Abstract
Gaucher disease causes pathologic skeletal changes that are not fully explained. Considering the important role of mesenchymal stromal cells (MSCs) in bone structural development and maintenance, we analyzed the cellular biochemistry of MSCs from an adult patient with Gaucher disease type 1 (N370S/L444P mutations). Gaucher MSCs possessed a low glucocerebrosidase activity and consequently had a 3-fold increase in cellular glucosylceramide. Gaucher MSCs have a typical MSC marker phenotype, normal osteocytic and adipocytic differentiation, growth, exogenous lactosylceramide trafficking, cholesterol content, lysosomal morphology, and total lysosomal content, and a marked increase in COX-2, prostaglandin E2, interleukin-8, and CCL2 production compared with normal controls. Transcriptome analysis on normal MSCs treated with the glucocerebrosidase inhibitor conduritol B epoxide showed an up-regulation of an array of inflammatory mediators, including CCL2, and other differentially regulated pathways. These cells also showed a decrease in sphingosine-1-phosphate. In conclusion, Gaucher disease MSCs display an altered secretome that could contribute to skeletal disease and immune disease manifestations in a manner distinct and additive to Gaucher macrophages themselves.
Introduction
Gaucher disease is a lysosomal storage disease caused by mutations in the gene encoding acid β-glucocerebrosidase (GBA).1 This leads to significant accumulation of glucocerebroside in cells of the phagocytic lineage, mostly in macrophages also known as “Gaucher” cells. Gaucher disease type 1 is the most frequent form of the disease and causes hepatosplenomegaly, bone marrow dysfunction, and skeletal disease and can be treated to different extents by enzyme replacement therapy and by substrate reduction therapy. There are also increased cytokines such as interleukin-1β (IL-1β), IL-1 receptor antagonist (IL-1Ra), soluble IL-2 receptor (sIL-2R), IL-6, IL-8, IL-10, IL-18,2 hepatocyte growth factor (HGF), macrophage colony-stimulating factor (M-CSF), macrophage-inflammatory protein 1 (MIP-1),3 CCL18/PARC (pulmonary and activation–regulated chemokine), soluble CD14 (sCD14), transforming growth factor-β1 (TGF-β1),2 and tumor necrosis factor-α (TNF-α) levels (see de Fost et al4 for more references). Some of these increases are inconstant between studies or patients and are not always correlated with clinical severity. It should be noted that not all these molecules seem to be secreted by the Gaucher cells.5 Bone involvement in Gaucher disease includes diffuse osteoporosis, epiphyseal collapse and osteoarthrosis, marrow expansion with cortical thinning, localized osteolysis, bone infarcts, and osteosclerosis.6 Marrow invasion with storage macrophages has been proposed to cause some of the changes such as vascular compromise, infarction, and scarring. The loss of bone mass is poorly explained, and proposed hypotheses include the fact that IL-10 could inhibit osteoblastic activity, that IL-6 and M-CSF could stimulate bone resorption,2,7 and that cysteine proteinases secreted by macrophages could cause lytic bone lesions.8 An enhanced capacity for osteoclast differentiation in Gaucher disease has also been proposed.9
Concerning cellular lipids in Gaucher disease, glucosylceramide and glucosylsphingosine are increased in Gaucher disease because they are upstream of GBA. However, other lipid abnormalities have been shown in Gaucher disease or the conduritol B epoxide (CBE) chemical model of Gaucher disease. Gaucher disease fibroblasts and macrophage chemical models accumulate ceramide, mostly in the lysosomes.10,11 Other sphingolipids also accumulate, and these accumulations are not limited to the lysosomes but are seen throughout the cells. Altered composition of the lipid rafts with secondary altered endosomal lipid trafficking is the preferred hypothesis to explain some of the secondary lipid accumulations.12,13 Ceramide and sphingosine-1-phosphate (S1P) have inflammation modulation properties and have been proposed to be involved in Gaucher disease pathophysiology.14,15 Moreover, S1P accumulation has recently been implicated in the neuronal pathology of Sandhoff disease,16 and decreased lysosomal sphingosine was implicated in the early pathophysiology of Niemann-Pick type C disease,17 rendering the study of these molecules compelling in sphingolipid storage disorders.
Bone marrow mesenchymal stromal cells (MSCs) are nonhematopoietic progenitor cells that possess immunomodulatory properties.18 They are bone marrow niche progenitor cells that function as a repository for precursor cells needed for bone development, growth, maintenance, and remodelling (reviewed in Phinney19 ). Their dysfunction has been linked to osteoporosis20 and osteoarthritis.21 In an effort to study the potential involvement of MSCs in Gaucher disease skeletal and immune pathophysiology, we have isolated MSCs from an adult patient with Gaucher type 1 disease (mutations N370S/L444P) and analyzed their cellular biochemistry to uncover potentially novel insights in bone and marrow anomalies associated with this ailment. We find that Gaucher MSCs display an altered cytokine secretome that may be important in the bone and immune manifestations of Gaucher disease.
Methods
MSC isolation, culture, immunophenotype, and differentiation
The patient with type 1 Gaucher disease here studied is of French-Canadian descent and was diagnosed at 45 years of age (after another family member was diagnosed). The patient was pauci-symptomatic with slight organomegaly and moderate thrombopenia. Total glucocerebrosidase enzymatic activity (without substraction of CBE-resistant activity) in lymphocytes was 1.8 nM 4-MU/min per mg protein, thus having 8% of the activity relative to a normal control with a GBA activity of 22.7 nM 4-MU/min per mg protein. Molecular analysis of the GBA gene showed that the patient carried the N370S and L444P common mutations. Radiologic investigations uncovered Erlenmeyer flask deformities at the metaphyses of femurs, extensive remote medullary bone infarcts with sparing of femoral heads, and severe osteoporosis. The patient was initially treated with enzyme replacement therapy (ERT; Cerezyme; Genzyme Therapeutics) with good clinical results. ERT was discontinued after plateau response (resolution of fatigue and improved platelet counts to 70 000-80 000/μL range) after 2 years of regular ERT. The patient's platelet count declined during a period of 18 months after cessation of ERT to levels less than 30 000/μL associated with some spontaneous bruising with a mild companion anemia (hemoglobin level, 125-130 g/L) without associated leukopenia or hepatosplenomegaly. Therefore, after informed consent, a bone marrow biopsy was performed (which confirmed only active marrow Gaucher disease) and a contemporaneous 2-mL sample of marrow aspirate was used to generate MSCs here under study. The normal control MSC samples were established from bone marrow aspirates in patients undergoing orthopedic surgery at the Montreal Jewish General Hospital, as described previously.22 All patients gave consent in accordance with the Declaration of Helsinki for their MSCs to be used for research, and all protocols were reviewed and approved by the Ethics Committee of the Montreal Jewish General Hospital. MSCs from a 24-year-old male (PT240) are a gift from Dr D. J. Prokop (Tulane University, New Orleans, LA). Human MSC growth medium was minimal essential medium α (MEM-α), 2 mM l-glutamine (Wisent Technologies), 16.5% fetal bovine serum (FBS; Atlanta Biologicals), and 100 U/mL Pen/Strep. MSCs were expanded and passed every 7 days (plated at 50-100 cells/cm2). MSC populations were tested for expression of CD31, CD34, CD44, CD45, CD73, CD90, and CD105 and ability to differentiate into adipocytes and osteocytes, as previously described.22 Cells were visualized in phosphate-buffered saline (PBS) using a Zeiss Axiovert 25 microscope (Carl Zeiss) with a 40×/0.55 NA objective, pictures were taken using a Sony VX-DSC-W5 Mpeg movie digital camera, and contrast was adjusted using Paint.NET Version 3.35.
Flow cytometric analysis
The following antibodies were used for flow cytometry analysis of MSCs (except when indicated, antibodies were from BD Biosciences): FITC-conjugated anti-CD105 (clone 8E11; Millipore); biotin-conjugated anti-CD90 (clone 5E10) and anti-CD45 (clone HI30); phycoerythrin-conjugated anti–β2-microglobulin (clone TU99), anti-CD11b (clone ICRF44), anti-CD31 (clone WM-59), anti-CD73 (clone AD2), anti-CD80 (clone L307.4), and anti-CD115 (clone 12-3A3-1B10; eBioscience); APC-conjugated anti-CD34 (clone 581), anti-CD44 (clone G44-26), and anti-CD206 (clone 19.2); and corresponding isotypic controls. Flow cytometric analysis was performed on 10 000 events with the use of a FACSCalibur cytometer, and data were analyzed with CellQuest software (BD Biosciences).
RT-PCR assays and arrays
DNA-free total RNA was prepared by homogenizing cells with a Qiashredder column and using the RNeasy kit with DNase digestion (QIAGEN). RNA (1 μg) was reverse transcribed with the MuLV reverse transcriptase (Applied Biosystems) with the use of the random hexamers in the presence of RNase inhibitor. Quantitative real-time polymerase chain reaction (RT-PCR) assays were performed in duplicate on an ABI 7500 Fast Real-Time PCR system thermal cycler with a SYBR Green RT-PCR Mastermix (Applied Biosystems). Primers used are described in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Data were analyzed with the standard curve method, using GADPH mRNA levels as internal standards. Dissociation curves were analyzed for all reactions. The lack of significant genomic DNA contamination was ensured by PCR analysis performed with total RNA (without reverse transcription) for each primer set for all samples. For RT-Arrays, 500 ng RNA was reverse transcribed with RT2 First Strand Kit and applied to PCR array plates (both from SABiosciences). Plates were processed in an Applied Biosystems 7500 Fast Real-Time PCR System, using automated baseline and threshold cycle detection. Data were analyzed by using the web-based PCR array data analysis tool from SABiosciences.
Immunoblots, enzyme-linked immunoabsorbent assays, and cytokine arrays
For immunoblots (Westerns blots), cell pellets were resuspended in Cell Lysis Buffer (Sigma-Aldrich) supplemented with protease inhibitors (Roche). Protein concentration of whole-cell extracts was determined with a Bradford Protein assay (Bio-Rad). Equal amounts of whole-cell extract (10 μg) were run on a 4% to 20% Tris-glycine gel (Invitrogen) and transferred unto a 0.45-μm polyvinylidene fluoride membrane (Millipore). Primary antibodies were specific for human COX-2 (also known as PTGS2, rabbit polyclonal antibody from Cell Signaling Technology), human APE-1 (mouse monoclonal antibody, clone 200913; R&D Systems), and GBA (mouse monoclonal antibody, clone 2E2; Abnova). Secondary antibodies were either HRP-conjugated sheep anti–rabbit IgG or rabbit anti–mouse IgG (GE Healthcare) and were revealed with the ECL Advance solution (GE Healthcare). Immunoblot were exposed to films (Kodak BioMax MR films) for 5 seconds to 1 minute. Figures show films with exposures below saturating levels of the chemiluminescent signal. Enzyme-linked immunoabsorbent assays for human IL-8, prostaglandin E2 (PGE2), and Osteopontin were from R&D Systems; enzyme-linked immunoabsorbent assays for human CCL2 and IL-1β were from eBioscience, and both were used according to the manufacturer's instructions. Human Cytokine Antibody Array 8 (screening 172 cytokines and cytokine receptors) from Raybiotech was used according to the manufacturer's protocol, to analyze 24-hour MSC supernatants (MEM-α medium supplemented with 0.5% bovine serum albumin instead of FBS) which was concentrated 50-fold with the use of Centricon centrifugal filter devices (3000 nominal molecular weight limit) from Millipore. Medium that had not been in contact with cells was used to evaluate background signal levels. The dot intensities of the array, although they represent a qualitative assay, were compared with the use of ImageJ (http://rsb.info.nih.gov/ij/).
Transmission electron microscopy
Cells were fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4), washed with sodium cacodylate buffer, and postfixed in 1% osmium tetroxide. The fixed samples were dehydrated through a graded series of ethanol/water solutions (30%-100%) and embedded in epoxy (Epon) resin. Serial, ultrathin sections were cut with a diamond knife with the use of a Reichert-Jung Ultracut E ultramicrotome. The sections were transferred onto copper TEM grids, conventionally stained with uranyl acetate and lead citrate, and imaged with a Philips CM200 200 kV transmission electron microscope (FEI Company) at the McGill Facility for Electron Microscopy Research. The electron microscope is equipped with an EDAX Genesis Energy Dispersive Spectroscopy system, and pictures were taken with an AMT 2k × 2k CCD camera.
GBA enzyme activity
GBA activity was measured with a standard protocol,23 modified for a 96-well plate. Briefly, per well, the reactions contained 2.5 mg/mL sodium taurocholate (Sigma-Aldrich) and 0.25 mg/mL oleic acid (Sigma-Aldrich; each separately suspended in 4:1 chloroform/methanol, then dried before mixing with the reaction buffer), 2.5 mM 4-methylumbelliferyl-β-d-glucopyranoside (Sigma-Aldrich) in pH 5.8 citric acid–phosphate buffer (mix of 0.1 M citric acid and 0.2 M Na2HPO4). The reaction was run for 30 minutes at 37°C and stopped using 4 volumes of 0.17 M glycine-carbonate pH 10. Results were read on a fluorometer at an emission wavelength of 450 nm and an excitation wavelength of 360 nm, using 4-methylumbelliferone (Sigma-Aldrich) as a standard, and normalized for protein content. The CBE-resistant β-glucosidase activity (measured in the presence of 2 mM CBE) was subtracted from the total activity. Negative controls consisted of Gaucher disease fibroblasts. The following human fibroblasts, from the McGill University Cell Bank (www.cellbank.mcgill.ca), were used: WG1090 (Gaucher disease type 1 fibroblasts [mutations N370S/ V394L]), WG0810 (Gaucher disease type 2 fibroblasts [mutations L444P and 2 base pair substitutions: 1483G > C and 1497G > C]), MCH047 (normal fibroblasts), MCH070 (normal fibroblasts). The integrity of the sample was ensured by measuring hexosaminidase activity with a similar protocol but with 1.5 mM 4-Methylumbelliferyl N-acetyl-β-d-glucosaminide (Sigma-Aldrich) instead of 4-Methylumbelliferyl-β-d-glucopyranoside.
Fillipin staining, cholesterol quantification, senescence assay, and cell proliferation assay
Filipin staining was performed by fixing cells with 1% paraformaldehyde in PBS for 5 minutes then staining for 15 minutes with 0.1 mg/mL filipin (Sigma-Aldrich) previously dissolved in dimethylsulfoxide. Cells were visualized in PBS with a Leica HCX PL FL 40×/0.75 oil objective, and images were taken using a Leica DFC350FX camera and Leica QWin software. Cholesterol and cholesteryl ester quantification was performed with a resofurin-based assay from BioVision according to the manufacturer's specifications and normalized for protein content. Senescence-associated β-galactosidase staining (at pH 6.0) was performed as previously described.24 Carboxyfluorescein succinimidyl ester (CFSE) analysis was performed with the CellTrace CFSE Cell Proliferation Kit from Invitrogen. Briefly, 5 × 105 cells were exposed to 10 μM CFSE for 5 minutes, washed, and were expanded in culture for 7 days (passing the cells once), before analyzing them by flow cytometry.
BODIPY-lactosylceramide and Lyso Tracker staining and confocal microscopy
Exogenously applied BODIPY-lactosylceramide trafficking was assessed essentially as previously.25 Briefly, cells were grown on Chamber slide coverslips (Thermo Fisher Scientific) at 50% confluence. Live cells were rinsed with MEM-α medium (FBS-free), incubated for 30 minutes at 10°C with 1 μM BODIPY-lactosylceramide (BSA-complexed; Invitrogen), rinsed with MEM-α medium at 10°C, incubated at 37°C for various times (30 minutes, 120 minutes, and 24 hours; 120 minutes used for the analysis), further rinsed with MEM-α medium at 10°C, and back-exchanged thrice for 10 minutes with MEM-α containing 5% fatty acid–free BSA at 10°C. Cells were then stained for 10 minutes with 50 nM Lyso Tracker Red (Invitrogen) and analyzed in PBS on an inverted Axiovert 200 M Zeiss microscope equipped with an LSM 5-Pa confocal imaging system (Carl Zeiss Canada). Confocal images (0.3 μm slices) were acquired with a Plan-Apochromat 63×/1.4 oil DIC objective, using the argon and HeNe laser lines (488 nm and 543 nm, respectively). Cells were stained with only one of the dyes used to ensure minimal “leakage” of the signal. Images from 2 distinct layers for each cell, for 15 cells per sample, were analyzed with ImageJ, with the Intensity Correlation Analysis plug-in to extract Manders and Pearson correlation coefficients (http://www.uhnresearch.ca/facilities/wcif/index.htm). For fluorescence-activated cell sorting analysis of Lyso Tracker-stained MSCs, 50 nM Lyso Tracker Yellow–HCK was used to stain MSCs for 15 minutes before fixing them in 1% paraformaldehyde and analyzing 10 000 events with a FACSCalibur cytometer.
Sphingolipid analysis
Accumulated sphingolipids were analyzed by high-performance liquid chromatography–tandem mass spectrometry (HPLC-TMS), in the Lipidomics Analytical Unit of the Medical University of South Carolina (directed by Dr Alicja Bielawska).
CBE treatment and microarrays
293T cells (clone 17) are from ATCC. Murine MSCs were isolated from female C57BL/6 mice from The Jackson Laboratory, immunophenotyped, and assessed for normal differentiation in adipocytes and osteoclasts as described previously.26 The 293T cells, murine MSCs, and human MSCs (from PT294) were grown in the presence or the absence of 100 mM CBE (Alexis Biochemicals) for 5 days before the RNA was extracted as described in “RT-PCR assays and arrays.” Microarray hybridization was performed by Phalanx Biotech Group. RNA integrity and quality was assessed with Agilent RNA 6000 Nano Assay, NanoDrop ND-1000, and agarose gel electrophoresis. RNA was labeled with Cy5 using aminoallyl RNA labeling and hybridized in duplicates on human and mouse full genome arrays (array versions HOA 4.3 and MOA 1.1, respectively). Filtered data were log2 transformed and corrected by quantile normalization before calculating average ratios and significance values. Data analysis was performed with MADNet, and the ontology of differentially regulated genes was analyzed and compared with FatiGO and DAVID. Human orthologs of regulated murine genes (identified with the use of g:Orth) were used for comparisons between the 2 species. See http://bioinformatics.ca/links_directory/nar/ for links and references to the bioinformatics programs mentioned here.
Statistical analysis
P values were calculated by the paired Student t test. All experiments were performed 3 times, once using normal MSCs from patient 302, a 48-year-old man, the 2 other times with other normal MSCs. Figures show data from representative experiments. RT-PCR array and microarray experiments were performed in duplicates.
Results
Characterization of Gaucher disease MSCs
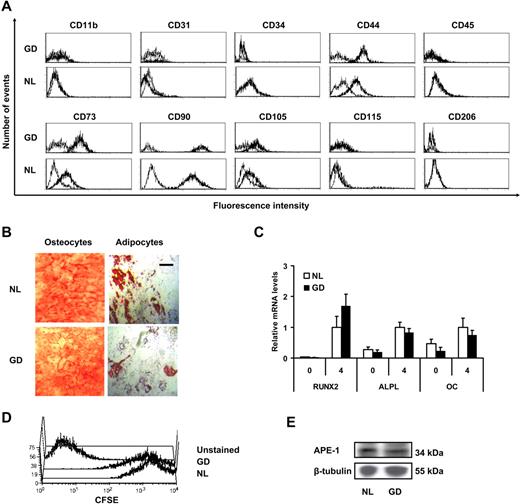
Marrow-derived MSCs from a patient with type 1 Gaucher disease were culture expanded and analyzed at passage 3 or 4, as specified. Gaucher MSCs were positive for CD44, CD73, CD90, and CD105 and negative for endothelial (CD31) and hematopoietic markers (CD11b, CD34, CD45, CD115, and CD206; Figure 1A). To test for mesenchymal plasticity, Gaucher MSCs were grown for 4 weeks in osteoblastic and adipocytic differentiation medium. After this period, the MSCs were stained with Alizarin Red to stain for mineralization and with Oil Red O to stain the lipid droplets (Figure 1B). Transcriptional analysis of selected genes involved in bone ontogeny was performed, and mRNA levels of RUNX2, bone alkaline phosphatase (ALPL), and osteocalcin (OC) were analyzed at 0 and 4 weeks by quantitative RT-PCR (Figure 1C). No significant difference was seen in the levels of these differentiation markers between Gaucher and normal human MSCs. To assess the growth and culture expansion potential of Gaucher MSCs, cells were labeled with CFSE and cultured for 1 week before analyzing them by flow cytometry, and a similar patterning of CFSE staining was observed with normal and Gaucher MSCs (Figure 1D). It has been reported that Gaucher disease fibroblasts have an abnormal antioxidant response with significantly up-regulated APE-1 expression.27 As a general screen for altered antioxidant response in Gaucher MSCs, we quantified the APE-1 protein content by Western blotting. This showed comparable APE-1 content in normal and Gaucher MSCs (Figure 1E).
MSC characterization. (A) Cells were analyzed for surface expression of MSC markers CD44, CD73, CD90, and CD105; endothelial marker CD31; hematopoietic markers CD34 and CD45; and phagocytic lineage markers CD11b, CD115, and CD206 by flow cytometry analysis (leftmost curves correspond to isotypic controls). (B) Photograph of differentiated MSCs grown in osteocytic differentiation medium and adipocytic differentiation medium for 4 weeks, stained, respectively, with Alizarin Red (left) and Oil Red O (right); scale bar = 50 μm. (C) Relative mRNA expression of osteoblastic differentiation markers runt-related transcription factor 2 (RUNX2), bone-specific alkaline phosphatase (ALPL), and osteocalcin (OC). Expression levels in normal MSCs at 4 weeks were used as a reference value. Error bars represent 1 SD. (D) Flow cytometry analysis of passage 3 MSCs stained with CFSE and allowed to proliferate for 7 days before flow cytometry analysis. (E) Scanned autoradiogram of a Western blot performed for APE-1 expression analysis in passage 3 MSCs, with β-tubulin expression used as a loading control.
MSC characterization. (A) Cells were analyzed for surface expression of MSC markers CD44, CD73, CD90, and CD105; endothelial marker CD31; hematopoietic markers CD34 and CD45; and phagocytic lineage markers CD11b, CD115, and CD206 by flow cytometry analysis (leftmost curves correspond to isotypic controls). (B) Photograph of differentiated MSCs grown in osteocytic differentiation medium and adipocytic differentiation medium for 4 weeks, stained, respectively, with Alizarin Red (left) and Oil Red O (right); scale bar = 50 μm. (C) Relative mRNA expression of osteoblastic differentiation markers runt-related transcription factor 2 (RUNX2), bone-specific alkaline phosphatase (ALPL), and osteocalcin (OC). Expression levels in normal MSCs at 4 weeks were used as a reference value. Error bars represent 1 SD. (D) Flow cytometry analysis of passage 3 MSCs stained with CFSE and allowed to proliferate for 7 days before flow cytometry analysis. (E) Scanned autoradiogram of a Western blot performed for APE-1 expression analysis in passage 3 MSCs, with β-tubulin expression used as a loading control.
Gaucher MSC lysosome morphology and glucocerebrosidase activity
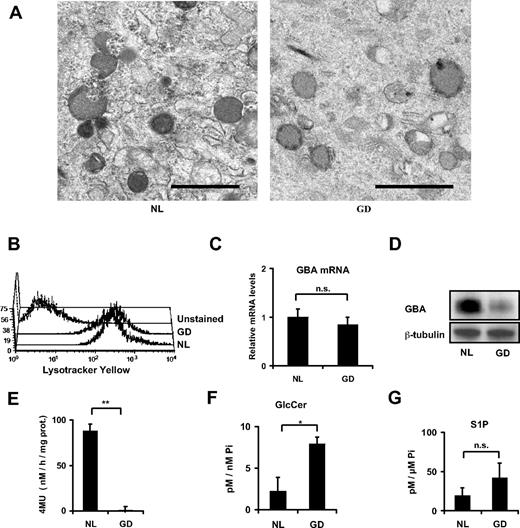
Gaucher MSC lysosomal size and morphology were normal by transmission electron microscopy (Figure 2A). The total lysosomal content of normal and Gaucher MSCs was similar as assessed by fluorescence-activated cell sorting analysis of Lyso Tracker Yellow–stained MSCs (Figure 2B). To assess GBA in these cells, we verified GBA mRNA levels that were comparable to that seen in normal MSCs (Figure 2C). However, the GBA protein content was significantly lower in the Gaucher MSCs (Figure 2D), compatible with the previously reported observation of a decreased stability of the protein with the L444P mutation.28,29 In addition, the specific GBA enzymatic activity was reduced to less than 1% of normal MSC activity (Figure 2E). The difference in activity from the peripheral leucocytes can be accounted for, partly by a difference in cell type residual activity and partly by a difference in the methodology, described in “Methods.” It is predicted that Gaucher disease MSCs would accumulate sphingolipids upstream of GBA in the sphingolipid degradation pathway. Indeed, we observed a net increase of glucosylceramide (Figure 2F). Because S1P is an important mediator of inflammation, we assessed its cellular levels and found there was no significant difference from controls (Figure 2G). In addition, no significant difference was observed in the sphingosine and the total ceramide molecular species (molar ratios [± SD] Gaucher MSCs vs normal MSCs, respectively, of 1.16 ± 0.51 and 2.82 ± 2.53).
Lysosomal and GBA studies. (A) Transmission electron microscope photography of passage 5 MSCs, the lysosomes being the dark gray organelles; scale bar = 1 μm. (B) Total lysosomal content assessed by flow cytometry of Lyso Tracker Yellow–stained passage 3 MSCs. (C) Relative mRNA levels for GBA in passage 4 MSCs. (D) Scanned autoradiogram of a Western blot performed for GBA expression analysis on passage 4 MSCs, with β-tubulin expression used as a loading control on the same samples. The large band for GBA is explained by differently glycosylated forms of GBA. (E) GBA enzymatic activity expressed in nanomoles of 4-MU formed per hour per milligrams of total protein content in the sample. **P < .001. (F-G) Sphingolipids were analyzed in normal and Gaucher disease MSCs by high-performance liquid chromatography–tandem mass spectrometry and were normalized for total cellular inorganic phosphate. Shown are glucosylceramide levels (F) and sphingosine-1-phosphate (G). *P < .05. All error bars represent 1 SD.
Lysosomal and GBA studies. (A) Transmission electron microscope photography of passage 5 MSCs, the lysosomes being the dark gray organelles; scale bar = 1 μm. (B) Total lysosomal content assessed by flow cytometry of Lyso Tracker Yellow–stained passage 3 MSCs. (C) Relative mRNA levels for GBA in passage 4 MSCs. (D) Scanned autoradiogram of a Western blot performed for GBA expression analysis on passage 4 MSCs, with β-tubulin expression used as a loading control on the same samples. The large band for GBA is explained by differently glycosylated forms of GBA. (E) GBA enzymatic activity expressed in nanomoles of 4-MU formed per hour per milligrams of total protein content in the sample. **P < .001. (F-G) Sphingolipids were analyzed in normal and Gaucher disease MSCs by high-performance liquid chromatography–tandem mass spectrometry and were normalized for total cellular inorganic phosphate. Shown are glucosylceramide levels (F) and sphingosine-1-phosphate (G). *P < .05. All error bars represent 1 SD.
MSC cholesterol content and exogenous BODIPY-lactosylceramide trafficking
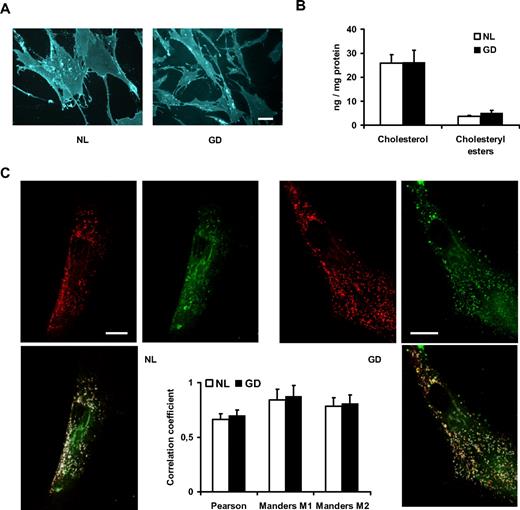
Chemical models of Gaucher disease macrophages have been reported to accumulate cholesterol.30 To evaluate MSC cholesterol content, we stained cells with filipin and observed grossly similar cholesterol content in Gaucher disease and normal cells (Figure 3A). To have a more precise assessment, we quantified cholesterol and cholesteryl ester content in MSCs with a resofurin-based assay and also observed similar cholesterol content (Figure 3B). BODIPY-lactosylceramide trafficking has been reported to be normal in Gaucher type 1 fibroblasts but abnormal in a chemically induced macrophage model30 (more diverted to the lysosomes), with a technique pioneered by Dr R. E. Pagano (Chen et al31 ). We have used this technique by applying BODIPY-lactosylceramide to the MSCs, allowing trafficking of lipids for 2 hours, and assessed colocalization with the lysosomal marker Lyso Tracker Red by confocal microscopy. This showed that Gaucher MSCs have a similar pattern of trafficking of exogenous BODIPY-lactosylceramide (Figure 3C), partly diverting the lipids to the lysosomes and partly to the Golgi apparatus, akin to what has been published for Gaucher disease fibroblasts. We have also observed a similar staining pattern in Gaucher disease fibroblasts (not shown).
Cholesterol content and lactosylceramide trafficking. (A) MSCs stained with filipin and observed under a fluorescence microscope; scale bar = 20 μm. (B) Total cholesterol and cholesteryl ester content expressed as nanograms per milligrams of total protein content in the samples. (C) Lysosomes (stained with Lyso Tracker Red) and BODIPY-lactosylceramide (green) after 2 hours of trafficking, with corresponding signal overlap in white, and correlation coefficients in the graph (Manders M1 corresponds to the correlation of the green signal on the red signal, Manders M2 is red on green); scale bar = 20 μm. All error bars represent 1 SD.
Cholesterol content and lactosylceramide trafficking. (A) MSCs stained with filipin and observed under a fluorescence microscope; scale bar = 20 μm. (B) Total cholesterol and cholesteryl ester content expressed as nanograms per milligrams of total protein content in the samples. (C) Lysosomes (stained with Lyso Tracker Red) and BODIPY-lactosylceramide (green) after 2 hours of trafficking, with corresponding signal overlap in white, and correlation coefficients in the graph (Manders M1 corresponds to the correlation of the green signal on the red signal, Manders M2 is red on green); scale bar = 20 μm. All error bars represent 1 SD.
Cytokine expression profile in Gaucher MSCs
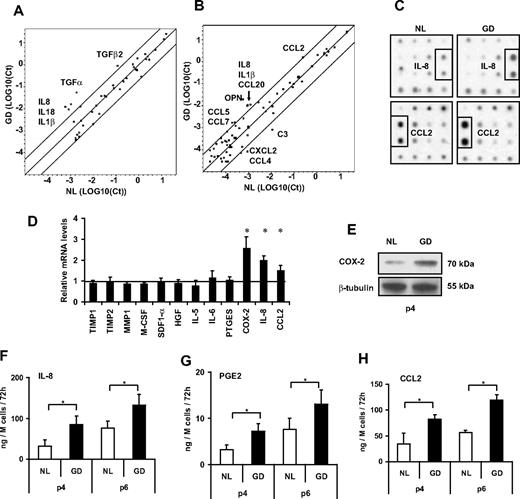
To study the physiologic consequences of GBA deficiency on MSC cytokine expression profile, we used RT-PCR array technology with dissociation curve analysis for each amplified product. When comparing normal and GD MSCs, Gaucher disease MSCs expressed higher levels of IL8 and CCL2 among other cytokine genes (Figure 4A-B). Increased IL8 and CCL2 expression was also confirmed by a cytokine antibody array analysis that assessed 172 proteins (Figure 4C). We confirmed the higher mRNA expression of IL8 and CCL2 by individual RT-PCR with the use of the standard curve method to analyze gene expression, and we have also assessed by this way other genes that could be of importance regarding the interaction of the MSCs with their surrounding cells, showing an increased expression of COX2 as well (Figure 4D). COX-2 protein expression of Gaucher disease MSCs relative to normal MSCs was assessed by Western blot analysis at passage 4 (Figure 4E). PGE2 (synthesized by COX-2), IL-8, and CCL2 are significantly oversecreted compared with passage-matched normal MSCs, and the effect is sustained over 6 passages in vitro (Figure 4F-H). For increased validity, we performed an assessment of senescence with the use of senescence-associated β-galactosidase staining at pH 6.0, which confirmed that there were no more senescent cells in the Gaucher disease MSC population. We have performed this assay because we have noticed that the expression of IL-8, COX-2, and PGE2 increases significantly with increased senescence (IL-8, PGE2, and CCL2 increase by 33-fold, 20-fold, and 3-fold, respectively, from passage 3 to 8; data not shown).
Inflammatory signal screening. (A) RT-PCR array results of genes encoding common cytokines. The log10 of the cycle threshold (Ct) were compared in a scatter plot, where circles in the top left represent genes with higher expression in Gaucher disease MSCs and circles in the bottom right represent genes with higher expression in normal MSCs. Cytokines not detected in either sample are not shown. (B) RT-PCR array results of genes encoding inflammatory cytokines. (C) Autoradiogram of relevant portions of “Human Cytokine Antibody Array 8” membranes, which screen 172 cytokines and cytokine receptors. (D) Relative mRNA levels of various genes important for MSC interactions with surrounding cells, in Gaucher disease MSCs compared with normal MSCs. (E) Scanned autoradiogram of a Western blot performed for COX-2 expression analysis on passage 4 MSCs, with β-tubulin expression used as a loading control on the same samples. (F-H) Secretion levels of IL-8, PGE2, and CCL2 by MSCs of different passages. *P < .05. All error bars represent 1 SD.
Inflammatory signal screening. (A) RT-PCR array results of genes encoding common cytokines. The log10 of the cycle threshold (Ct) were compared in a scatter plot, where circles in the top left represent genes with higher expression in Gaucher disease MSCs and circles in the bottom right represent genes with higher expression in normal MSCs. Cytokines not detected in either sample are not shown. (B) RT-PCR array results of genes encoding inflammatory cytokines. (C) Autoradiogram of relevant portions of “Human Cytokine Antibody Array 8” membranes, which screen 172 cytokines and cytokine receptors. (D) Relative mRNA levels of various genes important for MSC interactions with surrounding cells, in Gaucher disease MSCs compared with normal MSCs. (E) Scanned autoradiogram of a Western blot performed for COX-2 expression analysis on passage 4 MSCs, with β-tubulin expression used as a loading control on the same samples. (F-H) Secretion levels of IL-8, PGE2, and CCL2 by MSCs of different passages. *P < .05. All error bars represent 1 SD.
Corroboration of findings in CBE-treated normal MSCs
To corroborate the cell biochemical observations made with Gaucher disease MSCs in a normal genotype MSC background, we used a chemically induced Gaucher disease cellular model with CBE, an inhibitor of GBA. In addition, because we wanted to extend our observations beyond the realm of human MSCs, we also studied murine MSCs from a C57BL/6 mouse and 293T cells, a human embryonic kidney cell line selected for the purpose of reporter gene experiments (for their ease of transfection, see supplemental Figure 1). We assessed sphingolipids in the cells treated for 5 days with 100 μM CBE. As shown in Figure 5A to D, the cells accumulated up to twice the amount of glucosylceramide in this brief period. Interestingly, the amount of S1P was reduced by up to 50%. No significant difference was observed in the total amount of sphingosine, dehydrosphingosine, dehydrosphingosine-1-phosphate, sphingomyelin molecular species, ceramide molecular species, α-hydroxy-ceramide molecular species, and dehydroceramide molecular species (except for dehydro-C16-ceramide which was significantly increased 1.3- to 2-fold in CBE-treated cells). In addition, variable increases were observed in glucosylsphingosine, from below the detection limit (approximately 20 nM/nM cellular inorganic phosphate [Pi]), up to 246 nM/nM cellular inorganic phosphate in human MSCs.
Reproduction of findings in chemical model. (A) Glucosylceramide levels in untreated and CBE-treated normal hMSCs. (B) S1P levels in untreated and CBE-treated normal hMSCs. (C) Glucosylceramide levels in CBE-treated human and murine MSCs and 293T cells, expressed as molar ratios compared with untreated cells. (D) S1P levels in CBE-treated human and murine MSCs and 293T cells, expressed as molar ratios compared with untreated cells. (E) Scatter plot comparing log2 intensities of detected genes in arrays of untreated and CBE-treated hMSCs, to show the quality of the assay. (F) Relative mRNA levels of selected genes in CBE-treated normal MSCs compared with untreated normal MSCs. *P < .05. All error bars represent 1 SD.
Reproduction of findings in chemical model. (A) Glucosylceramide levels in untreated and CBE-treated normal hMSCs. (B) S1P levels in untreated and CBE-treated normal hMSCs. (C) Glucosylceramide levels in CBE-treated human and murine MSCs and 293T cells, expressed as molar ratios compared with untreated cells. (D) S1P levels in CBE-treated human and murine MSCs and 293T cells, expressed as molar ratios compared with untreated cells. (E) Scatter plot comparing log2 intensities of detected genes in arrays of untreated and CBE-treated hMSCs, to show the quality of the assay. (F) Relative mRNA levels of selected genes in CBE-treated normal MSCs compared with untreated normal MSCs. *P < .05. All error bars represent 1 SD.
Microarray experiments were performed with 5 μg Cy5-labeled RNA, and a sample scatter plot of log2-transformed, normalized signal intensity is shown in Figure 5E (for human MSCs) to show the quality of the samples. Our complete dataset is available on Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; series GSE13675). An analysis of gene ontology enrichment in significantly and consistently coregulated genes showed an up-regulation of genes involved in proteolysis (such as cathepsin Z; CTSZ), the inflammatory response (such as CCL2), lipid homeostasis, the actin cytoskeleton, and other genes encoding protein localized in the lysosome (such as ACP5, encoding tartrate-resistant acid phosphatase 5). Down-regulated genes were involved in the ubiquitin cycle, phosphoinositide biosynthetic processes, and transcriptional regulation (Table 1; supplemental Table for details). IL8 was increased in hMSCs (positive log2 ratio at 0.55; P = .025) but was not detected in 293T cells, and there is no murine ortholog of IL-8 to assess. These experiments thus corroborated our observations that pertained to increased cytokine expression in Gaucher disease MSCs. However, no increase in COX2 expression was observed. We validated selected gene expression results by real-time PCR in CBE-treated normal human MSCs, which corroborated the microarray results (Figure 5F). Genes selected for RT-PCR, in addition to CCL2, MMP2, and CTSZ, included genes involved in lipid transport (PLSCR3 and SCP2) and the actin cytoskeleton (GSN).
Differentially regulated groups of genes in CBE-treated cells
| Gene ontology group . | Trend . | Up-regulated (total 133)* . | Down-regulated (total 212)* . |
|---|---|---|---|
| Proteolysis (GO:0006508) other than ubiquitin cycle | ▴ | CTSZ, MMP2, SHFM1, CLPP, HTRA1, C1orf57 | PSMA1, BLMH |
| Ubiquitin cycle (GO:0006512) | ▾ | USP4, TSC1, FBXL5, NEDD8 | FBXL15, ANAPC7, FBXO42, BRAP, RNF34, FBXW7, USP1, APPBP1, CDC23, SKP2, USP16, SENP6, SPOP |
| Inflammatory response (GO:0006954) | ▴ | CCL2, IL8RA, TGM2, ANXA1, FN1 | ABCF1 |
| Lipid homeostasis (GO:0055088) | ▴ | NPC2, PLSCR3, SCP2 | |
| Phosphoinositide biosynthetic process (GO :0046489) | ▾ | PIGS, IMPA1, PIGT | |
| Lysosome compartment (GO:0005764) | ▴ | ACP5, NPC2, AGA, CTSZ, USP4, GABARAP, TMEM55A | LAMP2 |
| Actin cytoskeleton (GO:0015629) | ▴ | GSN, CLIC4, ACTR3, TSC1, GABARAP, MRCL3, VASP, MYL2, ACTR2, DYNLL2 | TNNC2 |
| Transcription factor activity (GO:0003700) | ▾ | FOSL2, DDIT3, ELF2, HNRPAB | HSF1, HOXB9, KLF9, RFX5, FBXW7, TP53, SUPT6H, KLF4, RB1, TCFL5, JUND, CREB3L1, ZNF449, ZFP36L1 |
| Gene ontology group . | Trend . | Up-regulated (total 133)* . | Down-regulated (total 212)* . |
|---|---|---|---|
| Proteolysis (GO:0006508) other than ubiquitin cycle | ▴ | CTSZ, MMP2, SHFM1, CLPP, HTRA1, C1orf57 | PSMA1, BLMH |
| Ubiquitin cycle (GO:0006512) | ▾ | USP4, TSC1, FBXL5, NEDD8 | FBXL15, ANAPC7, FBXO42, BRAP, RNF34, FBXW7, USP1, APPBP1, CDC23, SKP2, USP16, SENP6, SPOP |
| Inflammatory response (GO:0006954) | ▴ | CCL2, IL8RA, TGM2, ANXA1, FN1 | ABCF1 |
| Lipid homeostasis (GO:0055088) | ▴ | NPC2, PLSCR3, SCP2 | |
| Phosphoinositide biosynthetic process (GO :0046489) | ▾ | PIGS, IMPA1, PIGT | |
| Lysosome compartment (GO:0005764) | ▴ | ACP5, NPC2, AGA, CTSZ, USP4, GABARAP, TMEM55A | LAMP2 |
| Actin cytoskeleton (GO:0015629) | ▴ | GSN, CLIC4, ACTR3, TSC1, GABARAP, MRCL3, VASP, MYL2, ACTR2, DYNLL2 | TNNC2 |
| Transcription factor activity (GO:0003700) | ▾ | FOSL2, DDIT3, ELF2, HNRPAB | HSF1, HOXB9, KLF9, RFX5, FBXW7, TP53, SUPT6H, KLF4, RB1, TCFL5, JUND, CREB3L1, ZNF449, ZFP36L1 |
This table lists relevant groups of genes that are significantly and consistently differentially regulated.
Genes regulated significantly (log2 ratio > 0.25 or < −0.25, and P < .05) and consistently (at least 2 out of 3 cell types).
Discussion
To study the potential implication of bone marrow MSCs in Gaucher disease skeletal and immune pathophysiology, we have characterized MSCs from a patient with Gaucher disease type 1. These MSCs had a low-glucocerebrosidase activity and thus a significant increase in cellular glucosylceramide content. Nevertheless, in many respects, they are quite similar to normal MSCs with which they share an identical cell-surface marker profile, mesenchymal plasticity potential, growth kinetics in vitro, exogenous lactosylceramide trafficking, cholesterol content, lysosomal morphology, and total lysosomal content.
An important hypothesis-generating finding is that, relative to normal MSCs, Gaucher disease MSCs produce significantly more PGE2 (from COX-2), IL-8, and CCL2 as they are cultured in vitro. We have previously shown that CCL2 is up-regulated in the brain of a neuronopathic Gaucher disease mouse model.32 We currently do not have enough data to determine whether a specific sphingolipid alteration is responsible for the increase in CCL2. CCL2 has been shown to stimulate the formation of osteoclasts (although it is not sufficient for the osteoclasts to be fully active).33,34 Moreover, CCL2, also known as monocyte chemotactic protein-1, has long been known to recruit monocytes/macrophages.35 Thus, in Gaucher disease, an increased secretion of CCL2 by MSCs could contribute to the recruitment of monocytes and their transformation in osteoclasts at sites of bone disease. CCL2 could also be responsible for a general increase in osteoclast numbers and activity, thus promoting generalized osteopenia/osteoporosis. The role of MSCs in the homing of multiple myeloma is extensively documented.36 Patients with Gaucher disease have an increased incidence of gammopathies.4 Considering the fact that CCL2 acts as a chemoattractant for normal plasma cells37 and is important in the homing of multiple myeloma cells,38,39 it is tempting to propose that the increased CCL2 secretion by Gaucher disease MSCs might lead to a recruitment of normal plasmocytes and multiple myeloma cells to a pathologic bone marrow niche. IL-8 (which can be induced by COX-240 ) and PGE2 both stimulate osteoclastogenesis,41,42 and IL-8 has been shown to be elevated in the serum of patients with Gaucher disease. IL-10 has been shown to be increased in Gaucher disease,4 is a key cytokine involved in multiple myeloma,43 and has recently been shown that MSC-secreted PGE2 stimulates macrophages to produce IL-1044 , again providing a potential link between Gaucher MSCs and gammopathies. The lack of mouse models for Gaucher disease–associated gammopathies and skeletal disease limits the in vivo verification of a number of our hypotheses.
To validate our findings in a chemically induced model of Gaucher disease (neutral to genotype), we treated normal mesenchymal stromal cells with CBE.45 Accumulation of glucosylceramide and decreased S1P levels were noted. The decreased S1P levels in CBE-treated cells are puzzling because we had observed a trend for increased S1P levels in Gaucher disease MSCs (not significant, however). S1P has recently been implicated in the neuronal pathology of Sandhoff disease.16 In that condition, however, there is an increase in brain S1P which seems to lead to astrogliosis. Because S1P is known to modulate numerous signaling pathways via 5 known S1P receptors,46 a decreased S1P, if it is observed in brain of the severely affected CBE-induced neuronopathic Gaucher mouse model,47 could still be involved in the neuronal pathophysiology. Whether and how the decreased S1P relates to the gene expression profiles and the pathway activations we have observed is currently only hypothetical because we do not have data to show causative links between these. Our microarray experiments on CBE-treated MSCs show an increased expression of other genes involved in bone resorption, such as TRAP (ACP5)48 and proteases such as MMP249 . Moreover, TRAP50 and another cathepsin (cathepsin K8 ) are known to be overexpressed by Gaucher macrophages. Other up-regulated genes (such as cytoskeletal components) and down-regulated genes (such as those involved in ubiquitination) suggest interesting research avenues in the study of Gaucher disease pathophysiology. Indeed, given an increased expression of proteases in Gaucher disease,51 down-regulation of ubiquitin cycle genes could be hypothesized to be destined at equilibrating the proteolysis balance in cells. Up-regulation of cytoskeletal genes could be involved in endosomal trafficking and consist in a response to the altered endosomal lipid trafficking seen in Gaucher disease.12,13 Genes involved in lipid transport were also up-regulated, possibly in response to the accumulated glucocerebrosides; PLSCR3, encoding phospholipid scramblase 3, is involved in the trans-bilayer migration of membrane phospholipids,52 and SCP2, encoding sterol carrier protein 2, is a nonspecific lipid transfer protein with a broad ligand spectrum (including sphingolipids) thought to be involved in regulating lipid signaling pathways in lipid raft/caveolae.53 Another possibility is that these changes are not specific to GBA inhibition but could be caused by CBE directly affecting unrelated proteins and pathways. Further study of differentially regulated genes in other Gaucher models (such as shRNA-treated cells, cells from patients or from transgenic mice) and cell types (such as osteoblasts, osteoclasts, macrophages, or cell cocultures) will be necessary to further expand the pathophysiologic understanding of Gaucher disease.
In conclusion, MSCs derived from a patient with Gaucher disease display an altered cytokine and prostaglandin expression profile whose sum effect may explain in part the bone phenotype and immune anomalies associated with Gaucher disease. Pharmacologic targeting of these proteins may be of use in the long-term management and control of bone complications associated with Gaucher disease.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jeannie Mui for the electron microscopy sample processing, and training in using the transmission electron microscope. We also thank Judith Lacoste for training in using the confocal microscope.
This work was supported by the Canadian Gene Cure Foundation. G.A.G. continues to receive research support from the National Institutes of Health for basic research in Gaucher disease and other glycosphingolipidoses (DK36729 and NS36681) and epilepsy (NS45911), and a grant from the State of Ohio to support clinical services and a computational medicine center.
Authorship
Contribution: P.M.C. designed and performed the research, analyzed data, and wrote the paper; M.R. designed and performed the research and wrote the paper; M.-N.B. performed the research and wrote the paper; Y.S. and G.A.G. designed the research and wrote the paper; and J.G. provided clinical management, designed the research, and wrote the paper.
Conflict-of-interest disclosure: G.A.G. received travel expenses, honoraria, and speaker's fees in the past 3 years from Genzyme (consultant, basic research grant), Amicus (basic research grant), TKT/Shire (member of advisory board for Hunter disease, basic research grant), National Gaucher Foundation (member of the Medical Advisory Board), and Project Hope/Genzyme Gaucher Initiative (member of the expert committee, Chairman, Expert Committee, 1999-2008). The remaining authors declare no competing financial interests.
Correspondence: Jacques Galipeau, Division of Hematology/Oncology, Jewish General Hospital, 3755 Cote Ste-Catherine Rd, Montreal, QC, Canada, H3T 1E2; e-mail: jacques.galipeau@mcgill.ca.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal