Abstract
Two putative types of circulating endothelial progenitor cells have been recently identified in vitro: (1) endothelial colony-forming cell (ECFC) and (2) colony-forming unit–endothelial cell (CFU-EC). Only the former is now recognized to belong to endothelial lineage. We have used the ECFC and CFU-EC assays to readdress the issue of the clonal relation between endothelial progenitor cells and hematopoietic stem cells in patients with Philadelphia-positive and Philadelphia-negative chronic myeloproliferative disorders. Both ECFCs and CFU-ECs were cultured from peripheral blood mononuclear cells, and either BCR-ABL rearrangement or JAK2-V617F mutation were assessed in both types of endothelial colonies. We found that ECFCs lack the disease-specific markers, which are otherwise present in CFU-ECs, thus reinforcing the concept that the latter belongs to the hematopoietic lineage, and showing that in chronic myeloproliferative disorders the cell that gives rise to circulating ECFC has a distinct origin from the cell of the hematopoietic malignant clone.
Introduction
Recently, a role for circulating endothelial progenitor cells (EPCs) in the pathogenesis of bone marrow and spleen neoangiogenesis of patients with chronic myeloproliferative disorders (CMPDs) has been postulated, opening the way to the hypothesis that a common progenitor to both hematopoietic and vascular endothelial lineage, the so-called hemangioblast, could be the malignant stem cell of these disorders. The presence of a hemangioblast was initially suggested in chronic myeloid leukemia (CML),1 when the BCR-ABL fusion gene was detected in a proportion of endothelial-like cells generated in vitro from mononuclear cells (MNCs). Since then, this hypothesis was tested in CML and Ph-negative (Ph−) CMPDs,2,3 reinforcing the possibility that either the BCR-ABL rearrangement or the JAK2-V617F mutation, respectively, might happen at the level of a hemangioblastic progenitor cell.4
Several theories have been proposed about the origin and classification of EPCs.5–8 The scene has been further entangled by the finding that 2 types of EPCs, namely, colony-forming unit–endothelial cells (CFU-ECs) and endothelial colony-forming cells (ECFCs) can be detected in human peripheral blood.9–11 More recently, Yoder et al12–14 clearly settled that CFU-ECs, acknowledged by many authors as endothelial in origin,7,10 derive from the hematopoietic system, with no ability to form secondary endothelial colonies in vitro or new vessels in vivo. In contrast, ECFCs were shown to belong to the endothelial lineage.14 They have a robust proliferative potential, and vessel-forming capacity in vivo. Therefore, CFU-ECs and ECFCs are 2 distinct progenitor cells.
We have readdressed the issue of endothelial cell origin in CMPDs by investigating whether circulating EPCs, isolated as both CFU-ECs and ECFCs, bear the molecular clonality marker of the disease. For CML, the BCR-ABL fusion gene was used as such a marker, whereas for Ph− CMPDs the JAK2-V617F mutation and chromosomal aberrations detected by comparative genomic hybridization (CGH) were used.
Methods
Patients and healthy subjects
Seventy-six patients with CMPDs and 27 healthy subjects were studied. All patients were out of therapy, and their characteristics are reported in Table 1. The Institutional Review Board at Ospedale San Martino in Genoa and Istituto di Ricovero e Cura a Carattere Scientifico Policlinico San Matteo Foundation in Pavia approved all protocols. Informed consent was obtained according to the Declaration of Helsinki.
Patient data at the study time and MNC recovery
| Patients . | Age, y* . | M/F . | Hb, g/dL† . | WBC, ×103 μL† . | Ptl, ×103 μL† . | MNC recovery, ×106*‡ . |
|---|---|---|---|---|---|---|
| CML (n = 19) | 53 (27-79) | 12/7 | 10.8 ± 2.1 | 68.1 ± 73.4 | 299 ± 166 | 142.5 (5-860) |
| PMF (n = 28) | 59 (45-74) | 16/12 | 12.6 ± 2.3 | 9.1 ± 6.2 | 378 ± 268 | 41.7 (3.1-180) |
| PV (n = 22) | 68 (48-82) | 14/8 | 17.7 ± 0.9 | 10.3 ± 2.4 | 446 ± 141 | 67 (30-520) |
| ET (n = 7) | 72 (55-76) | 5/2 | 15.2 ± 0.3 | 11.8 ± 3.9 | 536 ± 125 | 52 (4.5-123.8) |
| Patients . | Age, y* . | M/F . | Hb, g/dL† . | WBC, ×103 μL† . | Ptl, ×103 μL† . | MNC recovery, ×106*‡ . |
|---|---|---|---|---|---|---|
| CML (n = 19) | 53 (27-79) | 12/7 | 10.8 ± 2.1 | 68.1 ± 73.4 | 299 ± 166 | 142.5 (5-860) |
| PMF (n = 28) | 59 (45-74) | 16/12 | 12.6 ± 2.3 | 9.1 ± 6.2 | 378 ± 268 | 41.7 (3.1-180) |
| PV (n = 22) | 68 (48-82) | 14/8 | 17.7 ± 0.9 | 10.3 ± 2.4 | 446 ± 141 | 67 (30-520) |
| ET (n = 7) | 72 (55-76) | 5/2 | 15.2 ± 0.3 | 11.8 ± 3.9 | 536 ± 125 | 52 (4.5-123.8) |
CML indicates chronic myeloid leukemia; PMF, primary myelofibrosis; PV, polycythemia vera; ET, essential thrombocythemia; Hb, hemoglobin; and Ptl, platelets.
Data are median (range).
Data are means ± SD.
Values refer to the median number of MNC recovered from 40 mL peripheral blood and plated for the ECFC and CFU-EC assays; the median yield of MNC from 40 mL PB from healthy subjects was 67 × 106 (30-225).
Spleen samples
Fresh tissue samples were obtained from the spleen of 3 patients with primary myelofibrosis (PMF) who underwent therapeutic splenectomy. After extensive washing, single-cell suspension was obtained by mechanical disaggregation. Cells were then used for the ECFC assay, as described in “EPC culture and characterization.”
EPC culture and characterization
Circulating ECFCs were isolated according to Ingram et al,11 whereas CFU-ECs were grown following the method of Hill et al.10 When appropriate, single ECFC-derived colonies were expanded for 2 or 3 passages to obtain an amount of cells suitable for further experiments, whereas CFU-ECs could not be expanded and were pooled. Immunophenotypic characterization was performed by flow cytometry analysis.11,15 Capillary formation assay was performed as described11 using ECFCs at passage 2, or pooled CFU-ECs.
BCR-ABL rearrangement by real-time quantitative RT-PCR
Total RNA was extracted using the PicoPure RNA Isolation Kit (Arcturus Bioscience). Quantitative reverse transcription–polymerase chain reaction (RT-PCR) for BCR-ABL and ABL were performed using the TaqMan system (7900 HT Fast Real-Time PCR System; Applied Biosystems).16
BCR-ABL fusion gene by FISH
ECFCs and CFU-ECs from 7 patients with CML were studied by interphase fluorescence in situ hybridization (FISH) with the LSI BCR-ABL dual-color, dual-fusion DNA probe from Vysis.
JAK2-V617F mutation detection by PCR
Array-CGH
Array-CGH was performed with the Human Genome CGH Microarray kit (Agilent Technologies)19 on 7 individual ECFC-derived colonies from 5 PMF patients. Every array-CGH experiment (ECFCs vs polymorphonuclear leukocytes [PMNs]) was performed 3 times to avoid possible artifacts. ECFC and PMN cells were hybridized against a normal control (Human Genomic DNA; Promega) to confirm the presence/absence of imbalances.
Results and discussion
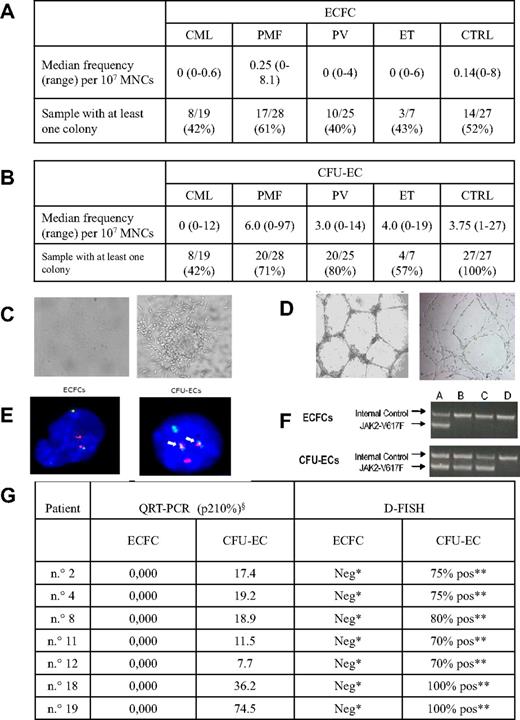
The frequency of ECFCs and CFU-ECs was highly variable both in CMPD patients and healthy subjects (Figure 1A-B). In both groups, ECFCs had a comparable endothelial phenotype (not shown) and were able to form capillary-like structures in vitro. CFU-ECs from patients and controls had also similar phenotypes, but they differed from ECFCs for the positive expression of CD45 and CD14 (not shown) and for the formation of an unshaped capillary network in vitro (Figure 1C-D). Quantitative RT-PCR on pooled CFU-ECs from CML patients invariably showed the presence of the specific p210 transcript, which, on the contrary, was never observed in 51 ECFC-derived colonies. In addition, ECFCs from 7 CML patients resulted negative for the presence of the t(9;22)(q34;q11) at interphase FISH, whereas all the CFU-EC-derived cells showed the fusion signal. The JAK2-V617F mutation was present in the genomic DNA from granulocytes of all Ph− CMPD samples. All ECFCs (n = 75) showed the JAK2 wild-type allele, whereas all CFU-ECs showed the JAK2-V617F allele (Figure 1E-G). In addition, 6 ECFCs, obtained from spleen samples of 3 PMF patients, showed the absence of the JAK-2V617F mutation. Consistent with these results, array-CGH showed the absence of chromosomal abnormalities in all ECFC-derived colonies from 5 PMF patients, although the PMN cells of 2 patients were characterized by chromosomal imbalances. One patient presented 3 deletions in his granulocytes: del(4)(21.21-q25) of 29 Mb, del(7)(pter-p13) of 47 Mb, and del(7)(q21.11-qter) of 75.3 Mb. The second patient was characterized by chromosome 9 trisomy. All anomalies were found with a medium log2 ratio representative of a cellular mosaic between an anomalous cancer clone and normal cells of approximately 90% and 50%, respectively. Thus, FISH and quantitative RT-PCR results obtained in ECFCs from CML patients as well as PCR and array-CGH results in ECFCs from Ph− CMPD patients clearly document that circulating EPCs are not clonally related to the cell giving rise to the hematopoietic malignancy. On the other hand, the presence of the disease marker in all CFU-ECs confirms that they belong to the malignant clone, as expected because they derive from myeloid cells.13,14 That the clone giving rise to CMPD does not encompass all lineages is known for a long time; indeed, often T cells in CML do not belong to the clone bearing the BCR-ABL transcript.20 Moreover, in keeping with our findings, Della Porta et al21 reported that myelodysplastic syndrome patients carrying chromosomal abnormalities in their hematopoietic cells had normal karyotype in their ECFCs both by FISH and by genotyping of polymorphic loci. Previously, Oppliger Leibundgut et al reported the presence of either trisomy 8 or JAK2-V617F mutation in the endothelial cells of CMPD patients with the same abnormalities in their granulocytes,3 whereas Gunsilius et al reported that endothelial-like cells grown in vitro from MNCs of CML patients had the BCR-ABL translocation.1 These results, interpreted as evidence that the endothelial lineage was part of the malignant clone, were probably obtained using an in vitro assay similar to that used for the growth of CFU-ECs. Taken together, our results challenge the hypothesis of a malignant common progenitor cell for hematopoietic and endothelial lineages and indicate that the transforming event occurs in a progenitor cell devoid of angiogenetic potential.
Frequency, morphology, and molecular characterization of ECFCs and CFU-ECs. (A) ECFC yield in patients and controls. In the first row, the median frequency (and range) of ECFCs obtained from patients and controls is reported, whereas the second row represents the proportion of cases in which at least 1 ECFC-derived colony was obtained. Frequency was calculated as: total number of colonies grown multiplied 107 and then divided by the total number of MNCs plated. (B) CFU-EC yield in patients and controls. In the first row, the median frequency (and range) of CFU-ECs obtained from patients and controls is reported, whereas the second row represents the proportion of cases in which at least 1 CFU-EC–derived colony was obtained. Frequency was calculated as: total number of colonies per 5 × 106 MNCs plated. (C) ECFC and CFU-EC colonies. Representative photomicrographs of an ECFCs cultured from peripheral blood MNCs of a PMF patient (left, original magnification ×4) and of a CFU-EC cultured from peripheral blood MNCs of a CML patient (right, original magnification ×5) are shown. Similar colonies were grown from other CMPD patients and controls. (D) ECFC-derived and CFU-EC–derived cell capacity of tube formation in matrigel. ECFC-derived cells (left, original magnification ×2.5) and CFU-EC–derived cells (right, original magnification ×4) from peripheral blood MNCs of patients and controls show different capacity of tube formation in matrigel. (E) FISH from ECFCs and CFU-ECs of a CML patient. ECFCs (left) and CFU-ECs (right) derived cells from a CML patient show, respectively, the lack (2 green and 2 red spots) and the presence (2 fusion signals, arrows) of BCR-ABL fusion gene by FISH reaction. The normal cutoff value used for this probe is 1%, as established in our laboratory using normal controls. (F) JAK2-V617F mutation in ECFC-derived and CFU-EC–derived cells. A seminested PCR was performed, in which the first round amplified a 364-bp product from both mutant and wild-type JAK2 allele and the second round a 203-bp product specific for the JAK2-V617F allele. Amplification products were analyzed by electrophoresis on an ethidium bromide 2% agarose gel. First line represents internal control; second line, JAK2-V617F. (A) Positive control. (B-C) Two representative polycythemia vera patients. (D) Negative control. (G) Quantitative RT-PCR for BCR-ABL fusion gene and FISH results in ECFCs and CFU-ECs from 7 CML patients. For each case, at least 200 nuclei were scored for the presence of the t(9;22)(q34;q11). §p210 values are expressed as a percentage of the ratio of BCR-ABL/ABL. *Two red and 2 green spots, no fusion spot. **Two fusion spots, one red and one green.
Frequency, morphology, and molecular characterization of ECFCs and CFU-ECs. (A) ECFC yield in patients and controls. In the first row, the median frequency (and range) of ECFCs obtained from patients and controls is reported, whereas the second row represents the proportion of cases in which at least 1 ECFC-derived colony was obtained. Frequency was calculated as: total number of colonies grown multiplied 107 and then divided by the total number of MNCs plated. (B) CFU-EC yield in patients and controls. In the first row, the median frequency (and range) of CFU-ECs obtained from patients and controls is reported, whereas the second row represents the proportion of cases in which at least 1 CFU-EC–derived colony was obtained. Frequency was calculated as: total number of colonies per 5 × 106 MNCs plated. (C) ECFC and CFU-EC colonies. Representative photomicrographs of an ECFCs cultured from peripheral blood MNCs of a PMF patient (left, original magnification ×4) and of a CFU-EC cultured from peripheral blood MNCs of a CML patient (right, original magnification ×5) are shown. Similar colonies were grown from other CMPD patients and controls. (D) ECFC-derived and CFU-EC–derived cell capacity of tube formation in matrigel. ECFC-derived cells (left, original magnification ×2.5) and CFU-EC–derived cells (right, original magnification ×4) from peripheral blood MNCs of patients and controls show different capacity of tube formation in matrigel. (E) FISH from ECFCs and CFU-ECs of a CML patient. ECFCs (left) and CFU-ECs (right) derived cells from a CML patient show, respectively, the lack (2 green and 2 red spots) and the presence (2 fusion signals, arrows) of BCR-ABL fusion gene by FISH reaction. The normal cutoff value used for this probe is 1%, as established in our laboratory using normal controls. (F) JAK2-V617F mutation in ECFC-derived and CFU-EC–derived cells. A seminested PCR was performed, in which the first round amplified a 364-bp product from both mutant and wild-type JAK2 allele and the second round a 203-bp product specific for the JAK2-V617F allele. Amplification products were analyzed by electrophoresis on an ethidium bromide 2% agarose gel. First line represents internal control; second line, JAK2-V617F. (A) Positive control. (B-C) Two representative polycythemia vera patients. (D) Negative control. (G) Quantitative RT-PCR for BCR-ABL fusion gene and FISH results in ECFCs and CFU-ECs from 7 CML patients. For each case, at least 200 nuclei were scored for the presence of the t(9;22)(q34;q11). §p210 values are expressed as a percentage of the ratio of BCR-ABL/ABL. *Two red and 2 green spots, no fusion spot. **Two fusion spots, one red and one green.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: G.P., V.R., F.F., and G. Barosi designed and performed research, collected and analyzed data, and wrote the paper; F.B., G. Bergamaschi, L.V., S.P., D.I., and A.G. performed quantitative RT-PCR analysis; R.G. and A.B. designed research and collected and analyzed data; M.C., E.B., and B.C. grew the endothelial cells; M.S. performed cytogenetic analysis; R.C., G.V., and M.M. performed FACS analysis; A.P. performed cytochemical studies; and F.N. and O.Z. performed CGH experiments.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Francesco Frassoni, Centro Cellule Staminali e Terapia Cellulare, Ospedale San Martino, Lgo R Benzi, 10, 16132 Genova, Italy; e-mail: francesco.frassoni@hsanmartino.it.
References
Author notes
*G.P. and V.R. should be considered equal first authors.
†F.F. and G. Barosi should be considered equal last authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal