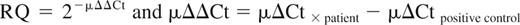Inhibitors of differentiation (Id) are a group of dominant inhibitors of basic helix-loop-helix transcriptional factors, which promote excessive proliferation, and also protect cells against drug-induced apoptosis in mammalians. Recently, Id1 has been identified as a common downstream target of several constitutively activated oncogenic tyrosine kinase, such as FLT3 internal tandem duplication, in leukemia cells. We analyzed Id1 expression as possible prognostic factor in 237 acute myeloid leukemia (AML) patients. High Id1 expression was associated with older age (P = .009) and with FLT3 internal tandem duplication (P = .003). However, 61% of the patients in the group of FLT3− AML were Id1+, suggesting that other tyrosine kinases are involved. In whole population, high Id1 expression independently predicted shorter disease-free survival (P = .05) and overall survival (P = .003). In young patients (age ≤ 60 years) with normal cytogenetics, Id1+ was, in multivariate analysis, associated with lower complete remission rates (P = .02), shorter disease-free survival (P = .02), and overall survival (P = .006). In conclusion, our data provide a new molecular marker for refining the risk classification of AML, especially in young patients with normal cytogenetic. Id1− patients with normal cytogenetic should be classified as favorable-risk leukemia. Id1, as a downstream target of constitutively activated tyrosine kinase, could be a suitable candidate for targeted therapy.
Introduction
Cytogenetic analysis at the time of diagnosis provides the most important prognostic information for therapeutic choices in adults with acute myeloid leukemia (AML). However, approximately 50% to 60% of patients do not present any cytogenetic aberrations, and are usually considered intermediate-risk patients.1,,–4 In this vast group, those patients are heterogeneous, and therapeutic results are highly variable. Therefore, in recent years, molecular alterations, such as acquired gene mutations and deregulation of gene expression, have been identified to help classifying this group and guiding therapeutic decisions.5,,,,,,,–13 These molecular alterations are observed in 2 complementation groups of genes. One group (class I) includes genes, such as c-KIT,14 FLT3,15 N-RAS, and K-RAS,16 in which mutations activate signal-transduction pathways and thereby increase proliferation or survival, or both, of leukemia cells. The other group (class II) includes genes, such as CEBP-α,17 MLL,18 and NPM1,19 in which mutations affect transcription factors or components of the transcriptional coactivation complex and cause impaired differentiation. The extensive analysis of nucleophosmin (NPM1) mutations, and FLT3 internal tandem duplication (FLT3/ITD), as well as other mutations with prognostic significance, such as CEBP-α, higher expression of BAALC, MLL, and N-RAS, has been reported in AML.5,,–8,10,,–13,16,18 According to these analyses, patients with mutant NPM1 or mutant CEBP-α without FLT3/ITD should be classified as favorable-risk leukemia. But patients with wild-type NPM1 and FLT3/ITD were difficult to classify because of contradictory results in those studies.11,–13,20
Inhibitors of differentiation (Id) or DNA-binding helix-loop-helix (HLH) proteins are a group of dominant inhibitors of basic HLH transcriptional factors, which promote cell differentiation.21,–23 Id proteins also act as positive regulators of cell growth or promote excessive proliferation in mammalian cells by direct interaction with pRB, p107, and p130 proteins,24,25 or through the inactivation of tumor suppressors.26,–28 Id proteins, and especially Id1, have been investigated as potential proto-oncogenes29 because overexpression of Id1 has been found in many types of human cancer, including breast, prostate, pancreas, and cervical cancer, and Id1 high expression is strongly correlated with advanced and metastatic tumor stages.30,,,,,,–37 Furthermore, overexpression of Id1 in human cancer cells induces cell proliferation and invasion, and also protects cells against drug-induced apoptosis.38,,,–42 Therefore, inactivation of Id1 in human cancer cells has been explored as a potential target to improve chemotherapy's efficiency. In vitro down-regulation of Id1 by small interfering RNA technology leads to increased sensitivity of cancer cells to chemotherapeutic drugs and decreases invasion of cancer cells.43,44
Recently, Id1 has been identified as a common downstream target of constitutively activated oncogenic tyrosine kinase, such as BCR-ABL, TEL-ABL, TEL-PDGF-βR, and FLT3/ITD, in primary leukemia cells.45 Moreover, Id1 down-regulation by small interfering RNA inhibited cell growth and increased sensitivity to Trail-induced apoptosis of Molm-14 and K562 leukemia cells, which are expressing, respectively, FLT3/ITD and BCR-ABL tyrosine kinase and both exhibiting high levels of Id1 expression.45 Furthermore, the role of Id1 in development of hematopoietic malignancies has been described; Id1 could immortalize hematopoietic progenitors in vitro, and especially common myeloid progenitors, and promote a myeloproliferative disease in vivo.46 Although overexpression of Id1 in the mammary epithelium was not sufficient for tumorigenesis, mice with both expression of Id1 and activated Ras developed metastatic cancer.47
Mutations in c-KIT receptor tyrosine kinases and in their downstream genes N-RAS or K-RAS occur, respectively, in 10% to 20% and 20% of AML.14,16 Whether Id1 is also a downstream target of c-KIT tyrosine kinase, N-RAS, and K-RAS has not been determined yet.
Those previous findings indicate a potential important role of Id1 in AML. Therefore, in this paper, for the first time, we analyzed Id1 expression and its possible prognostic impact in 237 patients with AML homogeneously treated in European Organisation for Research and Treatment of Cancer (EORTC) protocols. We compared Id1 expression with other well-known clinical, biologic, and cytogenetic prognostic factors, as well as molecular prognostic factors, such as NPM1, FLT3/ITD mutations, and BAALC expression. We also evaluated the association of high Id1 expression with treatment outcome and analyzed its role in guiding postremission therapy in patients with cytogenetically normal AML.
Methods
Patients and treatments
Bone marrow or peripheral blood samples were obtained from cell bank (Tumorothèque Leucémies Hôtel-Dieu no. 579) from 237 AML patients after their informed consent was obtained in accordance with the Declaration of Helsinki (Formulary EORTC study nos. 06931, 06991, and 06954) between 1996 and 2008. Patients had been included in either the EORTC protocols AML-10 or AML-12 trials for young patients or in AML-13 for older patients. Treatments of EORTC protocols AML-10, AML-12, and AML-13 have been described in detail elsewhere.48,–50 In AML-10 and AML-12 protocols, patients with myelosdysplasia or myeloproliferative diseases before AML were not included. The median follow-up time of living patients was 4 years. The availability of an HLA-matched donor was recorded for all young patients. Ethical approval for this study was obtained from Assistance Publique des Hopitaux de Paris.
RNA extraction and Id1 expression studies
Total RNA from AML patients was isolated by the guanidinium thiocyanate-phenol-chloroform method as described by Chomczynski and Sacchi.51 Complementary DNA (cDNA) was synthesized from 1 μg of total RNA using a high-capacity cDNA reverse-transcription kit (Applied Biosystems).
Id1, and ABL as endogenous internal control for each sample, were analyzed in duplicate in the same MicroAmp optical 96-well plates using a 7900HT Real-Time PCR System (Applied Biosystems). In case of a discrepancy greater than 1.0 in cycle of threshold (Ct) value between tests, a third test was performed. Comparative real-time reverse-transcribed polymerase chain reaction (RT-PCR) assays were performed following the manufacturer's (Applied Biosystems) instructions in a final reaction volume of 20 μL, containing 10 μL of 2× TaqMan Universal PCR Master Mix with AmpErase UNG, 50 ng cDNA, and l μL of 20× forward and reverse primers and probes (Applied Biosystems). Each specific probe was labeled with FAM dye and with nonfluorescent quencher (NFQ). Amplification was carried out at 50°C for 2 minutes, 95°C for 10 minutes, and followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The comparative Ct method was used to determine the relative expression level of Id1. The difference of Ct number (ΔCt = CtId1 − CtABL) was calculated for each replicate, and the mean of ΔCt from 2 replicates is μΔCt = ΣΔCt/2. Without detectable Id1 amplification within 40 cycles, Id1 expression values were set at 0. MV4-11 cell line (a human acute monocytic leukemia cell line, positive for FLT3/ITD) was used as positive control and set at 1, and was included in each run for standardization between runs. Relative Id1 expression values were calculated as relative quantification (RQ) = 2−μΔΔCt and μΔΔCt = μΔCt × patient − μΔCt MV4-11. Every 10 MicroAmp optical 96-well plates were analyzed by RQ Manager 1.2 software (Applied Biosystems). Patients were divided into 2 groups according to their Id1 expression status (high expression [≥ 1] considered as Id1+ and low expression [< 1] considered as Id1−).
To determine whether Id1 expression was influenced by the origin of the samples (blood or bone marrow) or by percentage of blast cells in the samples, and if expression has changed over time, we have shown that levels of Id1 expression in blast cells obtained from blood (43 samples, 4.8 ± 20) and blast cells obtained from marrow (193 samples, 4.3 ± 10, P = not significant) were concordant. Moreover, RNA expression is stable over time; similar mean Id1 expression was found in samples collected from 1996 to 2002, and in those collected from 2003 to 2008 (4.0 ± 9 vs 4.6 ± 3, respectively, P = not significant). Furthermore, Id1 expression was not correlated to percentage of blasts at diagnosis in bone marrow or blood samples used (63% ± 22% for Id1+ samples vs 59% ± 23% for Id1− samples, P = not significant).
DNA extraction and FLT3/ITD detection
DNA was extracted from frozen bone marrow or peripheral blood samples using Nucleon kits (GE Healthcare) according to the manufacturer's protocol.
FLT3 DNA (one part of exon 13, exon 15, and full exon 14) was amplified with 35 PCR cycles (94°C for 30 seconds, 60°C for 1 minute, and 72°C for 1 minute) using 250 ng genomic DNA, 0.3 μM each of primers ITD-f (TGGTGTTTGTCTCTTCTTCATTGT) and ITD-r (GTTGCGTTCATCACTTTTCCAA), 750 μM of deoxynucleotide triphosphate, 5 mM of MgCl2, 1.25 units of Taq GOLD polymerase, and 1× buffer GOLD 1 (PerkinElmer Life and Analytical Sciences) in a total of 50 μL. Amplicons were migrated in 2% agarose gels, allowing the detection of a 240-bp fragment for wild-type alleles and/or longer fragments for FLT3-ITD alleles. Two categories were distinguished: samples showing an absence or a weak detection of the mutated allele were considered negative (ITD−) for this study, and samples with marked or predominantly mutated bands were considered positive (ITD+).52
Analysis of type A, type B, and type D mutations of NPM1
Specific forward and reverse primers for exon 12 of NPM1, and 3 specific probes for detecting type A, type B, and type D mutations of NPM1 were designed by Applied Biosystems. NPM1-forward (5′-GGATGACTGACCAAGAGGCTATTC-3′) and NPM1-reverse (5′-CAGAAATGAAATAAGACGGAAAATTTTTTAACAAATTGT-3′) primers are common, wild-type probe is FAM-5′-CTCCACTGCCAGAGAT-NFQ-3′, and specific type A probe is FAM-5′-CACTGCCAGACAGAGAT-NFQ-3′, specific type B probe is FAM-5′-ACTGCCATGCAGAGAT-NFQ-3′, specific type D probe is FAM-5′-CTGCCAGGCAGAGAT-NFQ-3′. Absolute real-time RT-PCR quantification assays were performed following the manufacturer's instructions (Applied Biosystems) in a final reaction volume of 20 μL, containing 10 μL of 2× TaqMan genotyping Master Mix, 10 ng cDNA, and l μL of 20× forward, reverse primers and probes (Applied Biosystems). Amplification condition was as described in “RNA extraction and Id1 expression studies” (TaqMan 7900HT; Applied Biosystems).
BAALC expression analysis
BAALC expression analysis was carried out with AML patients' cDNA samples used in Id1 expression studies. BAALC and ABL as endogenous internal control for each sample were analyzed in duplicate in the same MicroAmp optical 96-well plates using a 7900HT Real-Time PCR System (Applied Biosystems). Comparative real-time RT-PCR assays were performed as described in Id1 expression studies. One patient sample was used as positive control, and set at 1, and was included in each run for standardization between runs. Relative BAALC expression values were calculated as follows:
Statistical analyses
In our 237 samples, Id1 expression levels represented a continuum ranging from 0.034 to 152.106 (mean = 4.41, median = 1.57, quartile 3 = 3.32). To evaluate the impact of Id1 expression levels on clinical outcome without seeking an optimal cut-point, patients were divided into low and high expressers using Id1 expression of MV4-11 cell line as reference. Id1 expression as continuous variable was also used. Complete remission (CR) was defined as recovery of morphologically bone marrow and blood count (ie, neutrophil count ≥ 109/L, and platelet count ≥ 100 × 109/L) and no evidence of extramedullary leukemia. Disease-free survival (DFS) was measured from the date of CR until the date of relapse or death from any cause, with observation censored for patients last known to be alive without report of relapse. Overall survival (OS) was measured from the date of diagnosis until the date of death from any cause, with observation censored for patients last known to be alive.
Association between high and low Id1 groups and baseline clinical and biologic features were analyzed using Fisher exact test for categorical variables and Mann-Whitney or Kruskal-Wallis tests for continuous variables. Estimated probabilities of DFS and OS were calculated using the Kaplan-Meier method, and differences between survival distributions were evaluated by the log-rank test. Proportional hazards models were constructed to determine whether Id1 expression was associated with outcome when adjusting for other prognostic variables. Full model used the variables significant in univariate analysis. Logistic regression was used to model the impact of Id1 expression on the achievement of CR. We used StatView software (Version 5.0) for statistical analysis (SAS Institute Inc).
Results
Correlations of Id1 expression with clinical and biologic characteristics
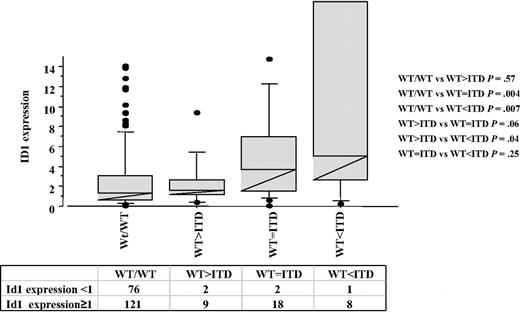
A total of 237 patients were enrolled in this study. The clinical and biologic characteristics of the patients are listed in Table 1. Id1+ patients were significantly older than Id1− patients (P = .009), but no significant differences were found among French-American-British subtypes, sex, leukocyte count, karyotype, NMP1 mutations (NPM1+), and BAALC expression (Table 1). Screening for molecular markers NPM1+ and FLT3/ITD was performed in all samples of blood or bone marrow at the time of diagnosis. Fifty of 237 (21%) presented NPM1+, 40 of 237 were FLT3/ ITD+ (17%), and 156 of 237 showed high level Id1 expression (65.8%). Id1 expression was significantly associated with FLT3/ITD+ (P = .003; Table 1; Figure 1). For further determination of correlation between Id1 and FLT3/ITD, patients were divided into 4 groups according to FLT3/ITD quantity: wild-type (WT) only (WT/WT), ITD fragment weak (WT > ITD), ITD fragment equal to wild-type (WT = ITD), and ITD fragment stronger than wild-type (WT < ITD). Significant differences in Id1+ frequency were found between these 4 groups: 61.2% for wt/wt, 81.8% for WT > ITD, 90% for WT = ITD, and 88.9% for WT < ITD (Figure 1). Significant differences were also observed in Id range and mean by Mann-Whitney analysis (WT/WT vs WT > ITD, P = .57; WT/WT vs WT = ITD, P = .004; WT/WT vs WT < ITD, P = .007; WT > ITD vs WT = ITD, P = .06; WT > ITD vs WT < ITD, P = .04; WT = ITD vs WT < ITD, P = .25). In our statistic analysis, we have considered WT > ITD as FLT3/ITD−.
Comparison of clinical and biologic variables of all patients according to Id1 expression
| Characteristic . | All patients (n = 237) . | Id1+ (n = 156) . | Id1− (n = 81) . | P . |
|---|---|---|---|---|
| Sex, no. (%) | NS | |||
| Female | 107 (46) | 73 | 34 | |
| Male | 130 (54) | 83 | 47 | |
| Median age, y (range) | 54 (16-80) | 54 | 47 | .009 |
| Median leukocytes, 109/L (range) | 25 (1-284) | 27 | 22 | NS |
| FAB subtypes, no. (%) | NS | |||
| M0 | 10 (4.2) | 7 (70) | 3 | |
| M1 | 58 (24.4) | 39 (67.2) | 19 | |
| M2 | 84 (35.4) | 51 (60.7) | 33 | |
| M4 | 36 (15.2) | 21 (58.3) | 15 | |
| M5 | 43 (18.1) | 32 (74.4) | 11 | |
| M6 | 6 (2.5) | 6 (100) | 0 | |
| Karyotype,* no. (%) | NS | |||
| Good | 23 (9.7) | 14 (60.9) | 9 | |
| Intermediate | 171 (72) | 112 (65.5) | 59 | |
| Normal | 138 (81) | |||
| Other | 33 (19) | |||
| Poor | 38 (16) | 27 (71) | 11 | |
| Not done or not assessable | 5 (2.1) | 3 (60) | 2 | |
| NPM1 mutation status, no. (%) | NS | |||
| NPM1+ | 50 (21.1) | 35 (70) | 15 | |
| NPM1− | 187 (78.9) | 121 (30) | 66 | |
| FLT3/ITD status, no. (%) | .003 | |||
| FLT3/ITD+ | 40 (16.9) | 35 (87.5) | 5 | |
| FLT3/ITD− | 197 (83.1) | 121 (61.4) | 76 | |
| BAALC expression | 1.51 (0.001-16.8), median (range) | 1.67 ± 2.9, mean ± SD | 1.24 ± 2.23, mean ± SD | NS |
| Characteristic . | All patients (n = 237) . | Id1+ (n = 156) . | Id1− (n = 81) . | P . |
|---|---|---|---|---|
| Sex, no. (%) | NS | |||
| Female | 107 (46) | 73 | 34 | |
| Male | 130 (54) | 83 | 47 | |
| Median age, y (range) | 54 (16-80) | 54 | 47 | .009 |
| Median leukocytes, 109/L (range) | 25 (1-284) | 27 | 22 | NS |
| FAB subtypes, no. (%) | NS | |||
| M0 | 10 (4.2) | 7 (70) | 3 | |
| M1 | 58 (24.4) | 39 (67.2) | 19 | |
| M2 | 84 (35.4) | 51 (60.7) | 33 | |
| M4 | 36 (15.2) | 21 (58.3) | 15 | |
| M5 | 43 (18.1) | 32 (74.4) | 11 | |
| M6 | 6 (2.5) | 6 (100) | 0 | |
| Karyotype,* no. (%) | NS | |||
| Good | 23 (9.7) | 14 (60.9) | 9 | |
| Intermediate | 171 (72) | 112 (65.5) | 59 | |
| Normal | 138 (81) | |||
| Other | 33 (19) | |||
| Poor | 38 (16) | 27 (71) | 11 | |
| Not done or not assessable | 5 (2.1) | 3 (60) | 2 | |
| NPM1 mutation status, no. (%) | NS | |||
| NPM1+ | 50 (21.1) | 35 (70) | 15 | |
| NPM1− | 187 (78.9) | 121 (30) | 66 | |
| FLT3/ITD status, no. (%) | .003 | |||
| FLT3/ITD+ | 40 (16.9) | 35 (87.5) | 5 | |
| FLT3/ITD− | 197 (83.1) | 121 (61.4) | 76 | |
| BAALC expression | 1.51 (0.001-16.8), median (range) | 1.67 ± 2.9, mean ± SD | 1.24 ± 2.23, mean ± SD | NS |
NS indicates not significant.
Good karyotypes included t(8;21) and inv(16); poor karyotypes included 3q26rearangements, t(6;9), del(7)/−7, del(5)/−5 and 3 or more than 3 abnormalities.
Id1 expression according to FLT3/ITD. Wild-type only (WT/WT), ITD fragment weak (WT > ITD), ITD fragment equal to wild-type (WT = ITD), and ITD fragment higher than wild-type (WT < ITD). P value is according to Mann-Whitney test.
Id1 expression according to FLT3/ITD. Wild-type only (WT/WT), ITD fragment weak (WT > ITD), ITD fragment equal to wild-type (WT = ITD), and ITD fragment higher than wild-type (WT < ITD). P value is according to Mann-Whitney test.
Clinical outcome and prognostic significance of Id1 expression for all patients
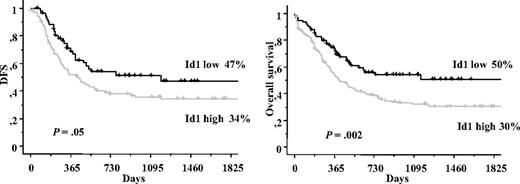
Prognostic significance of Id1 expression analysis was first performed in the whole population (237 patients). High Id1 expression was not a poor prognostic factor for CR achievement (P = .34). In contrast, DFS and OS were significantly shorter in the patients with high Id1 expression (156 patients) compared with the patients with low Id1 expression (81 patients). Median DFS was 445 days (34% ± 5% at 5 years) for Id1+ patients versus 1184 days for Id1− patients (47% ± 7% at 5 years; P = .05). The estimated 5-year OS rates of Id1+ patients were 30% plus or minus 3% (median = 454 days) versus 50% plus or minus 6% (median = 1140 days) for Id1− patients (P = .002; Figure 2).
In continuous variable Id1 value analysis, Id1 expression has also a prognostic impact for CR achievement (P = .09), DFS (P = .05), and OS (P = .003).
Clinical outcome and prognostic significance of Id1 expression according to age
Among 83 patients older than 60 years, Id1 expression status was not a prognostic factor for CR achievement (P = .12), DFS (23%, 95% confidence interval [CI]: 5%-58% for Id1− vs 15%, 95% CI: 5%-34% for Id1+, P = .15), and OS (25%, 95% CI: 11%-48% for Id1− vs 13%, 95% CI: 5%-24% for Id1+; P = .19).
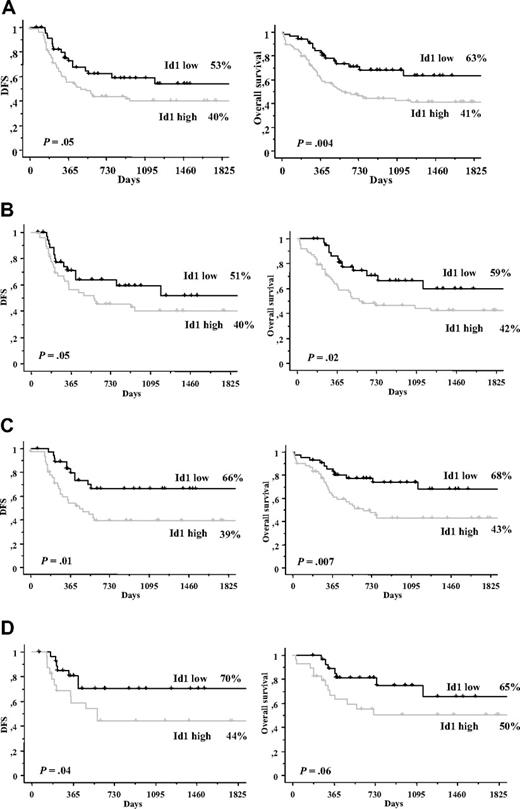
A total of 154 patients younger than 60 years were separately analyzed. Patients expressing high Id1 levels had a lower CR achievement (76% vs 88%, P = .05), a shorter DFS (40% ± 6%, median = 464 days vs 53% ± 8%, median = not reached, P = .05), and OS (41% ± 5%, median = 560 days vs 63% ± 7%, median = not reached, P = .004) than patients expressing low Id1 levels (Figure 3A).
Disease-free and overall survival in young patients. (A) DFS and OS in young patients (≤ 60 years) according to Id1 expression. (B) DFS and OS in young patients with normal cytogenetics according to Id1 expression. (C) DFS and OS in young patients with both normal cytogenetics and NPM1−/FLT3/ITD−. (D) DFS and OS in young patients with both normal cytogenetics and NPM1+/FLT3/ITD−.
Disease-free and overall survival in young patients. (A) DFS and OS in young patients (≤ 60 years) according to Id1 expression. (B) DFS and OS in young patients with normal cytogenetics according to Id1 expression. (C) DFS and OS in young patients with both normal cytogenetics and NPM1−/FLT3/ITD−. (D) DFS and OS in young patients with both normal cytogenetics and NPM1+/FLT3/ITD−.
In continuous variable Id1 value analysis, Id1 expression has also a strong prognostic impact for CR achievement (P = .05), DFS (P = .04), and OS (P = .008).
Clinical outcome and prognostic significance of Id1 expression according to cytogenetics
Id1 expression was not a prognostic factor in patients with a poor cytogenetic (CR, P = .21; DFS [33%, 95% CI: 4%-82%, for Id1− patients vs 0%, 95% CI: 0%-40% for Id1+ patients, P = .10]; OS [22%, 95% CI: 6%-58% for Id1− patients vs 6%, 95% CI: 1%-28% for Id1+ patients, P = .63] or a good cytogenetic; CR, P = .15; DFS [58%, 95% CI: 22%-85% for Id1− patients vs 72%, 95% CI: 42%-92% for Id1+ patients, P = .51]; OS [77%, 95% CI: 43%-94% for Id1− patients vs 70%, 95% CI: 43%-87% for Id1+ patients, P = .78]).
By contrast, patients with intermediate prognostic cytogenetic, including normal cytogenetics, with high Id1 expression have a poorer prognosis (intermediate cytogenetics: CR, P = .02, and P = .04; DFS, P = .08; and P = .05; OS, P = .02 and P = .02, respectively) than patients with low Id1 expression.
Clinical outcome and prognostic significance of Id1 expression in young patients with normal cytogenetics
Id1 expression being a prognostic factor in young patients and patients with normal cytogenetics, we have analyzed its prognostic value in young patients with normal cytogenetics. Among 154 patients younger than 60 years, 101 patients have a normal karyotype. Achievement rate of CR was significantly more important in Id1− patients than in Id1+ patients (94% vs 79%, P = .04). In the same way, 5-year DFS rates were 51% plus or minus 10% (median = not reached) versus 40% plus or minus 8%, median = 555 days, P = .05; and 5-year OS rates were 59% plus or minus 9% (median = not reached) versus 42% plus or minus 6% (median = 566 days), P = .02 (Figure 3B).
In continuous variable Id1 expression, the value has also shown that Id1 was effectively a prognostic factor for achievement of CR (P = .02), for DFS (P = .04), and for OS (P = .005).
Clinical outcome and prognostic significance of Id1 expression in young patients with normal cytogenetics according to NPM1 mutation and FLT3/ITD status
Mutational status of NPM1 and FLT3/ITD has been shown to be associated with the outcome of treatment for patients with cytogenetically normal AML in several different studies and our own studies. Therefore, we analyzed the prognostic impact of Id1 expression in patients younger than 60 years with normal karyotype, and with NPM1−/FLT3/ITD− or with NPM1+/FLT3/ITD−.
There were 51 patients in NPM1−/FLT3/ITD− subgroup. A higher CR achievement, longer DFS, and longer OS at 5 years were found in patients with Id1− status than in patients with Id1+ status, respectively: 96% versus 79% (P = .03), 66% plus or minus 8% (median = not reached) versus 39% plus or minus 7%, median = 415 days (P = .01) and 68% plus or minus 8% (median = not reached) versus 43% plus or minus 6%, median = 690 days (P = .007; Figure 3C). Prognosis of this Id1− subgroup is comparable with prognosis of good risk cytogenetic patients. When Id1 expression was analyzed in continuous variable, Id1 expression was also a good prognostic factor for achievement of CR (P = .01), DFS (P = .05), and OS (P = .006).
In the NPM1+/FLT3/ITD− subgroup, 30 patients were studied. Patients with low Id1 expression had a higher CR achievement (96% vs 79%, P = .04), a longer DFS (70% ± 9%, median = not reached vs 44% ± 10%, median = 556 days, P = .04), and a longer OS (65% ± 11%, median = not reached vs 50% ± 9%, median = 729 days, P = .06; Figure 3D). When Id1 expression was analyzed in continuous variable, Id1 expression was also a good prognostic factor for achievement of CR (P = .01), DFS (P = .05), and OS (P = .006).
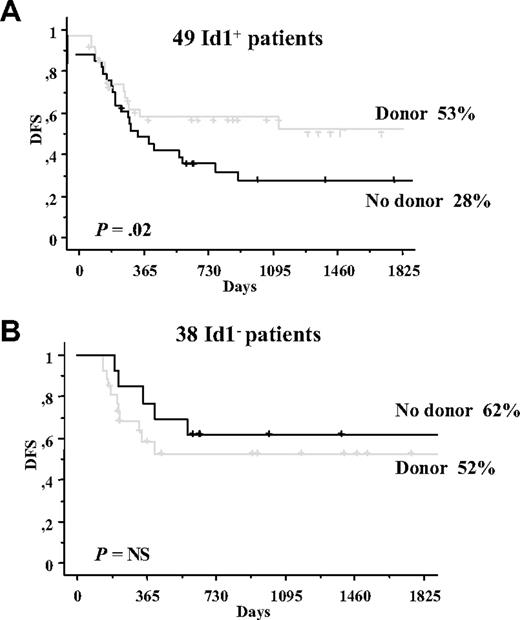
Allogeneic transplantation analysis according to Id1 expression in young patients with normal cytogenetics in CR
A univariable analysis of data for 87 patients with a CR, comparing the donor group with the no-donor group, revealed a significantly longer DFS (P = .02) in the donor group, but this difference did not translate into a significant difference in OS. The allogeneic transplantation analysis according to Id1 expression is shown in Figure 4. Figure 4 shows a better DFS curve for patients with a donor in patients with high Id1 expression (53% ± 9% in donor group, median = not reached, vs 28% ± 8% in no-donor group, median = 317 days, P = .02), but not in patients with low Id1 expression (52% ± 10% in donor group vs 62% ± 11% in no-donor group, median = not reached in both groups, P = not significant; Figure 4).
DFS among the patients achieving a complete remission according to the availability of donor status. (A) Patients with high expression of Id1. (B) Patients with low expression of Id1.
DFS among the patients achieving a complete remission according to the availability of donor status. (A) Patients with high expression of Id1. (B) Patients with low expression of Id1.
Multivariate analysis and Id1 expression
According to the prognostic value of Id1 expression in the 101 young patients with a normal karyotype, Id1 expression was entered into a multivariate model in addition to factors significantly associated with prognosis in univariate analysis for this population. Id1 expression (P = .02), BAALC expression (P = .03), and NPM1 mutation (P = .03) were independent prognostic parameters for CR achievement. Flt3/ITD (P = .008), donor versus no donor (P = .01), Id1 expression (P = .02), CR achievement in 1 versus 2 courses (P = .02), and NPM1 mutation (P = .03) were independent prognostic parameters for DFS. Id1 expression (P = .006), CR achievement in 1 versus 2 courses (P = .009), BAALC expression (P = .01), Flt3/ITD (P = .02), and NPM1 mutations (P = .03) were independent prognostic parameters for OS (Table 2).
Multivariate analysis of associations between prognostic parameters and clinical outcome in 101 young patients (< 60 years) with normal cytogenetics
| Endpoint/parameter . | Univariate analysis, P . | Multivariate analysis, P . |
|---|---|---|
| CR* | ||
| Id1 expression (continuous variable) | .01 | .02 |
| BAALC expression (continuous variable) | .01 | .03 |
| NPM1+ vs NPM− | .04 | .03 |
| Flt3/ITD+ vs Flt3/ITD− | .10 | .19 |
| AML10 vs AML12 | .31 | .45 |
| DFS† | ||
| Flt3/ITD+ vs Flt3/ITD− | .009 | .008 |
| Donor vs no donor | .01 | .01 |
| Id1 expression (continuous variable) | .02 | .02 |
| CR achieved in 1 vs 2 courses | .01 | .02 |
| NPM1+ vs NPM− | .06 | .03 |
| WBC count (continuous variable) | .07 | .60 |
| AML10 vs AML12 | .46 | .80 |
| OS‡ | ||
| Id1 expression (continuous variable) | .001 | .006 |
| CR achieved in 1 vs 2 courses | .008 | .009 |
| BAALC expression (continuous variable) | .01 | .01 |
| Flt3/ITD+ vs Flt3/ITD− | .01 | .02 |
| NPM1+ vs NPM− | .04 | .03 |
| AML10 vs AML12 | .56 | .87 |
| Endpoint/parameter . | Univariate analysis, P . | Multivariate analysis, P . |
|---|---|---|
| CR* | ||
| Id1 expression (continuous variable) | .01 | .02 |
| BAALC expression (continuous variable) | .01 | .03 |
| NPM1+ vs NPM− | .04 | .03 |
| Flt3/ITD+ vs Flt3/ITD− | .10 | .19 |
| AML10 vs AML12 | .31 | .45 |
| DFS† | ||
| Flt3/ITD+ vs Flt3/ITD− | .009 | .008 |
| Donor vs no donor | .01 | .01 |
| Id1 expression (continuous variable) | .02 | .02 |
| CR achieved in 1 vs 2 courses | .01 | .02 |
| NPM1+ vs NPM− | .06 | .03 |
| WBC count (continuous variable) | .07 | .60 |
| AML10 vs AML12 | .46 | .80 |
| OS‡ | ||
| Id1 expression (continuous variable) | .001 | .006 |
| CR achieved in 1 vs 2 courses | .008 | .009 |
| BAALC expression (continuous variable) | .01 | .01 |
| Flt3/ITD+ vs Flt3/ITD− | .01 | .02 |
| NPM1+ vs NPM− | .04 | .03 |
| AML10 vs AML12 | .56 | .87 |
Only independent parameters are sorted in order of significance.
Other parameters, such as age, WHO performance status, and CD34 expression, were not significantly associated with CR in univariate analysis and were not included in the multivariate model.
Other parameters, such as age, WHO performance status, CD34 expression, and BAALC expression, were not significantly associated with DFS in univariate analysis and were not included in the multivariate model.
Other parameters, such as age, WBC count, WHO performance status, and CD34 expression, were not significantly associated with OS in univariate analysis and were not included in the multivariate model.
Discussion
Patients with AML frequently show aberrant activation of members of the receptor tyrosine kinase family that serve as mediators of intracellular signal transduction by extracellular growth factors. Two closely related cytokine receptors with important roles in normal hematopoiesis, FLT3 and c-KIT, are frequently deregulated. Tam et al45 recently identified Id1 as a common target gene of several oncogenic tyrosine kinase-dependent signaling in proliferation and cell survival. This strongly suggests that Id1 may be important for pathogenesis of hematologic malignancies caused by oncogenic tyrosine kinases and may serve as a novel therapeutic target in leukemia involving oncogenic tyrosine kinase. Therefore, Id1 expression level may be a more pertinent prognostic factor than mutations of individual tyrosine kinases. In addition of its role as a strong prognostic factor, down-regulation of Id1 may be a potential target to increase sensitivity of chemotherapy. It has been shown that inactivation of the Id1 gene in different cancer cell lines leads to an increased sensitivity to commonly used anticancer drugs.43,53
Recently, there has been substantial progress in using molecular markers, such as gene mutations or aberrant gene expression, to risk-stratify patients with AML, especially with normal cytogenetic AML. In this report, we analyzed 237 patients with AML for the expression levels of Id1 and tested its prognostic value.
The major role of Id1 in AML is strengthened by our work. In a large cohort of AML, patients with high Id1 expression were less likely to achieve CR and had poorer DFS and OS than patients with low Id1 expression, independently from the FLT3/ITD, NPM1 mutation status, and BAALC expression, especially in young patients with normal cytogenetics. In other cytogenetic subgroups and in older patients, the 95% CI indicates that there was a possible difference of prognosis between Id1+ and Id1− groups.54 But further studies will be required in these subgroups to confirm whether Id1 expression is a prognostic factor or not, especially in favorable cytogenetic AML, whether tyrosine kinase receptor activity has an impact value. Therefore, new observations will also be valuable to improve current molecular models for risk stratification and to design novel approaches to treat patients more effectively.
Id1 is a common target gene of several oncogenic tyrosine kinases.45 In our study, Id1 expression correlated imperfectly to Flt3/ITD. A total of 61% of the patients with FLT3 wt/wt have an Id1 expression more than 1, suggesting perhaps that other oncogenic tyrosine kinases are involved in Id1 overexpression, such as c-Kit, N-Ras, and K-Ras mutations, or other tyrosine kinases, or may be undetected Flt3/ITD because of our method: our primers detect all Flt3/ITD in JM domain, but not all in tyrosine kinase domain. Further study will be required to examine this hypothesis. In the other extreme, in FLT3+ patients, only 3 patients (10%) have low Id1; these patients were all NPM1+. These results raise interesting questions about the underlying interaction between FLT3/ITD, NPM1, and Id1. In other studies, patients could be stratified into 3 prognostic groups using their FLT3 and NPM1 mutant status: good (NPM1+/FLT3/ITD−), intermediate (FLT3/ITD−/NPM1− or FLT/ITD+/NPM1+), or poor (NPM1−/FLT3/ITD+),55 as NPM1 blast cells show a high in vivo sensitivity toward induction chemotherapy irrespective of the FLT3/ITD mutation status. But it is clear that further studies are required to elucidate the biologic consequences of NPM1 mutation on FLT3/ITD activity.
In our study, we compared Id1 subgroups (Id1+ or Id1−) for the result of postinduction therapy in young patients with normal cytogenetics. The probability of DFS of Id1− AML did not differ according to whether a donor was available, and was similar with core-binding factor leukemias. In contrast, an allogeneic transplantation performed during the first remission was beneficial for Id1+ patients.
In conclusion, our data provide a new molecular marker for refining the risk classification of AML and the design of novel approaches to treat patients more effectively, especially in young patients with normal cytogenetic. Id1− patients with normal cytogenetics should be classified as favorable-risk leukemia, such as the core-binding factor AML. Further study will be required to examine the role of Id1 in other cytogenetic subgroups, especially in core-binding leukemias. Id1 is also a suitable candidate for targeted therapy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by Tumorothèque Leucémies, Hôpital Hôtel-Dieu, in part by Inserm, and in part by Université Pierre et Marie Curie.
Authorship
Contribution: R.T. designed research, performed experiments, and wrote the manuscript; P.H. collected clinical data and participated in manuscript review; S.L. collected clinical data; F.F., C.M., and I.T. performed RT-PCR analysis; J.P. assisted with cell preparation; J.-P.M. designed research and participated in manuscript review; and O.L. designed research, performed statistical analysis, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ollivier Legrand, Hôtel-Dieu, Département d'Hématologie, 1 Place du Parvis Notre Dame, 75181 Paris, France; e-mail: ollivier.legrand@htd.aphp.fr.