta-analysis of clinically relevant ins/del mutations,22 the authors showed that certain sequence motifs are statistically associated with ins/del mutagenesis, and several of these motifs were found surrounding the mutations in exon 2. In our own samples, these included a DNA polymerase α frameshift hotspot (TCCCCC), an Ig heavy-chain switch repeat hotspot (TGGGG), and a palindrome (CTGGGG/CCCCAG), between which one insertion and one duplication occurred. In the analysis of all studies, additional Ig heavy-chain switch repeat hotspots (CCCCA, GGGGT) and a FragileX hotspot (CCG…CCG) were present. Thirty-seven mutations (accounting for those that may arise as a result of more than one of the hotspots) arise at known ins/del hotspot sites (44% of all ins/del/dup mutations).
Six tandem duplications were detected in our patient samples (23% of all our mutations). This finding is intriguing because the accumulation of uracil can generate tandem duplications through strand-break intermediates,23 providing evidence that uracil may be an initiating lesion in the mutagenesis of DS. It is known that uracil incorporation in Escherichia coli generates base substitutions (more in “Base substitutions”) as well as ins/del/dup mutations.24 A specific tetramer, CTGG, was associated with ins/del/dup mutations in response to uracil accumulation in these studies. Although the importance of this sequence is unclear, it is intriguing that our clinical samples demonstrated a similar sequence.
Four of our observed mutations directly involved the CTGG site, and 5 of 6 duplications were in close proximity, suggesting a role for uracil in the mutagenesis of DS. Among the different studies, 16 duplication events (19% of all ins/del/dup mutations) could be definitively identified, and 13 of these occur in a 47-bp fragment of exon 2 (Table 4). In addition, 16 more mutations within this short region may also be duplication events because many reports list only “insertion” as the type of mutation without specifying whether the insertion is a duplication. This 47-bp stretch is very cytosine rich: 42.5% of all bases in this stretch are cytosine (even distribution would be 25% of all bases). Cytosine and/or CpG sites are involved in either the flanking sequence or the duplicated sequence in 16 of 16 duplication mutations. In every duplication event, cytosine and/or CpG sites are involved in either the flanking sequence or the duplicated sequence.
Duplication mutations across studies
| Number . | Mutation . | Duplicated sequence . |
|---|---|---|
| 7 | 33 dup18-33 (Rainis et al7 ) | GGG GTCCCTGGGG ACC (cytosine at 3′ end of duplication) |
| 12 | 45 dup 41-44 (Cabelof et al) | CCCT (cytosine repeat within duplication) |
| 19 | 90 insAGGG (Xu et al9 ) (dup) | cAGGG (cytosine flanks 5′ end of duplication) |
| 38 | 147 (dup 147-181) (Groet et al8 ) | gcCCCG AGCACAGCCA CCGCTGCAGC TGCGGCACTGgc (CpG flanks duplication) |
| 45 | 154-155 insCAGC (Hitzler et al6 ) (dup) | CAGC (cytosine at 5′ and 3′ ends of duplication) |
| 46 | 158-159 insACAG (Hitzler et al6 ) (dup) | gcACAGc (CpG flanks duplication) |
| 51 | 161-162 insAGCACAGCCAC (Hitzler et al6 ) (dup) | cgAGCACAGCCACcg (CpG flanks duplication) |
| 60 | 172 dup166-171delG (Cabelof et al) | gctGCA(g)CTgc (CpG flanks duplication) |
| 63 | 174 dup159-173insGC (Cabelof et al) | gcCA CCGCTGCAGC TGCGC (CpG flanks and additional gc added in duplication) |
| 68 | 182 dup172-181insGGG (Cabelof et al) | GCGGCACTG GGGGc (CpG at duplication endpoints) |
| 70 | 184 dup84-183 (Cabelof et al) | ccACAATCA…GCCT (tandem CC flank 5′ end and are duplicated at 3′ end) |
| 72 | 186 dup177-186 (Rainis et al7 ) | gcACTG GCCTAC (CpG flanks 5′ end; cytosine last duplicated base) |
| 75 | 187 dup166-187 (Rainis et al7 ) | GCAGC TGCGGCACTG GCCTACT (CpG at 5′ end of duplication) |
| 76 | 187-188 ins TACT (Weschler et al4 ) (dup) | ccTACT (tandem CC flank 5′ end of duplication) |
| 77 | 187-188 ins TACT (Weschler et al4 ) (dup) | ccTACT (tandem CC flank 5′ end of duplication) |
| 80 | 192 dup187-191 (Cabelof et al) | cTACA G (cytosine flanks 5′ end of duplication) |
| Number . | Mutation . | Duplicated sequence . |
|---|---|---|
| 7 | 33 dup18-33 (Rainis et al7 ) | GGG GTCCCTGGGG ACC (cytosine at 3′ end of duplication) |
| 12 | 45 dup 41-44 (Cabelof et al) | CCCT (cytosine repeat within duplication) |
| 19 | 90 insAGGG (Xu et al9 ) (dup) | cAGGG (cytosine flanks 5′ end of duplication) |
| 38 | 147 (dup 147-181) (Groet et al8 ) | gcCCCG AGCACAGCCA CCGCTGCAGC TGCGGCACTGgc (CpG flanks duplication) |
| 45 | 154-155 insCAGC (Hitzler et al6 ) (dup) | CAGC (cytosine at 5′ and 3′ ends of duplication) |
| 46 | 158-159 insACAG (Hitzler et al6 ) (dup) | gcACAGc (CpG flanks duplication) |
| 51 | 161-162 insAGCACAGCCAC (Hitzler et al6 ) (dup) | cgAGCACAGCCACcg (CpG flanks duplication) |
| 60 | 172 dup166-171delG (Cabelof et al) | gctGCA(g)CTgc (CpG flanks duplication) |
| 63 | 174 dup159-173insGC (Cabelof et al) | gcCA CCGCTGCAGC TGCGC (CpG flanks and additional gc added in duplication) |
| 68 | 182 dup172-181insGGG (Cabelof et al) | GCGGCACTG GGGGc (CpG at duplication endpoints) |
| 70 | 184 dup84-183 (Cabelof et al) | ccACAATCA…GCCT (tandem CC flank 5′ end and are duplicated at 3′ end) |
| 72 | 186 dup177-186 (Rainis et al7 ) | gcACTG GCCTAC (CpG flanks 5′ end; cytosine last duplicated base) |
| 75 | 187 dup166-187 (Rainis et al7 ) | GCAGC TGCGGCACTG GCCTACT (CpG at 5′ end of duplication) |
| 76 | 187-188 ins TACT (Weschler et al4 ) (dup) | ccTACT (tandem CC flank 5′ end of duplication) |
| 77 | 187-188 ins TACT (Weschler et al4 ) (dup) | ccTACT (tandem CC flank 5′ end of duplication) |
| 80 | 192 dup187-191 (Cabelof et al) | cTACA G (cytosine flanks 5′ end of duplication) |
In the left column, the number of the mutation (as in Table 3) is indicated. In the middle column, the mutation is described. In the far right column duplicated sequences and important flanking sequences are indicated. The duplicated sequences are in uppercase letters; mutations within the duplication are in uppercase italics; flanking sequences that are not part of the duplicated sequence are in lowercase.
Because deamination of cytosine yields uracil, which drives duplication events, these data support a role for uracil in the initiation of duplication mutations in DS. Mechanistically, cytosine deamination increases to a rate 140-fold greater in single-stranded DNA than in double-stranded DNA,25 and single-strand DNA arises during transcription. GATA1 is an actively transcribed gene, increasing the presence of single stranded DNA and the likelihood of cytosine deamination.
The 4 sequence elements discussed (mononucleotide repeats, direct repeat sequence, indel hotspots, and cytosine-rich regions) potentially account for 72 of the 85 mutations. Although the sequence elements are important in permitting mutagenesis, the end product of this mutagenesis (GATA1s) may be the critical factor providing a selective growth advantage that drives leukemogenesis. The accumulation of mutations in exon 2 provides a locus to study the underlying mechanisms which lead to the generation of the mutations, which ultimately may account for the 500-fold greater risk for DS children to develop AMkL.
Base substitutions
A total of 6 base substitutions were identified in our patient population (Table 5). Half of the base substitutions (3/6) occurred in patients also expressing ins/del/dup mutations responsible for the generation of GATA1s, and these base substitutions would not generate GATA1s. Three patients expressed only base substitution mutations (patients 3, 8, and 10), which would be expected to generate GATA1s (Table 5), implicating these in the development of the leukemia phenotype. Analysis of the mutational specificity of the base substitutions revealed that of the 6 base substitutions identified, 4 were transition mutations and 2 were transversion mutations.
Single nucleotide substitutions at GATA1/exon 2 in DS AMkL and TMD patients: Characterization of single-nucleotide substitutions
| Nucleotide change . | Amino acid change . | Type . | Potential source . | |
|---|---|---|---|---|
| Oxidative stress . | Uracil . | |||
| 5 A>G | glu2gly | ms | Yes | |
| 49 C>T | gln17ter | ns | Yes | |
| 115 G>T | glu39ter | ns | Yes | |
| 160 A>G | thr54ala | ms | Yes | |
| 163 G>T | ala55ser | ms | Yes | |
| 221 G>A | gly74asp | spl | Yes | |
| Nucleotide change . | Amino acid change . | Type . | Potential source . | |
|---|---|---|---|---|
| Oxidative stress . | Uracil . | |||
| 5 A>G | glu2gly | ms | Yes | |
| 49 C>T | gln17ter | ns | Yes | |
| 115 G>T | glu39ter | ns | Yes | |
| 160 A>G | thr54ala | ms | Yes | |
| 163 G>T | ala55ser | ms | Yes | |
| 221 G>A | gly74asp | spl | Yes | |
ms indicates missense mutation; ns, nonsense mutation; and spl, splice variant.
Oxidative stress and mutations
DS is characterized by increased oxidative stress secondary to increased copy number and expression of several key redox-related genes encoded on chromosome 21 (eg, cystathionine-β-synthase [CBS], zinc-copper superoxide dismutase [SOD1], amyloid precursor protein). A signature mutation induced by oxidative stress is the G:C>T:A transversion. A total of 2 of 6 base substitutions were G:C>T:A transversions (Table 5), which may result from the increased oxidant load in DS. Table 6 summarizes the published base substitutions of GATA1 mutations in combination with our findings, which indicate that the G:C>T:A transversion is the predominant base substitution observed in DS TMD and AMkL, consistent with oxidative stress as a potential significant contributor to mutagenesis in DS.
Comparison of single-nucleotide substitutions across studies
| . | G:C>T:A . | G:C>A:T . | A:T>G:C . | C:G>G:C . |
|---|---|---|---|---|
| Number of previously reported base substitutions | 8 | 6 | 4 | 2 |
| Number of base substitutions reported in this study | 2 | 2 | 2 | 0 |
| Total number of base substitutions reported | 10 | 8 | 6 | 2 |
| . | G:C>T:A . | G:C>A:T . | A:T>G:C . | C:G>G:C . |
|---|---|---|---|---|
| Number of previously reported base substitutions | 8 | 6 | 4 | 2 |
| Number of base substitutions reported in this study | 2 | 2 | 2 | 0 |
| Total number of base substitutions reported | 10 | 8 | 6 | 2 |
A compilation of all base substitution mutations reported in previous studies6-9 are compared with and added to the base substitution mutations found in this study.
The other predominant base substitutions observed across studies (Tables 5–6) were the A:T>G:C and G:C>A:T mutations. These mutations are known to arise in response to uracil substitution in E coli DNA, both as a result of uracil misincorporation during semiconservative DNA synthesis23 and as a result of cytosine (or 5-methyl cytosine) deamination.24 Uracil accumulation occurs in response to altered folate metabolism within the cell.26,27 In DS, it is hypothesized that thymidylate synthesis is inhibited by the trapping of folate in the 5-methyl tetrahydrofolate (5-meTHF) form.28 Genetic manipulation of uracil metabolism, or thymidine starvation, induces G:C>A:T and A:T>G:C transition mutations in E coli, as well as in mammalian systems.23,27,29,30 A total of 4 of 6 of the identified base substitutions were represented by these transition mutations (Table 5). Further, 14 of 26 base substitutions reported across studies (Table 6) were represented by transitions, suggesting a role for uracil and altered folate metabolism in the mutagenesis of DS.
All of the base substitutions reported in this study, and 24 of 26 reported by other groups, can be explained by a combination of oxidative stress and uracil accumulation. Oxidative DNA damage and uracil are repaired through the DNA BER pathway. The mutational spectrum of DS suggests that chromosome 21-related factors that drive mutagenesis (and thereby leukemogenesis) are oxidative stress, aberrant folate metabolism, and reduced DNA repair capacity. We propose that constitutional trisomy 21 in DS induces an increase in mutation frequency and that the resulting GATA1s protein generated as a consequence of the mutations imparts a survival advantage, allowing this heterogeneous group of mutations to drive a specific leukemia phenotype, namely TMD and AMkL. The mutational evidence supports multiple mechanisms of mutagenesis, all of which may be explained by altered biologic pathways in DS, including increased oxidative stress, altered folate metabolism/uracil substitution in the DNA, and reduced DNA repair capacity.
DNA repair in DS
Under conditions of constitutive oxidative stress, a corresponding increase in steady-state levels of DNA damage is anticipated. Mutations accumulate when DNA damage is inadequately repaired, as observed at GATA1/exon 2 in DS. There are multiple studies that provide evidence of DNA repair defects in DS cells. Lymphocytes from persons with DS show lower baseline DNA repair31 and exhibit increased sensitivity to phytohemagglutinin stimulation,32 N-methyl-N′-nitro-N-nitrosoguanidine31 and γ-irradiation,33 indicating an increased sensitivity to DNA oxidation, methylation, and strand breaks. Although more than one DNA repair pathway might be impacted by the DS phenotype, BER deficiency is a compelling candidate because it repairs these types of DNA damage.
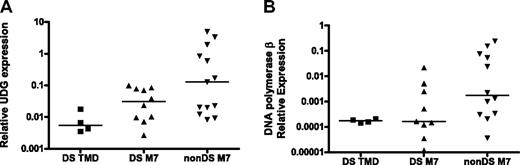
On the basis of this premise, the relationship between 2 key BER gene products involved in the repair of uracil in DNA, UDG and β-pol, and DS phenotype were evaluated in DS tissues. UDG is a monofunctional glycosylase that excises uracil from DNA to initiate BER. Loss of UDG in E coli and in mouse models induces mutations characterized predominantly by the G:C>A:T transition, which is similar to our observations in DS.26,27,30 The DS patient samples used for analysis of GATA1 mutations were used to compare UDG expression levels between DS TMD/AMkL (n = 14) and non-DS (n = 13) AMkL blasts (5 DS samples had no cDNA available for analysis). DS samples exhibited 75% lower UDG expression than the non-DS AMkL samples (P < .01, not shown). When DS TMD samples were analyzed separately from the DS-AMkL and non-DS AMkL groups, UDG expression in the DS TMD samples (median, 0.005) were 90% lower than expression levels in the non-DS AMkL patients (median, 0.127; Figure 2A, P < .01). The difference between DS and non-DS AMkL samples (median, 0.03; Figure 2A, P = 0.1) was not statistically significant, possibly because of the small sample size. These data present a scenario in which DS may predispose to mutagenesis through a uracil intermediate as a result of reduced UDG expression.
Reduced expression of BER genes in DS patient samples. RNA was isolated and reverse transcribed from blast cells obtained from DS and non-DS (nonDS) patients as described in “Clinical TMD and AMkL samples.” Median gene expression levels were determined by quantitative real-time RT-PCR and normalized to 18S expression. (A) UDG expression. DS TMD vs non-DS AMkL, P < .01; DS AMkL vs non-DS AMkL, P = .1. (B) DNA polymerase β expression. DS TMD vs non-DS AMkL, P = .02; DS AMkL vs non-DS AMkL, P = .07. DNA polymerase β expression in 1 DS AMkL patient sample fell below the level of detection and was excluded from analysis.
Reduced expression of BER genes in DS patient samples. RNA was isolated and reverse transcribed from blast cells obtained from DS and non-DS (nonDS) patients as described in “Clinical TMD and AMkL samples.” Median gene expression levels were determined by quantitative real-time RT-PCR and normalized to 18S expression. (A) UDG expression. DS TMD vs non-DS AMkL, P < .01; DS AMkL vs non-DS AMkL, P = .1. (B) DNA polymerase β expression. DS TMD vs non-DS AMkL, P = .02; DS AMkL vs non-DS AMkL, P = .07. DNA polymerase β expression in 1 DS AMkL patient sample fell below the level of detection and was excluded from analysis.
Interestingly, DS samples (TMD and AMkL together) showed a 90% reduction in β-pol expression (the rate-determining enzyme in the BER pathway) compared with non-DS AMkL samples (not shown, P < .001). When analyzed separately, β-pol expression was significantly lower in the DS TMD samples (median, 0.000 174; Figure 2B; P = .02) and also lower (P = .07) in the DS AMkL samples (median, 0.000 16) in comparison with non-DS AMkL samples (median, 0.00 177; Figure 2B). This finding is striking because we have previously demonstrated that a 50% reduction in β-pol expression predisposes mice to cancer.17 Further, these findings confirm previous reports of reduced β-pol expression in lymphocytes of healthy persons with DS.31 The tight clustering of expression for β-pol in the TMD sample group implies that the onset of leukemia may distort our ability to evaluate β-pol expression in leukemic samples. To this end, important predisposing factors should be evaluated in a preleukemia sample population if possible, which is addressed to some extent by the analysis of TMD samples.
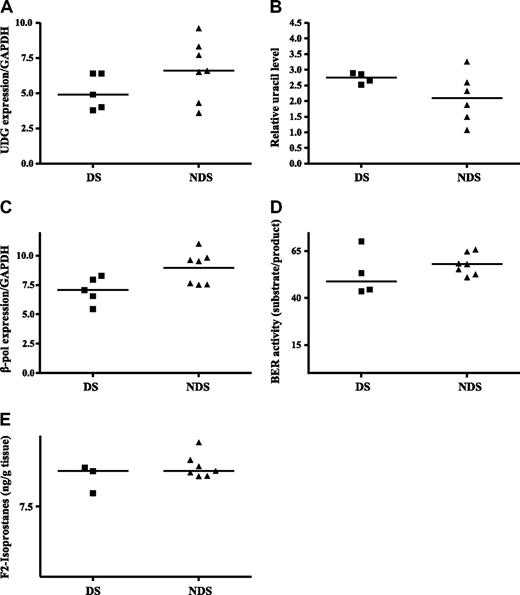
An ideal tissue in which to study mechanisms of mutagenesis in DS is the fetal liver for several reasons: (1) liver is the primary site for fetal hematopoiesis from the 6th to 22nd week of gestation (including the time period when we previously detected GATA1 mutations in fetal liver samples)11 ; (2) fetal hydrops and hepatosplenomegaly (markers suggestive of TMD) were reported in 11 (14%) of 79 DS fetuses34 ; and (3) hydrops could be detected by ultrasound in a 25-week gestation DS fetus and megakaryoblasts were found at autopsy.35 To address this question further, gene expression was analyzed in fetal liver samples. Because of the difficulty in obtaining these samples, only a small number of samples (5 DS and 7 non-DS for most analyses) were available for analysis. Despite this, gene expression differences similar to the expression patterns in TMD samples were observed. UDG expression in the DS fetal liver samples was 35% lower than that in the non-DS fetal liver samples (median, 4.9 vs 6.6, respectively; P = .07, Figure 3A). Loss of the UDG gene product should generate an accumulation of uracil in the DNA. Sufficient amounts of DNA were isolated from 4 DS samples and 6 non-DS samples to quantify uracil accumulation. In Figure 3B a trend toward greater uracil accumulation (32%) in the DS fetal tissue compared with non-DS tissues (median, 2.75 μg vs 2.09 μg of DNA, respectively; P = .1) was demonstrated. For both UDG gene expression and uracil accumulation, all DS values were either above (uracil) or below (UDG expression) the median values for the non-DS tissues. Our inability to detect statistically significant differences was likely attributable to the small sample size and the wide range in values in the heterogeneous non-DS group because uracil accumulation and UDG expression in non-DS tissue showed no consistent pattern, whereas the DS tissues exhibited tightly clustered values for both these base excision repair factors.
Altered BER and increased oxidative stress in DS fetal liver. (A) UDG expression. RNA was isolated and reverse transcribed from DS and non-DS (NDS) fetal liver tissues as described in “Fetal liver tissue.” Median gene expression levels were determined by quantitative real-time RT-PCR and normalized to GAPDH expression. (B) Uracil accumulation in DNA. DNA was isolated from DS and non-DS fetal tissue and analyzed for uracil incorporation as described in “Uracil detection.” Relative uracil levels were determined and median values are presented. (C) DNA polymerase β expression. RNA was isolated and reverse transcribed from DS and non-DS fetal liver tissue as described in “Base excision repair activity.” Median gene expression levels were determined by quantitative real-time RT-PCR and normalized to GAPDH expression. (D) Base excision repair activity. Nuclear proteins were isolated, and BER capacity was determined as described in “Methods.” BER capacity is calculated as the percentage of probe repaired (16-mer/30-mer), and median values are presented. (E) F2 isoprostane detection. F2 isoprostanes were measured by gas chromatography–mass spectrometry in fetal liver tissues as described in “Fetal liver tissue.” Median values are presented.
Altered BER and increased oxidative stress in DS fetal liver. (A) UDG expression. RNA was isolated and reverse transcribed from DS and non-DS (NDS) fetal liver tissues as described in “Fetal liver tissue.” Median gene expression levels were determined by quantitative real-time RT-PCR and normalized to GAPDH expression. (B) Uracil accumulation in DNA. DNA was isolated from DS and non-DS fetal tissue and analyzed for uracil incorporation as described in “Uracil detection.” Relative uracil levels were determined and median values are presented. (C) DNA polymerase β expression. RNA was isolated and reverse transcribed from DS and non-DS fetal liver tissue as described in “Base excision repair activity.” Median gene expression levels were determined by quantitative real-time RT-PCR and normalized to GAPDH expression. (D) Base excision repair activity. Nuclear proteins were isolated, and BER capacity was determined as described in “Methods.” BER capacity is calculated as the percentage of probe repaired (16-mer/30-mer), and median values are presented. (E) F2 isoprostane detection. F2 isoprostanes were measured by gas chromatography–mass spectrometry in fetal liver tissues as described in “Fetal liver tissue.” Median values are presented.
β-pol expression in the DS fetal liver samples was 35% lower than that in the non-DS fetal liver samples (median, 7.06 vs 9.53, respectively, P = .07; Figure 3C). As mentioned previously, expression in every DS sample fell below the median value in the non-DS samples. To determine whether the lower UDG and β-pol expression translated to reduced activity of the BER pathway, BER capacity was evaluated in 4 DS and 7 non-DS samples with adequate amounts of tissue to isolate nuclear proteins. A trend toward lower BER activity (20%) was detected in the DS compared with non-DS tissues (48.86 μg vs 57.98 μg of protein, respectively, P = .02; Figure 3D). Collectively, these data support the hypothesis that reduced DNA repair capacity in the DS prenatal environment predisposes to mutagenesis.
Oxidative stress in DS
More than 20 genes involved in oxidative metabolism are localized to chromosome 21, including SOD1 and CBS. Accordingly, increased levels of oxidative stress are reported for persons with DS for a variety of oxidation parameters, including lipid peroxidation, protein oxidation, and DNA oxidation.36-40 Recently Perrone et al41 reported that amniotic fluid in DS-diagnosed pregnancies exhibited a 9-fold increase in isoprostane levels compared with non-DS pregnancies. On the basis of these observations, the analysis of the prenatal environment vis-à-vis oxidative stress in fetal hematopoietic tissue also was performed in 3 DS fetal livers and 7 non-DS fetal livers in which there was sufficient tissue for analysis. Isoprostanes accumulate as a result of free radical attack on membrane phospholipids and are a marker of lipid peroxidation. To determine the extent of lipid peroxidation in fetal livers from DS and non-DS pregnancies, levels of F2-isoprostane were measured. There was no difference in median F2 isoprostane levels in the DS (11.25 g of tissue) compared with non-DS fetal tissues (11.25 g of tissue; Figure 3E). It is possible that other measures of oxidative stress (eg DNA 8-hydroxydeoxyguanosine) may be more informative as well as an analysis of a greater number of samples, to better define the role of oxidative stress in the generation of G:C>T:A mutations, a signature mutation of oxidative stress.
CBS- and SOD-driven model of genomic instability in DS
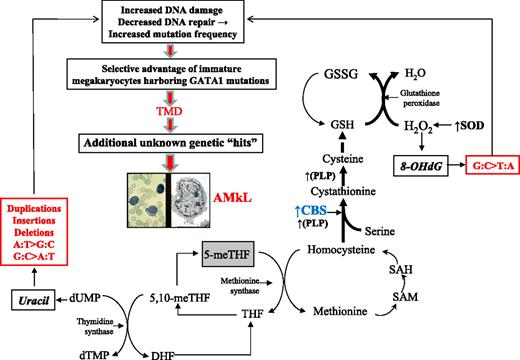
We propose a model to explain the mutational spectrum of DS as a function of CBS activity. CBS catalyzes the irreversible condensation of serine and homocysteine to form cystathionine, which directs homocysteine toward cystathionine synthesis and away from methionine remethylation, creating a folate trap and thymidylate imbalance (Figure 4). The latter will result in uracil misincorporation into DNA. Left unrepaired, uracil will initiate a pattern of G:C>A:T, A:T>G:C, and ins/del/dup mutations, as seen in our analyses. The impact of CBS is greater than can be explained by the DS “gene dosage” effect (1.5-fold) alone, on the basis of our earlier finding that CBS transcripts are a median 12-fold greater in DS AMkL blasts than in AML blasts from non-DS children.14 This finding suggests an adaptive response by CBS to the increased reactive oxygen species environment in DS. Reports in the literature support a role for CBS as a key factor driving metabolic flux in response to oxidative stress, and establish that both oxidative stress and DS amplify CBS activity.42-47 Our model hypothesizes that the increased copy number of SOD in trisomy 21 will drive increased H2O2 production that is quenched by glutathione peroxidase. One impact of increased H2O2 production is the oxidation of DNA 8-hydroxydeoxyguanosine, which becomes fixed as G:C>T:A mutations when DNA repair capacity is compromised. The effects of CBS in trisomy 21 cells accommodate the pro-oxidant effects of trisomy 21 by accelerating glutathione production and causing a corresponding trap of folate in its fully reduced form, a biochemical dead-end. We propose this mechanism is the source of mutagenesis in DS that generates GATA1 mutations as an early initiating step in the process of leukemogenesis.
Model: Trisomy 21 accelerates CBS activity and drives mutagenesis of GATA1 toward TMD/AMkL. This model proposes that the mutational spectrum at GATA1 can be explained by increased CBS activity, occurring as a combined result of both copy number increase and an adaptive response to oxidative stress in DS. Bold letters indicate enzymes known to be up-regulated in DS. Bold arrows indicate the primary direction of the reaction. The box around 5-meTHF depicts the trapping of this metabolite in this fully reduced form. CBS indicates cytathionine-β-synthase; DHF, dihydrofolate; dTMP, deoxythymidinemonophosphate; dUMP, deoxyruidinemonophosphate; GSH, reduced glutathione; GSSG, oxidized glutathione; 8-OHdG, 8-hydroxyguanosine; PLP, pyridoxal-l-phosphate; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; SOD, Cu/Zn superoxide dismutase; 5-meTHF, 5-methyltetrahydrofolate; 5,10-meTHF, 5,10-methyltetrahydrofolate; THF, tetrahydrofolate.
Model: Trisomy 21 accelerates CBS activity and drives mutagenesis of GATA1 toward TMD/AMkL. This model proposes that the mutational spectrum at GATA1 can be explained by increased CBS activity, occurring as a combined result of both copy number increase and an adaptive response to oxidative stress in DS. Bold letters indicate enzymes known to be up-regulated in DS. Bold arrows indicate the primary direction of the reaction. The box around 5-meTHF depicts the trapping of this metabolite in this fully reduced form. CBS indicates cytathionine-β-synthase; DHF, dihydrofolate; dTMP, deoxythymidinemonophosphate; dUMP, deoxyruidinemonophosphate; GSH, reduced glutathione; GSSG, oxidized glutathione; 8-OHdG, 8-hydroxyguanosine; PLP, pyridoxal-l-phosphate; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; SOD, Cu/Zn superoxide dismutase; 5-meTHF, 5-methyltetrahydrofolate; 5,10-meTHF, 5,10-methyltetrahydrofolate; THF, tetrahydrofolate.
Conclusions
DS children with AML, and specifically AMkL, display some of the most unique biologic and clinical features that can potentially be exploited to identify factors linked to leukemogenesis and genes linked to treatment outcome. The prenatal origin of TMD cases11 and the prenatal generation of somatic GATA1 mutations provide credence to the notion of a multistep process of leukemogenesis in DS. This finding is similar to the prenatal generation of leukemia-associated translocations and the detection of leukemic/preleukemic clones on Guthrie newborn screening cards of non-DS children.48 In an effort to identify factors linked to leukemogenesis in DS, both the mutational specificity of DS and aspects of DS that may impact mutagenesis were evaluated in clinically relevant human tissues.
On the basis of our data, we hypothesize that oxidative stress, uracil accumulation, and reduced DNA repair capacity all interact to generate the mutations observed in GATA1 exon 2 of DS TMD and AMkL cases. Oxidative stress can damage DNA bases and oxidize nucleotide pools, accounting for G:C>T:A mutagenesis. It can also drive glutathione production, thereby diverting metabolic intermediates away from folate metabolism, creating a functional folate deficiency that in turn would drive uracil accumulation in the DNA. Evidence for abnormal folate metabolism in DS includes the following: (1) excessive systemic toxicity (myelosuppression, mucositis, hepatotoxicity) in DS children with ALL treated with the antifolate methotrexate49,50 ; (2) decreased methotrexate toxicity in DS lymphocytes accompanying administration of folic acid in vivo or in vitro51 ; and (3) the increased methotrexate toxicity observed when folate is deficient.52 The DNA damage induced by oxidative stress and uracil incorporation in DNA is processed by the BER pathway, and we provide evidence for a BER defect in DS, suggesting that this defect is a critical factor driving the mutator phenotype of DS. Reduced repair capacity in the face of increasing DNA damage will drive increased rates of mutagenesis.
Our study is the first to examine possible mechanisms of mutagenesis in DS tissues including the fetal liver, which may lead to the generation of GATA1 mutations. Several recent studies identified that erythromegakaryocytic progenitor frequency was increased in DS fetal livers compared with gestation-matched non-DS control samples53 and megakaryocyte hyperproliferation was detected in the fetal liver of a DS fetus with hydrops fetalis in the absence of GATA1 mutations,53,54 indicating that abnormal hematopoiesis occurs in the DS fetal liver with the background of constitutional trisomy 21. Hence, DS fetal liver cells appear to be primed for the acquisition of GATA1 mutations as early steps in the development of TMD and/or AMkL.
One limitation of our study was the availability of number and quantity of fetal liver samples, and planned future studies will be performed on hematopoietic progenitor cells (eg, CD34+) isolated from freshly obtained tissue as opposed to cryopreserved tissue.
We propose mechanisms linking chromosome 21–localized genes (CBS and SOD1) to the generation of mutations in the GATA1 gene as an early or initiating step in the DS leukemogenesis. The sequence alterations of GATA1 in DS TMD/AMkL cases demonstrate patterns that suggest ins/del/dup and base substitution mutations can arise from altered folate metabolism and oxidative stress in trisomy 21 cells, which prevent adequate repair of DNA damage. The generations of the GATA1s protein as an end result of the mutations may provide a selective growth advantage, allowing for the survival of preleukemic clones, which may ultimately lead to the development of TMD and AMkL.
Additional studies are critical to further confirm the mechanisms in both clinically relevant cell line models and a comprehensive prospective analysis of GATA1 mutations of patients enrolled on the Children's Oncology Group AAML0431 clinical trial, “Treatment of Down Syndrome Children with AML and MDS Under the Age of 4 Years” and AAML 08B1 “Biology Study of Transient Myeloproliferative Disorder in Children with Down Syndrome.” These studies will also better elucidate whether the sequence characteristics of GATA1 exon 2, themselves play a specific role in the mutagenesis process in DS.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Supported by grants R01-CA92308 and R01-CA120772 from the National Cancer Institute, National Institute of Environmental Health Sciences Center grant P30 ES06639 (Wayne State University), the Leukemia & Lymphoma Society, The Elana Fund, Justin's Gift Charity, the Ring Screw Textron Chair in Pediatric Cancer Research. D.C.C. is a recipient of a Research Grant from the American Federation for Aging Research and Children's Research Center of Michigan.
National Institutes of Health
Authorship
Contributions: D.C.C., H.V.P., Q.C., Y.G., and H.v.R. performed the research; and D.C.C., Y.G., L.H.M., and J.W.T. contributed to experimental design, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jeffrey W. Taub, MD, Children's Hospital of Michigan, 3901 Beaubien Blvd, Detroit, MI 48201; e-mail: jtaub@med.wayne.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal