The human HIN-200 gene cluster and its mouse counterpart, the interferon inducible-200 (Ifi200) family, both on Chr 1, are associated with several diseases, including solid tumors and lupus. Our study was initiated to identify the modifier gene(s) encoded by the Pctm locus, in which mouse B-cell plasmacytomas induced by pristane are associated with heterozygosity of Chr 1 genes near the Ifi200 cluster. A screen for differentially expressed genes in granulomatous tissues induced by pristane in resistant and susceptible strains identified a new Ifi200 member whose expression was 1000-fold higher in the strain carrying the resistant allele of Pctm and was the most highly expressed Ifi200 gene. The gene, designated Mndal (for MNDA-like, myeloid nuclear differentiation antigen-like), was absent in the susceptible genome, as were genomic sequences upstream of Ifi203, the gene adjacent to Mndal. Ectopic expression of MNDAL suppressed cell growth, which, together with the disease susceptibility of heterozygotes at the Pctm locus, suggests that Mndal, perhaps with Ifi203, acts as a tumor suppressor and display(s) haploinsufficiency. Mndal is highly polymorphic among inbred mouse strains, because it is absent in 10 of 24 strains. This polymorphism may have implications for other disease modifiers mapping to the same region.
Introduction
B-cell tumor development is a complex genetic trait controlled by multiple low penetrance susceptibility genes modified by macro/microenvironmental influences. The pristane-induced mouse plasmacytoma (PCT) system represents an experimental tumor model, because multiple genes regulate susceptibility to PCT.1 Pristane (2, 6, 10, 14-tetramethylpentadecane [TMPD]), produced by zooplankton and found in high concentrations (in micrograms per gram) in some fish species,2 is used in the production of monoclonal antibodies and can induce a lupus-like disease in mice.3 In the PCT system, intraperitoneal TMPD injection induces chronic inflammation within 14 to 21 days and produces mesenteric oil granulomas in response to cytokines elicited from TMPD-containing macrophages.4,5 TMPD has recently been shown to elicit type I interferon through a TLR7-dependent pathway.3
In BALB/cAnPt(BALB) mice, PCTs develop in 60% of TMPD-treated mice, whereas in DBA/2N(DBA) mice, they remain tumor free (0%) for longer than a year.6,7 Resistance to PCTs is genetically dominant; however, 2% of (BALBxDBA)F1 hybrids develop tumors with a latency period longer than a year.1 Tumor susceptibility is less penetrant in NZB/BlNJ compared with BALB; only 30% of NZB/BlNJ mice develop PCTs.6,7 Different numbers of susceptibility loci in the strains probably to contribute to the difference in incidence. It should be noted that any given strain is likely to carry both tumor resistance and susceptibility alleles, and it is the relative effect of these genetic alleles that will determine the degree to which the strain is phenotypically resistant or susceptible.
Plasmacytoma susceptibility is linked to homozygosity of BALB(C/C) alleles of genes on mouse Chr 4 as determined by a linkage study involving the backcross BALBx(BALB × DBA)F1.1,8 This genetic cross, in conjunction with development of congenic lines,8 led to identification of loci, designated Pctr or Pctm, that contribute to PCT susceptibility.1,9,–11 Genes identified for Pctr1 and Pctr2, respectively, are, Cdkn2a (p16), identified by a candidate gene approach, and Frap (mTOR), by positional cloning.9,–11 The BALB alleles of both p16 and mTOR encode efficiency alleles, whose functional activities are much less active than the DBA allele.9,–11 PCT-susceptible NZB/BlNJ mice carry the same high-efficiency allele of p16 as that of DBA10 and, as such, carry a PCT-resistance allele at Pctr1 despite their 30% incidence of PCTs. In contrast, NZB mice carry a PCT-susceptibility allele at Pctr2 because its mTOR allele is the less efficient BALB allele,9 defective in phosphorylating p53.
Although BALB is the most PCT-susceptible strain, not all of its potential modifier genes encode susceptibility alleles. Similarly, the resistance of DBA to PCTs does not mean that it only carries resistance alleles. In the cross between BALB and DBA that led to identification of p16 and mTOR as Pctr loci, a locus on Chr 1 was also detected1 whereby DBA (D) is predicted to have a susceptibility allele and BALB (C) a resistance allele. This PCT modifier of resistance/susceptibility (Pctm), linked to Chr 1 near the Fcgr2 locus, was the only susceptibility locus in the backcross linked to heterozygosity (C/D alleles), suggesting that DBA carries a susceptibility gene for plasmacytomagenesis on Chr 1 that displays a susceptible phenotype when Pctm is heterozygous; these same progeny were C/C at p16 and mTOR. The interval containing the Pctm locus also includes the hematopoietic interferon-inducible 200 (Ifi200) gene cluster, whose human counterpart is HIN-200. This family, consisting of a series of related genes sharing various degrees of homology, has been linked to modifiers for several diseases, including systemic lupus12 and solid tumors.13,–15 Malignancy has also been associated with somatic mutation and epigenetic silencing of alleles within these clusters.16
In this study, gene expression profiling identified a new member of the Ifi200 cluster, which shares homology with human MNDA (myeloid nuclear differentiation antigen) and whose level of expression was found to have the greatest difference in inflammatory tissues between BALB and DBA mice. We have designated the murine gene Mndal (myeloid nuclear differentiation antigen-like), and it maps to the region of mouse Chr 1 also implicated as the site of the Pctm susceptibility locus in DBA. Our analysis indicates that Mndal is expressed in BALB, whereas its entire open reading frame is absent from DBA.
Methods
Gene expression
With the use of animal protocols approved by the Institutional Animal Care and Use Committee of the National Institutes of Health (NIH), mice were given 0.5 mL TMPD intraperitoneally. Three and 18 days later, total RNA was extracted from mesentery and spleen of experimental/control animals, with the use of TRIzol (Invitrogen). Labeled aRNA prepared from 1 μg RNA (MessageAmpII aRNA Amplification kit; Ambion) was hybridized to mouse genome 430 2.0 array chips, processed on Workstation 450, and analyzed with Gene Chip Operating Software (Affymetrix). Differential expression was assessed by analysis of variance (Partek Genomics Suite). Expression levels (average difference; GeneChip 3.1) are presented for probe sets targeting EST sequences that overlap the Mndal full-length cDNA. Reverse transcription–polymerase chain reaction (RT-PCR) methods are described in the supplemental Methods section (available on the Blood website; see the Supplemental Materials link at the top of the online article).
cDNA cloning
To identify, clone, and sequence the most differentially expressed gene, target sequences of 2 probe sets (Affy 1426906AT [labeled as Mnda-like] and 1452231xAT) were used to search the National Center for Biotechnology Information (NCBI) expressed sequence tag (EST) database.17 EST sequences were identified and used to build a virtual transcript of Mndal. A cDNA clone recognizing one of the ESTs, BE68641, was obtained from Invitrogen, and the full-length double-strand sequence analyzed contained an 1829-base pair (bp) cDNA from a C57BL/6 mouse thymus; it includes the entire coding sequence, 5′ and 3′ untranslated regions, and polyA tail. First-strand cDNA was synthesized with polyA RNA from BALB spleen (SuperScriptIII first-strand kit; Invitrogen). PCR amplification was performed with Mndal-specific primers 1, 2 (supplemental Table 1). Plasmid construction, genomic sequence analysis, and genetic linkage analyses are all described in supplemental Table 1.
Cellular localization assay
Cells grown on coverslips were transiently transfected with V5-His–tagged Mndal, V5-His–tagged Ifi205, or control plasmid, with the use of Lipofectamine (Invitrogen) or AMAXA Buffer 5 with a Nucleofector II. Cells were fixed with 3.5% paraformaldehyde 24 hours after transfection, permeabilized with 0.5% Triton X-100, blocked with 10% fetal bovine serum (FBS), and blotted with anti–V5-fluorescein isothiocyanate (FITC) antibody (Invitrogen). Stained cells were mounted with Vectashield with 4′,6′-diamidino-2-phenylindole (DAPI), and examined using a fluorescence microscope. Western blot methods are described in supplemental Methods.
Interferon treatment of cell lines
Cells were seeded in 100-mm plates and grown for 24 hours in Dulbecco modified Eagle medium with 10% FBS. At 90% confluence, cells were treated with or without interferon-α at 1000 units/mL for 18 hours. Total RNA was prepared with RNeasy mini kit (QIAGEN), and expression of Ifi200 genes was examined by RT-PCR.
NIH3T3 colony-forming assay
Cells were grown in Dulbecco modified Eagle medium, 10% FBS at 37°C, and 5% CO2. For the colony-formation assay, 300 ng of either pEV-15 or pEV19-Mndal or Ifi-203 plasmid was cotransfected with 50 ng pSV2Neo into 3 × 105 NIH3T3 cells in 35-mm plates with the use of calcium phosphate precipitation. Cells were split into five 100-mm plates 24 hours after transfection. Colonies of transfected cells were selected by growing in medium with G418 at a concentration of 0.5 mg/mL. Colonies were counted 2 weeks after selection, and efficiency of colony formation was determined by comparing the number of Mndal- or Ifi203-transfected colonies with those of empty vector–transfected cells. Eight independent experiments were performed with at least 2 different plasmid preparations for control Mndal and Ifi203 plasmids.
Supplemental data
Materials provided online include details about the analysis of the genomic structure of Mndal, including the 30 primer sequences used in PCRs (supplemental Table 1). Alignments of EST sequences used to identify and clone Mndal are presented (supplemental Figure 1). Sequence comparisons of Mndal between C57BL/6J and BALB/cAnPt are made (supplemental Figure 2). GEP datasets at GEO/NCBI were mined for Mndal expression in B-cell populations. supplemental Figure 4 shows Mndal expression from a NP-CGG immunization study,18 and in B and plasma cell tumors from XBP1s transgenic mice.19 The alignment of the HIN “a” domains of mouse IFI205, MNDAL, and human MNDA and AIM2 proteins are also presented (supplemental Figure 3). Detailed methods for RT-PCR, genomic organization, and linkage analysis; Western blotting; and isolation of cell populations are provided online.
Results
Gene discovery by microarray analyses
Gene expression profiling was used to identify genes differentially expressed between BALB and DBA strains, by comparing profiles from the pristane-primed mesenteries of mice and searching for differentially expressed genes mapping to Chr 1. Microarray analysis was performed with RNA isolated from at least 3 mesenteries and spleens from each strain, as well as RNA from 2 to 3 different samples of each of 4 BALB PCT cell lines, using the whole-genome Affymetrix mouse chip. Affy probe set 1426906_AT (originally annotated as Ifi205-like) was the most differentially expressed gene, with greater than 1000-fold difference between the 2 strains (Figure 1). This probe set gave a strong signal in the mesenteries of BALB mice and, furthermore, mapped to Chr 1 at position 175 694 010 within the Pctm interval. The same oligo-pair, 1426906_AT, was not expressed in DBA, as scored by Gene Chip Operating Software analysis. Another differentially expressed gene mapping to the Pctm region was Ifi203, at position 175 850 538; its expression in BALB mesenteries was 19-fold higher than DBA. Reevaluation of 1426906_AT by RT-PCR confirmed that it was not expressed in DBA but was highly expressed in BALB; Ifi203 was also more highly expressed in BALB than in DBA, but the difference was approximately 3- to 4-fold (Figure 2A). The microarray data are available in the GEO public database under accession no. GSE17297.
Differential expression of Mndal (Affy 1426906_AT) in BALB versus DBA before and after TMPD treatment. The signals from individual RNA samples from the mesenteries of BALB (blue), BALB × DBA F1 hybrids (CDF1; green), DBA (red), and (PCT) cell lines (black) are shown.
Differential expression of Mndal (Affy 1426906_AT) in BALB versus DBA before and after TMPD treatment. The signals from individual RNA samples from the mesenteries of BALB (blue), BALB × DBA F1 hybrids (CDF1; green), DBA (red), and (PCT) cell lines (black) are shown.
Expression of Ifi200 genes. (A) Expression of Ifi200 genes in the mesenteries of BALB and DBA mice at days 0, 3, and 18 after TMPD. Mndal (dark blue) was not expressed in DBA; similarly low levels of Ifi203 (medium blue) were also seen in DBA relative to BALB at all time points. Ifi202 (red) and Ifi205 (yellow) levels were higher in day 18 tissues in DBA relative to BALB. (B) Tissue-specific expression of Ifi200 genes in untreated adult BALB mice. Mndal expression (dark blue) is higher in BALB tissues than that of other Ifi genes. Splenic and thymic tissues showed the highest levels of expression. Other bars represent expression levels of Ifi202 (red), Ifi203 (medium blue), Ifi204 (light blue), and Ifi205 (yellow).
Expression of Ifi200 genes. (A) Expression of Ifi200 genes in the mesenteries of BALB and DBA mice at days 0, 3, and 18 after TMPD. Mndal (dark blue) was not expressed in DBA; similarly low levels of Ifi203 (medium blue) were also seen in DBA relative to BALB at all time points. Ifi202 (red) and Ifi205 (yellow) levels were higher in day 18 tissues in DBA relative to BALB. (B) Tissue-specific expression of Ifi200 genes in untreated adult BALB mice. Mndal expression (dark blue) is higher in BALB tissues than that of other Ifi genes. Splenic and thymic tissues showed the highest levels of expression. Other bars represent expression levels of Ifi202 (red), Ifi203 (medium blue), Ifi204 (light blue), and Ifi205 (yellow).
Clones and sequence analysis of Mndal
Analysis of the region surrounding Affy 1426906_AT determined that this region is highly repetitive; direct cloning and sequencing of several BALB and DBA genomic fragments was required to characterize the gene recognized by the probe set (GenBank bankit 1163875, 1165658, 1165663, 1165660). Sequence analysis showed that 1426906_AT targets a sequence that does not match any previously reported gene in the database.
To identify the gene, the 193-bp target sequence of 1426906_AT was used to search the dbEST database.17 Several EST clones were identified that matched the 1426906_AT sequence (supplemental Figure 1). A 459-bp EST, BE686461A, was recognized by Invitrogen clone (3377079) isolated from a C57BL/6J (B6) mouse thymus cDNA library, and our sequence data showed it contains an 1839-bp cDNA insert, including the 3′ end polyA tail (GenBank bankit 1165668). The target sequence of 1426906_AT matched the cDNA sequence isolated from the B6 thymus 100% near the 3′ end (supplemental Figure 1).
Further analysis showed the cDNA is from a novel gene with high sequence homology to members of the human HIN-200 and murine Ifi200 families. We designated the gene as Mndal (Mnda-like), because of its high degree of homology with human MNDA. The full-length Mndal cDNA was also isolated and sequenced (GenBank bankit 1156570) from the spleen of BALB/cAn mice. We found that the first 2 exons of the Mndal open reading frame (ORF) are 100% identical to exons 1 to 2 of Ifi203 (exons 1 of Mndal and Ifi203 are both noncoding), and the last 4 exons of Mndal are 97% to 99% identical to the last 4 exons of Ifi204 and Ifi205; these exons encode the 200 amino acid “a” domain. Only one exon in the ORF had sequences totally unique to Mndal (Figure 3A). By virtue of its chromosomal location and homology to Ifi genes, Mndal is a member of the Ifi200 family, whereas the unique exon verifies it is a new gene.
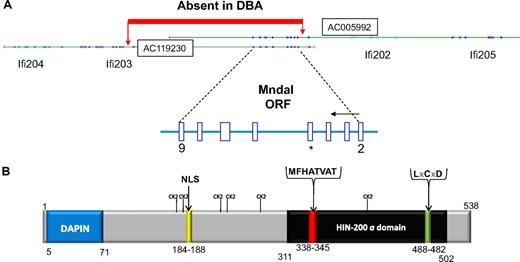
Protein and gene structure for Mndal. (A) Two BAC clones harbor Mndal sequences. BAC clones are oriented to show gene order on Chr 1 from centromere to telomere. The ORF of Mndal starts at exon 2 and finishes in exon 9. Sequence analysis of genomic DNA showed a segment absent in DBA from within intron 1 of Ifi203 to intron 2 of Mndal. *Exon 5 harbors sequences unique to Mndal. (B) Predicted structural domains of MNDAL. The canonical HIN “a” domain is highlighted in black and contains the highly conserved MFHATVAT motif thought to mediate dimerization and protein-protein interactions. The protein also contains a nuclear localization signal (NLS), a possible Rb-binding site (LxCxD), and several weak (S/T-P) Cdk2 sites.
Protein and gene structure for Mndal. (A) Two BAC clones harbor Mndal sequences. BAC clones are oriented to show gene order on Chr 1 from centromere to telomere. The ORF of Mndal starts at exon 2 and finishes in exon 9. Sequence analysis of genomic DNA showed a segment absent in DBA from within intron 1 of Ifi203 to intron 2 of Mndal. *Exon 5 harbors sequences unique to Mndal. (B) Predicted structural domains of MNDAL. The canonical HIN “a” domain is highlighted in black and contains the highly conserved MFHATVAT motif thought to mediate dimerization and protein-protein interactions. The protein also contains a nuclear localization signal (NLS), a possible Rb-binding site (LxCxD), and several weak (S/T-P) Cdk2 sites.
Gene structure and genomic location
By screening the NCBI database17 with Mndal sequence, 2 overlapping mouse BAC clones (AC005992, AC119230) were identified that contained sequences encompassing the Mndal locus (Figure 3A). The structure of the Mndal gene was determined by comparing cDNA and BAC clone sequences and confirmed by PCR and sequencing of genomic DNA from BALB. The resulting gene structure, with its ORF of 1614 bp predicting a putative protein of 538 amino acids (Figure 3B) distributed over 8 exons (exons 2-9), is consistent with that predicted by Mapview software and Murine Genome Build 33-35 (Figure 3A; supplemental Figure 1). The ORF spans approximately 23 kb and is located in the distal area of chromosome 1 in the Ifi200 cluster, between Ifi202 and Ifi203 (Figure 3A).
The Ifi200 cluster lies within the same interval, D1Mit16 to Mtv7, as the Pctm locus.1 The estimated distance between D1Mit16 and Mtv7 is 8.8 cM and corresponds to 8.7 Mb in the current build of the mouse genome map (NCBI). Analyses of 163 backcross progeny for an additional 4 D1Mit markers lying on either side of Fcgr2 continue to support linkage of Pctm to each of these markers with the same probability (P < .005). The Pctm tumor susceptibility phenotype still has the fewest recombinants with Fcgr2 (C/D) genotypes. D1Mit 206 has only 1 additional recombinant from that observed for Fcgr2 (supplemental Figure 2). Analysis of the recombinants suggests that Pctm lies between D1Mit145 and D1Mit206. These data would estimate an interval of approximately 5.2 Mb.
Genetic polymorphism of Mndal among mouse strains
To understand what accounts for the lack of Mndal expression in the DBA mouse, we compared the genomic structure of Mndal and its neighboring sequences between BALB and DBA. Genomic DNA from the kidney of DBA mice was used for PCR and sequence analysis. When the sequence obtained (GenBank bankit 1165663, 1165660) was aligned with that of the BALB strain, it indicated that the Mndal gene is absent in the DBA mouse (Figure 3A). Sequence data also showed that the genomic segment between Mndal and intron 1 of Ifi203 is also missing in DBA.
To determine the extent to which Mndal is conserved or absent in mouse strains, genomic DNA from 24 inbred strains was examined with 2 sets of allele-specific primers for Mndal (Table 1; supplemental Table 1). Some pairs of these strains are known to differ in their responses to cancer induction or autoimmunity, with the modifier gene in question mapping to the region of Chr 1 that includes Mndal. Fourteen strains show the expected PCR product that indicates the presence of Mndal. However, the gene is missing in the other 10 inbred strains, indicating that its absence represents a frequent polymorphism. In addition, comparison of Mndal sequences from 2 strains with an intact Mndal gene, BALB and B6, showed several single nucleotide polymorphisms, resulting in amino acid changes in MNDAL between the 2 strains (supplemental Figure 2). This comparison suggests that Mndal sequences among the inbred mouse strains harboring the gene are probably polymorphic.
Presence versus absence of Mndal gene in mouse strains and cell lines
| Mouse strain, BAC clone, and cell line distribution pattern . | |
|---|---|
| BALB/cAnPt variant (Mndal present) | |
| Mouse strains | BALB/cJ, BALB/cHeA, CBA/N, C57L/J, C57BL/6J, C57BL10/SnJ,129/J, C3H/HeJ, C3H/HeN, NOD, NZW/LacJ, Mus castaneous, Mus molossinus, Mus spretus |
| BAC clones | AC005992 (129X1/SvJ), AC119230 (C57BL/6J) |
| Cell lines | BALB3T3 fibroblasts, XRPC24 PCT (BALB-derived), TEPC1165 PCT (BALB-derived) |
| DBA/2NPt variant (Mndal absent) | |
| Mouse strains | AKR/N, A/J, DBA/2J, NON, NZB/BlNJ, P/J, SENCARA/Pt, SWR/J,STS/A, Mus caroli |
| BAC clone | AC006944(129S6/SvEvTac) |
| Cell lines | NIH3T3 fibroblasts, D210–403 PCT (DBA-derived), D281–288 PCT (DBA-derived) |
| Mouse strain, BAC clone, and cell line distribution pattern . | |
|---|---|
| BALB/cAnPt variant (Mndal present) | |
| Mouse strains | BALB/cJ, BALB/cHeA, CBA/N, C57L/J, C57BL/6J, C57BL10/SnJ,129/J, C3H/HeJ, C3H/HeN, NOD, NZW/LacJ, Mus castaneous, Mus molossinus, Mus spretus |
| BAC clones | AC005992 (129X1/SvJ), AC119230 (C57BL/6J) |
| Cell lines | BALB3T3 fibroblasts, XRPC24 PCT (BALB-derived), TEPC1165 PCT (BALB-derived) |
| DBA/2NPt variant (Mndal absent) | |
| Mouse strains | AKR/N, A/J, DBA/2J, NON, NZB/BlNJ, P/J, SENCARA/Pt, SWR/J,STS/A, Mus caroli |
| BAC clone | AC006944(129S6/SvEvTac) |
| Cell lines | NIH3T3 fibroblasts, D210–403 PCT (DBA-derived), D281–288 PCT (DBA-derived) |
The presence/absence of the Mndal gene was based on the presence of PCR products using two different primer pairs (supplemental Table I). Determination that 129S6/SvEvTac has the same structure as that of DBA comes from sequence analysis of BAC clone AC006944.
Ifi200 mRNA expression levels in BALB versus DBA
The mRNA expression of Mndal and other Ifi200 family members was compared in BALB and DBA by RT-PCR in the mesentery, with and without TMPD (Figure 2A), and in other tissues from BALB (Figure 2B). Mesentery from unprimed BALB mice contained approximately 200 copies of Mndal mRNA per 1 ng total RNA. This value increased approximately 5-fold after 3 days of TMPD treatment and was still at that level 15 days later (Figure 2A). The levels of Mndal also increased in the lung but decreased in spleen in response to TMPD (data not shown). As noted in Figure 1, there was no detectable signal of Mndal in the mesenteries of DBA mice. In BALB, basal levels of Ifi202, Ifi203, Ifi204, and Ifi205 were detectable in the mesentery and were induced further by TMPD (Figure 2A). In DBA, this was also true for Ifi202, Ifi204, and Ifi205, although the response appeared to be slower than in BALB. By contrast, Ifi203 was not detectably expressed in untreated mesenteries of DBA mice, and TMPD treatment led only to its low-level expression by day 18 (Figure 2A). This result correlates with the absence of 5′ sequences in Ifi203 noted in Figure 3, indicating possible impairment of the DBA Ifi203 promoter. Another possibility is that promoter sequences used by Mndal, which are present in DBA, may be driving expression of the Ifi203 gene. In either case, the induction of Ifi203 expression is lower in DBA than in BALB. The low-to-absent expression of Mndal and Ifi203 in DBA suggests that the defect in either or both genes could contribute to the plasmacytomagenesis associated with Pctm.
Examination of basal mRNA expression in multiple tissues of BALB mice showed that Mndal expression is tissue specific and that it is the most highly expressed Ifi200 gene in BALB tissues (Figure 2B). The highest levels of Mndal were observed in spleen and thymus (2000 copies/ng total RNA). Bone marrow, lung, skin, and heart expressed more moderate levels (approximately 1000 copies/ng total RNA), whereas low levels were seen in muscle, liver, and intestine. The other Ifi200 genes were expressed in a similar pattern, but all at a lower level; as with Mndal, brain and pancreas had little or no expression of the other Ifi200 genes. Although spleen and thymus express all members of the family, Mndal is clearly the predominant gene, because it is expressed at levels at least 5 times higher than Ifi203 and 20 to 30 times higher than Ifi202, Ifi204, and Ifi205 (Figure 2B). These results suggest that Mndal may be controlled by upstream signals distinguishable from those controlling some of the other Ifi200 genes and/or that the Mndal promoter is more sensitive to the same signals.
Expression in B cells
CD19+ B cells, isolated from control versus 18-day pristane-primed mesenteric tissues, showed consistent increases in Mndal expression after pristane. Mndal expression, as assessed by RT-PCR increased after pristane approximately 3-fold (2021 vs 5859 molecules/ng RNA) in splenic B cells, approximately 5-fold (993 vs 4784 molecules/ng RNA) in bone marrow cells, and 103-fold (14 vs 1451 molecules/ng RNA) in mesenteric B cells. Mndal expression also increased 1.5-fold (2772 vs 4282 molecules/ng RNA) in splenic T cells. Relatively high levels of Mndal were also found in mesenteric macrophages (3522 molecules/ng RNA). In addition to our experimental data, NCBI GEO datasets containing microarray data for sorted B cells were also mined for Mndal expression. A time-course study involving NP-CGG immunization of C57BL/Ka (a strain capable of sustaining myeloma bone lesions) mice showed high levels of Mndal in naive (nonimmunized B220+) B cells and (immunized 10 weeks) memory B cells.18 In contrast, Mndal expression was modest in plasma cells (immunized 1 week) and even lower to very little expression in germinal center B cells (immunized 2 weeks18 ; supplemental Figure 4A).
Expression in plasma cell tumors
As expected, the 2 PCT cell lines derived from DBA mice (Table 1; Figure 1 lane 3, D2107-403; lane 4, D281-288) did not express Mndal, because the gene is absent from the DBA genome. Unexpectedly, there was also no detectable signal in the BALB-derived PCT cell lines (Table 1; Figure 1A lane 1, XRPC24; lane 2, TEPC1165). The absence of signal in the BALB PCT cell lines is not due to deletion, because the gene remained intact in the lines (Table 1). A similar observation was made in a study of myeloma tumors arising in XBP1s mice in which gene expression of Mndal was lower in the tumors than in XBP1s B cells19 (supplemental Figure 4B).
Protein structure and cellular localization
MNDAL contains a conserved 200–amino acid motif at its C-terminus referred to as the “a domain” (Figure 3B). MNDAL also has a conserved N-terminal basic region, including a DAPIN (Domain in APoptosis and Interferon Response) motif, a consensus nuclear localization signal, a MFHATVAT motif thought to be important in mediating dimerization and protein-protein interactions, a potential Rb-binding site (LxCxD), and several weak (S/T-P) Cdk2 binding sites. Sequence alignment showed that MNDAL is most closely aligned with IFI205, with which it shares 55% identity in full-length and 86% identity in the “a” domain. Compared with the human HIN-200 genes, MNDAL is most homologous to MNDA, sharing 25% and 57% identity to the full-length and “a” domain alignment, respectively (supplemental Figure 3). MNDAL also shares 22% and 42% identity with full-length and “a” domain AIM2 sequences, respectively (supplemental Figure 3).
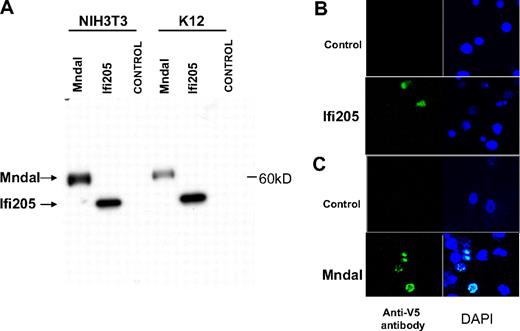
To confirm the production of MNDAL protein from its transcript, the Mndal ORF was cloned into the pcDNA3/V5His vector in-frame with the V5-His tag. When the resulting plasmid was transfected into NIH3T3 and mouse K12 osteosarcoma cells, it expressed a single band of approximately 60 kDa, which was larger than the V5-His–tagged IFI205 control (Figure 4A). To examine the subcellular location of MNDAL, V5-His–tagged Mndal was expressed in NIH3T3 and mouse XRPC24 PCT cells by transient transfection. V5-His–tagged Ifi205 was also transfected into NIH3T3 cells, as a control. The proteins encoded by other members of the Ifi200 family have a predominantly nuclear localization.20 As expected, IFI205 protein was localized to the nucleus (Figure 4B), and a similar localization was also found for MNDAL (Figure 4C, data shown for XRPC24).
MNDAL protein expression. (A) Western blot analysis of cells transfected with V5-His–tagged IFI205 (detected at ∼ 50 kDa) and MNDAL (detected at ∼ 60 kDa). (B-C) Immunofluorescent staining of NIH3T3 cells transfected with V5-His–tagged IFI205 (B) and XRPC24 PCT cells transfected with V5-His–tagged MNDAL (C). Staining with V5 antibody (green) showed overlap with DAPI (blue) stained nuclei. Cells were examined at 25°C with a Zeiss LSM 510 NLO confocal system (Carl Zeiss Inc) with an Axiovert 200M inverted microscope and operating with a 2-photon laser tuned to 750 nm, 25 mW argon laser tuned to 488 nm, and 1 mW HeNe laser tuned to 543 nm. Cells were imaged with a 63×/1.4 NA Zeiss Plan-Apochromat oil-immersion objective. Digital images (512 × 512 pixels, 8 bit) were collected with the use of the Zeiss AIM software with a scan zoom from 2 to 4 and a multitrack configuration in which the FITC, Cy3, and DAPI signals were collected sequentially with a BP 500- to 530-nm filter, with BP 565- to 615-nm filter, and BP 390- to 465-nm filter after excitation with 488-nm, 543-nm, and 750-nm laser lines, respectively.
MNDAL protein expression. (A) Western blot analysis of cells transfected with V5-His–tagged IFI205 (detected at ∼ 50 kDa) and MNDAL (detected at ∼ 60 kDa). (B-C) Immunofluorescent staining of NIH3T3 cells transfected with V5-His–tagged IFI205 (B) and XRPC24 PCT cells transfected with V5-His–tagged MNDAL (C). Staining with V5 antibody (green) showed overlap with DAPI (blue) stained nuclei. Cells were examined at 25°C with a Zeiss LSM 510 NLO confocal system (Carl Zeiss Inc) with an Axiovert 200M inverted microscope and operating with a 2-photon laser tuned to 750 nm, 25 mW argon laser tuned to 488 nm, and 1 mW HeNe laser tuned to 543 nm. Cells were imaged with a 63×/1.4 NA Zeiss Plan-Apochromat oil-immersion objective. Digital images (512 × 512 pixels, 8 bit) were collected with the use of the Zeiss AIM software with a scan zoom from 2 to 4 and a multitrack configuration in which the FITC, Cy3, and DAPI signals were collected sequentially with a BP 500- to 530-nm filter, with BP 565- to 615-nm filter, and BP 390- to 465-nm filter after excitation with 488-nm, 543-nm, and 750-nm laser lines, respectively.
Interferon induction of the Ifi200 genes in vitro
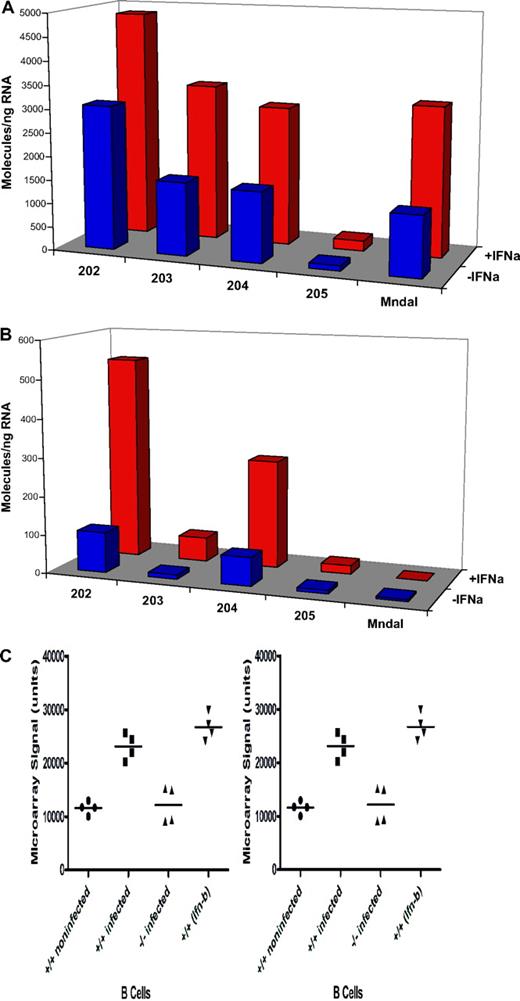
To examine whether the expression of endogenous Mndal is induced by interferon, NIH3T3 and BALB3T3 cell lines were treated with interferon-α. In BALB3T3 cells, which harbor the intact gene, this treatment induced higher levels of Mndal expression than in untreated cells, and a similar response was seen with the other Ifi genes examined (Figure 5A). By contrast, Mndal was not induced in NIH3T3 cells, which correlated with its absence in those cells (Figure 5B). As controls, interferon treatment of NIH3T3 did induce other Ifi genes. Mndal expression was also shown to increase in isolated B cells stimulated with Ifnβ21 (Figure 5C NCBI GEO). This same study shows that active influenza infection results in a 2-fold increase in Mndal expression in lymph node B cells, in wild-type mice, but no change in B cells from interferon receptor (Ifnr) null mice21 (Figure 5C).
BALB3T3 and NIH3T3 cells were treated with interferon-α for 18 hours. Expression levels of Ifi202, Ifi203, Ifi204, Ifi205, and Mndal were determined by RT-PCR in the BALB3T3 (A) and NIH3T3 (B) cells. The Mndal gene was found to be absent by sequencing genomic DNA from NIH3T3 cells. (C) Microarray data (mined from the GEO database)21 collected from isolated B cells from mice that were either treated with type I interferon or infected with influenza. Mndal expression in B cells isolated from wild-type (+/+) mice that were either infected with influenza or treated with type I interferon-β (Ifn-β) were compared with control. Mndal expression was also examined in B cells isolated from interferon receptor knockout (−/−) mice that had also been infected with influenza.
BALB3T3 and NIH3T3 cells were treated with interferon-α for 18 hours. Expression levels of Ifi202, Ifi203, Ifi204, Ifi205, and Mndal were determined by RT-PCR in the BALB3T3 (A) and NIH3T3 (B) cells. The Mndal gene was found to be absent by sequencing genomic DNA from NIH3T3 cells. (C) Microarray data (mined from the GEO database)21 collected from isolated B cells from mice that were either treated with type I interferon or infected with influenza. Mndal expression in B cells isolated from wild-type (+/+) mice that were either infected with influenza or treated with type I interferon-β (Ifn-β) were compared with control. Mndal expression was also examined in B cells isolated from interferon receptor knockout (−/−) mice that had also been infected with influenza.
Growth inhibition
Previous studies have suggested that other Ifi200 genes are candidate tumor suppressors, because they are inactivated or down-regulated in tumors and their overexpression in a variety of cells can suppress proliferation. We evaluated the effect of Mndal expression in NIH3T3 cell growth in colony formation assays. This line was chosen because, as noted above, it does not contain endogenous Mndal. Full-length Mndal was inserted into a mammalian expression vector, the resulting plasmid DNA was cotransfected with an expression plasmid containing neo resistance into NIH3T3 cells, and the G418-resistant colonies were counted 3 weeks after transfection. Consistent with a possible tumor suppressor function, the expression of Mndal reproducibly (8 independent experiments) decreased colony-forming efficiency by approximately 80% compared with the control vector (Figure 6). In contrast Ifi203 decreased colony formation by approximately 40% compared with the control vector (Figure 6).
Colony formation in NIH3T3 cells is suppressed when transfected with Mndal or Ifi203 compared with empty vector. The results of 8 separate experiments are shown. SEs are indicated, and P values represent t test comparisons with the control plasmid.
Colony formation in NIH3T3 cells is suppressed when transfected with Mndal or Ifi203 compared with empty vector. The results of 8 separate experiments are shown. SEs are indicated, and P values represent t test comparisons with the control plasmid.
Discussion
We have identified a new interferon-inducible gene, Mndal, linked to Chr 1 in the same interval containing the Pctm modifier of PCT susceptibility, by screening for differentially expressed genes in inflammatory tissues derived from the mesenteries of TMPD-primed BALB and DBA mice. Among the differentially expressed genes, Mndal had the greatest difference between the 2 strains. Mndal shares homology with the MNDA.
MNDA is a member of the HIN-200 family of hematopoietic interferon-inducible nuclear antigens, which includes AIM2, IFIX, and IFI16.22,–24 This multigene family is found on human chromosome 1q21-2425 and derives its name from a unique 200–amino acid domain conserved among its members. MNDAL is similar to MNDA and AIM2 in that it contains only one 200 amino acid “a” domain, has a nuclear localization sequence, has several putative Cdk2 sites, and is found in the nucleus. MNDAL has slightly greater homology to MNDA than to AIM2. The murine counterpart of the HIN-200 family is the Ifi200 family (members p202-p206), which reside on the distal portion of mouse Chr 1, in the same region as Mndal. As with other Ifi200 members, Mndal expression can be induced by interferon. Ifi205 is the only member with the same predicted structure as Mndal, and it shares even greater homology to Mndal than to human MNDA.
In normal tissues from mice carrying the intact gene, Mndal was expressed at high levels in hematopoietic tissues and in isolated B cells. Although Mndal is highly expressed in the tissues of TMPD-primed BALB mice, PCT cell lines do not express it, and primary plasma cell tumors in XBP1s transgenic mice also have low levels. Because the gene remains intact in the BALB-derived tumor cell lines, it is possible that it has been silenced epigenetically or transcriptionally at some point during the tumorigenic process. Promoter hypermethylation of AIM2, another Ifi gene, has been found in colon cancer.15 In addition, overexpression of Mndal and to a lesser extent Ifi203 in NIH3T3 cells reduced their growth in vitro. Similar results with other Ifi's, including Ifi205, have shown that they are also growth inhibitory and they have been proposed as tumor suppressors. Mndal and/or Ifi203, therefore, become a candidate gene(s) for the Pctm locus that modifies PCT susceptibility.
Further studies will be needed to confirm Mndal's role in plasmacytomagenesis. However, its membership in the Ifi200/HIN-200 family increases the plausibility of Mndal being a tumor suppressor gene. Overexpression of several Ifi200/HIN-200 members has been reported to inhibit growth, by cell cycle arrest, apoptosis, or senescence, and some members have been reported to interact with Rb and p53.16,20,26 Members of the HIN-200 family are somatically inactivated in several solid tumors, including breast,14 prostate,13 melanoma,22 and colon.15 Mndal is the first murine Ifi200 gene implicated in B-cell tumor formation in vivo either as a modifier gene or possibly by some form of somatic inactivation.
Little is known about the role of the Ifi/HIN genes in B-cell neoplasia. Interferon-α treatment has been shown to be effective in inducing apoptosis in patients with myeloma and other B-cell malignancies.27 Treatment of Daudi Burkitt lymphoma cells with interferon-α induced both MNDA and IFI16, which led to increases in p21 and Rb and decreases in c-Myc levels.28 More recently, high expression of MNDA has been associated with good outcome in patients with B-cell chronic lymphocytic leukemia.29
Because of the high levels of Mndal expression relative to the other Ifi family members in tissues from adult BALB mice, Mndal may be the predominant Ifi200 gene in those strains that harbor an intact gene. The presence or absence of Mndal could have important implications for phenotypes associated with the Ifi200 cluster, especially because Ifi200 genes may have partially overlapping functions.
Our RT-PCR studies determined that Ifi202 to Ifi205 and Mndal expression in BALB was induced by TMPD, with a possible increase in Ifi205 and Mndal relative to the other members. This observation is congruent with the finding that lymphoid tissue from TMPD-induced granulomas contains activated dendritic cells,30 as well as Ly6Chigh monocytes,31 both of which produce type I interferon which in turn would induce Ifi genes. In DBA mesenteries, both Ifi202 and, to a lesser extent, Ifi204/Ifi205, levels increased by day 18; it is possible that the enhanced expression of these genes compensates for the absence of Mndal and low levels of Ifi203 seen in DBA. Given the nature of the genomic segment missing in DBA and the substantially lower expression of Ifi203 in DBA relative to BALB, it appears that the Ifi203 promoter is partially defective in DBA mice and thus produces a compound defect affecting 2 genes simultaneously in the cluster.
Our data indicate that many inbred mouse strains resemble BALB in having an intact Mndal gene, whereas others resemble DBA in the absence of the gene. The absence of Ifi genes from the germline has not been documented before; human AIM2 sequences have been found missing in tumors but not in the germline.15 Although we have not examined Ifi203 in strains other than BALB and DBA, it seems likely that the Ifi203 promoter deletion we have identified in DBA that is contiguous with the absence of Mndal may also be polymorphic.
The 2-megabase region of mouse Chr 1, harboring the Ifi200 genes as well as Spna1, Apcs, Slam, and Fc receptor genes, has been proposed to represent a highly conserved antigen presentation cluster, reminiscent of the 6p MHC region, which may be dedicated to lipid/glycolipid antigens rather than antigen-derived peptides.32 Asefa et al20 and Ludlow et al33 have both hypothesized that this region of Chr 1 arose by gene duplication,20,33 and Ludlow et al33 predicted an additional 4 genes (p207-210) in the cluster from the then current builds of the human and mouse genomes. From analyzing the predicted protein structures of IFI207 to IFI210,33 that encoded by Mndal is distinct from the predicted p207 to p210 proteins.
It is tempting to speculate that Mndal (and/or Ifi203) could play a role in diseases, such as lupus, in which susceptibility loci have been mapped near this interval. The mouse Ifi202 and human IFI16 genes have been implicated in the development of this B cell–mediated autoimmune disease.12,34,,–37 Increased levels of IFI16 have been reported in patients with lupus.36,38 In mouse models of lupus, it has been hypothesized that overexpression or high levels of Ifi202 lead to an increase in the development of autoantibodies in response to apoptosis.12,39 NZB mice are susceptible to lupus, and one of the lupus susceptibility genes in NZB, designated Nba2, was mapped to mouse Chr1, in the same interval as the Ifi200 gene cluster.12 In fact, gene expression profiling studies examining a limited 11 000 genes/probe set, uncovered Ifi202 as being overexpressed in the congenic strain carrying the NZB Nba2 locus.12,40 The Affy probe set for Mndal was not included in this original screen. Because Mndal is absent in the NZB strain, examining its potential role in lupus susceptibility could be worthwhile.
In summary, Mndal, a new interferon inducible gene with tumor suppressor activity that is expressed at higher levels than other family members, has been found to reside in a highly polymorphic region of the genome implicated in several autoimmune and cancer phenotypes. Determining the various biologic activities and the conditions that are likely to induce these various diseases has the potential to yield new insights into their pathogenesis and treatment.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank our fellow laboratory members Karen Reidy, Matt Schlough, Justin English, all from the Laboratory of Cancer Biology and Genetics, for their suggestions and contributions. We also thank Susan Garfield for her contributions to our confocal analyses. Comments and suggestions from Pam Schwartzberg, Howard Young, John O'Shea, Kent Hunter, Doug Lowy, Glenn Merlino, and 2 anonymous reviewers contributed significantly to the manuscript.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
National Institutes of Health
Authorship
Contribution: K.Z. designed and performed research, collected and analyzed data, and wrote the manuscript; D.K. collected and analyzed data; W.D., R.R., W.C.V., and S.Z. performed research; V.B. analyzed data; and B.A.M. designed research, analyzed data, and wrote manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Beverly A. Mock, Laboratory of Cancer Biology and Genetics, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bldg 37, Rm 3146, 37 Convent Dr, MSC 4258, Bethesda, MD 20892-4258; e-mail: bev@helix.nih.gov.