Abstract
The transformation from monoclonal gammopathy of undetermined significance (MGUS) to multiple myeloma (MM) is thought to be associated with changes in immune processes. We have therefore used serologic analysis of recombinant cDNA expression library to screen the sera of MGUS patients to identify tumor-associated antigens. A total of 10 antigens were identified, with specific antibody responses in MGUS. Responses appeared to be directed against intracellular proteins involved in cellular functions, such as apoptosis (SON, IFT57/HIPPI), DNA and RNA binding (ZNF292, GPATCH4), signal transduction regulators (AKAP11), transcriptional corepressor (IRF2BP2), developmental proteins (OFD1), and proteins of the ubiquitin-proteasome pathway (PSMC1). Importantly, the gene responsible for the oral-facial-digital type I syndrome (OFD1) had response in 6 of 29 (20.6%) MGUS patients but 0 of 11 newly diagnosed MM patients. Interestingly, 3 of 11 (27.2%) MM patients after autologous stem cell transplantations showed responses to OFD1. We have confirmed T-cell responses against OFD1 in MGUS and observed down-regulation of GLI1/PTCH1 and p-β-catenin after OFD1 knock-down with specific siRNA, suggesting its functional role in the regulation of Hh and Wnt pathways. These findings demonstrate OFD1 as an important immune target and highlight its possible role in signal transduction and tumorigenesis in MGUS and MM.
Introduction
Multiple myeloma (MM) is a plasma cell (PC) malignancy that remains incurable for the majority of patients by conventional antitumor therapy, with a median survival of 7 years.1 Importantly, allogeneic bone marrow (BM) transplantation2,3 and donor lymphocyte infusion4 have been shown to be effective therapies in MM by inducing an alloimmune graft-versus-myeloma response, strongly suggesting that the immune system may control progression of MM supporting further research for new immunotherapeutic strategies. These strategies aim to induce tumor-specific immunity by immunizing patients with tumor cells or their antigenic components, known as tumor-associated antigens (TAAs), which may be mutated or selectively expressed or overexpressed in malignant, but not normal, cells. In the case of MM, however, no ideal TAA currently exists. The idiotype protein has been the main target for immunotherapy as it is the only MM-restricted antigen, but idiotype vaccinations have not induced significant clinical response.5
Retrospective studies have reported that the majority of MM patients have preexisting monoclonal gammopathy of undetermined significance (MGUS).6 Identifying those patients with MGUS who will go on to develop MM, however, remains a challenge. Thus, predicting the evolution from the preneoplastic condition MGUS to MM by the identification of genes involved in the initiation and/or transformation of this disease is of great prognostic importance and may also lead to development of successful targeted therapies and vaccination strategies. Genetic alterations,7 increased angiogenesis,8 as well as PCs and BM microenvironment interactions9,10 are implicated in progression from MGUS to MM, although their specific roles are still unknown. The expression of surface molecules of PCs and soluble factors associated with increased antigen presentation,10 the ability to mount vigorous immune responses against autologous premalignant cells11 or against specific antigens such as SOX212 and MICA13 in MGUS, is in marked contrast to cellular immune defects and other immunologic dysfunctions in MM.10,14 Taken together, the evidence suggests that the immune system in patients with MGUS and in the initial stages of MM plays an important role in controlling tumor proliferation and that disease correlates with attenuation of this response. In this context, the identification of antigens recognized in patients with MGUS may allow for the development of innovative immunotherapeutic approaches to delay or prevent transformation to MM.
Our objective here was to identify immunologically recognized antigens in MGUS and MM for translational application. With this aim, we applied a serologic analysis of recombinant cDNA expression library (SEREX) approach to identify tumor antigens able to induce a B-cell response in MM patients.15 Many antigens that are relevant to the etiology, diagnosis, and therapy of the cancer have been identified by SEREX; the tumor suppressor gene p53 and the oncogene HER-2/neu are classic examples.16-18 This approach additionally provides the opportunity to identify the immune responses to intracellular cancer antigens. Using SEREX, we identified a total of 10 antigens (OFD1, ZNF292, AKAP11, GPATCH4, SON, FAM50A, SSSCA1, IFT57/HIPPI, IRF2BP2, and PSMC1) whose sequences encode proteins associated with apoptosis, DNA and RNA binding, cilium-related signaling, and the ubiquitin-proteasome pathways. Based on immune responses in MGUS and in MM patients in remission after transplantation, we focused our attention on the gene responsible for oral-facial digital type 1 syndrome (OFD1), a developmental antigen involved in primary cilium formation and functional regulation of the Hedgehog (Hh)19 and Wingless int (Wnt) genes20 in mice models, and recently involved in promoting the switch from canonical to noncanonical Wnt signaling in zebrafish models.21 Primary cilium is a sensory organelle with a key role in development and intracellular signaling transduction; therefore, defects in ciliary proteins are associated with a series of pathologies, including developmental disorders and cancers.22-28 Here we demonstrate, for the first time, in a human system and in a tumor model, that the aberrant expression of OFD1 may mediate, at least in part, the constitutive activation or inhibition of Hh and Wnt pathways, respectively, in MM. The involvement of OFD1 in the functional regulation of Hh- and Wnt-related genes highlights that ciliary proteins may have a role in signal transduction and tumorigenesis in MM. Finally, the presence of OFD1-specific T-cell activity in MGUS compared with MM suggests it as an important immune target. Overall, these identified TAAs both improve our understanding of the transformation from MGUS to MM and provide the framework for novel targeted immunotherapies in MM to improve patient outcome.
Methods
Patient serum samples
Serum samples from peripheral blood (PB) and/or bone marrow (BM) aspirates were obtained during routine diagnostic procedures from MGUS and MM patients. Normal sera were collected from 25 age-matched healthy donors. Approval for these studies was obtained from the Review Board of the Dana-Farber Cancer Institute, Harvard Medical School. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki protocol. All serum samples were processed in exactly the same way and were stored at −80°C until use. Patients with MGUS (n = 3) individually used for screening the MM cDNA library were in a stable state of disease from 1 to 4 years with paraprotein immunoglobulin G (IgG) and κ light chain. The additional 26 MGUS patients had stable disease with IgG and non-IgG paraprotein and κ light chain. Of 34 MM patients analyzed, 12 were newly diagnosed and untreated, 11 had refractory MM, and 11 patients were in partial or complete remission 3 to 6 months after transplantation.
Primary cells and MM cell lines
Primary tumor cells from BM aspirates of MM patients were purified by Ficoll-Hypaque separation (GE Healthcare), followed by CD138+ microbead selection29 (Miltenyi Biotec) and used for immunoblotting experiments. The purity of MM cells was more than 95%. BM mononuclear cells (MNCs) obtained by selection from healthy donors were used to establish long-term bone marrow stromal cells (BMSCs)29 and also for immunoblotting. Peripheral blood mononuclear cells (PBMCs) from healthy donors were isolated by Ficoll-Hypaque and used for immunoblotting; PBMCs isolated from HLA-A2+ subjects have been also used for studying T-cell responses directed to OFD1. MM cell lines U266, NCIH929, RPMI8226, MM1R, HEK293, and COS-7 were obtained from American Type Culture Collection; KMS12BM and KMS12PE MM cells were obtained from German Collection of Microorganisms and Cell Cultures (DSMZ); MM1S cells were a gift of Steven Rosen (Northwestern University); OPM1 and OPM2 cells were provided by Dr P. Leif Bergsagel (Mayo Clinic–Tucson); S6B45 was provided by Dr T. Kishimoto (Osaka University); KMS11 was provided by the Kawasaki Medical School; and INA-6 were obtained from Dr Renate Burger (University of Kiel). All cells were cultured at 37°C in RPMI 1640 containing 10% heat-inactivated fetal bovine serum (Sigma-Aldrich), 2 mM/L l-glutamine, 100 units/mL penicillin, and 100 mg/mL streptomycin (Invitrogen).
Myeloma cDNA library screening by SEREX methodology
The cDNA MM library was constructed, as previously described,30 from total RNA isolated from BM CD138+ MM cells from a patient with IgG paraprotein. For antigen identification, serum samples from 3 MGUS patients were individually used to screen this cDNA MM library by SEREX for the presence of IgG antibody. Serum was diluted 1:10, preabsorbed with transfected Escherichia coli lysate, and further diluted to 1:500. E coli bacteria transfected with recombinant λZAPII phages were plated on NZY agar plates and cultured at 42°C for 4 hours. After emergence of visible plaques, expression of recombinant protein was induced by incubation with isopropyl β-D-thiogalactoside (IPTG)-treated nitrocellulose membranes for 3.5 hours at 37°C. To exclude IgG clones in the primary screening, an identical second set of IPTG-treated nitrocellulose membranes was incubated for another 3.5 hours at 37°C. After washing and blocking with 2% nonfat dry milk in Tris-buffered saline (TBS), one set of membranes was incubated overnight with MGUS patient sera. The next day, the filters were washed and then incubated with alkaline-phosphatase-conjugated goat antihuman IgG antibody (Jackson ImmunoResearch Laboratories) diluted 1:2000 in TBS/Tween20 (TBST). The second set of membranes was only incubated with the secondary antibody. Antigen-antibody complexes were visualized by staining with 5-bromo-4-chloro-3-indolylphosphatase and nitroblue tetrazolium (Promega). Individual clones identified in both sets of membranes were presumed to express human IgG and were not further processed. All other clones were plated for secondary and tertiary screening until single plaques were isolated.
Sequence analysis of the reactive clones and phage plaque assay
The clones reactive with the sera but not with secondary antibody alone were subcloned, purified, and the phagemid cDNA insert excised from the λZAP vector with an ExAssist helper phage system (Stratagene). Plasmid DNA was prepared using the Quiaprep Spin Miniprep kit (QIAGEN). Clones were sequenced by the Molecular Biology Core facility at the Dana-Farber Cancer Institute using an ABI Prism 373 DNA sequencer (Applied Biosystems). Sequences were further analyzed for homology with known genes and proteins and searched for mutations or single nucleotide polymorphisms using public databases (http://www.ncbi.nlm.nih.gov). To investigate the antigenicity of detected clones in MGUS and MM patients compared with healthy donors, a phage plaque assay was performed. Phages from positive clones were mixed with nonreactive phages of the cDNA library as internal negative controls at 1:10 ratio; 200 μL of XL1-blue E coli bacteria was transfected with this mix and plated in NZY agar plates for 4 hours at 42°C. Recombinant protein expression was induced by incubation with IPTG-treated nitrocellulose membranes for an additional 3.5 hours at 37°C. After washing in TBST and blocking overnight with 2% nonfat dry milk in TBS, filters were incubated overnight with sera from healthy donors, MGUS, and MM patients (1:500), followed by incubation with alkaline phosphatase-conjugated goat antihuman IgG antibody diluted 1:2000 in TBST. The reactive phages were visualized by staining with 5-bromo-4-chloro-3-indolylphosphatase and nitroblue tetrazolium, and the intensity of positive staining was visually graded on a scale of 1 to 4 and photographed using a Nikon Coolpix 5000 camera.
Evaluation of OFD1 expression by microarray analysis and Western blotting
A gene expression analysis was performed using data deposited in the National Institutes of Health Gene Expression Omnibus (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE5900, which included data from 22 healthy donors, 44 MGUS, and 12 smoldering myeloma (SMM) patients.31 Genes were identified as significant if their probeset level expression value differed from the comparison groups with a minimum fold change of 1.2 and a minimum signal value difference of 100, with P < .05. Myeloma cell lines (MM1S, MM1R, OPM1, OPM2, RPMI8226, U266, INA6, NCIH929, KMS12BM, KMS12PE, KMS11, S6B45), primary tumor cells from MM patients, and PBMCs and BMSCs from healthy donors were harvested, lysed, and used for immunoblotting experiments. Cell lysis and Western blot analysis were done as previously described.29 For detection of OFD1 protein, we used the rabbit polyclonal anti-OFD1 antiserum directed against the C-terminus (residues 996-1011; Neosystem). As a positive control for OFD1 protein expression, we used HEK29332 and COS7 cell lines.33
Effect on Hh- and Wnt-related genes after OFD1 knockdown by specific siRNA
For OFD1-specific knockdown experiments, MM1S cells were transiently transfected with small-interfering RNA (siRNA) SMART pool for OFD1 (100-200 nM) and nonspecific control duplexes (pool of 4; Dharmacon RNA Technologies) using the Cell Line Nucleofector Kit V according to the manufacturer's instructions (Amaxa Biosystems). We then evaluated the effects of the OFD1 knocked down on the Hh- and Wnt-regulated genes, GLI1, PTCH1, and [beta]-catenin, respectively. For immunoblotting, we used the following antibodies: Gli1 from Santa Cruz Biotechnology, anti–p-β-catenin S33-37 from Cell Signaling Technology, and PTCH1 from Abcam.
Screening for peptide-specific HLA-restricted CD8+ T cells in MGUS and MM patients
HLA-A2–specific peptides were identified within the sequence of the immunogenic clones of OFD1, using 4 independent standard software programs (BIMAS, RANKPEP, NetMHC, and MULTIPRED). Based on the scores, 15 specific peptides were chosen and synthesized for evaluation. All selected peptides were synthesized (Proimmune) using standard fluorenylmethoxycarbonyl method (purity > 90%) and validated with mass spectrometry. On evaluation of binding ability of these peptides to HLA-A2 molecules, one peptide (peptide-3) was selected to make pentamers (Proimmune). After isolation of PBMCs from healthy persons as well as MGUS and MM patients, cells were incubated with pentamers for 10 to 15 minutes at the room temperature and stained with CD8 antibody (BD Biosciences). Pentamer-positive cells within the CD8 population were analyzed by flow-cytometric evaluation.
T-cell proliferation is induced by OFD1-derived specific peptides
PBMCs of HLA-A2+ subjects were isolated by Ficoll-Hypaque density gradient centrifugation. We analyzed MGUS (n = 9), MM (n = 5), and healthy donors (n = 4). Of 15 peptides synthesized, one peptide with a high binding affinity was selected (peptide-3). Cells (4 × 105) were incubated with or without peptide at 2 μg/mL as a final concentration for 7 days, and the proliferation was determined, adding [3H] thymidine (NEN Life Science Products) to the last 8 hours of culture. Candida antigen and, in some experiments, pooled viral peptides were used as a positive control to confirm that the cells were able to respond to common antigens. Cells alone were used as a negative control.
Results
Identification of immunoreactive antigens by SEREX in MGUS
To identify antigens recognized by the immune system and potentially involved in the progression from MGUS to MM, we individually used serum from 3 MGUS patients to screen, by SEREX, a cDNA expression library constructed from CD138+ MM cells from a patient with IgG-secreting MM.30 As shown in Table 1, all 3 MGUS patients at the time of analysis had stable disease from 1 to 4 years with IgG paraprotein. Library screening with these sera using SEREX methodology identified 18 clones. After restriction enzyme digestion and DNA sequencing, these clones were found to correspond to 10 different gene products, which are reported in Table 1 together with MGUS patient serum that specifically recognized each one of them. As summarized in Table 2, all clones have high homologies with previously described genes; 2 of these genes have unknown molecular functions. All genes encoded for intracellular proteins involved in functions, including ciliary proteins (OFD1, IFT57/HIPPI), DNA and RNA binding (ZNF292, GPATCH4), transcriptional corepressors (IRF2BP2), proteins of the ubiquitin-proteasome pathways (PSMC1), signal transduction regulators (AKAP11), and apoptosis (SON, IFT57/HIPPI).
Clinical characteristics of MGUS patients used to screen MM cDNA library and immunogenic antigens individually recognized
| Patient no. . | Age, y/sex . | Isotype . | Time since diagnosis, y . | Immunogenic antigens . |
|---|---|---|---|---|
| 1 | 68/M | IgG κ | 1 | SON |
| AKAP11 | ||||
| SSSCA1 | ||||
| FAM50A | ||||
| 2 | 81/M | IgG κ | 1.5 | PSMC1 |
| IRF2BP2 | ||||
| GPATCH4 | ||||
| 3 | 75/F | IgG κ | 4 | OFD1 |
| ZNF292 | ||||
| HIPPI |
| Patient no. . | Age, y/sex . | Isotype . | Time since diagnosis, y . | Immunogenic antigens . |
|---|---|---|---|---|
| 1 | 68/M | IgG κ | 1 | SON |
| AKAP11 | ||||
| SSSCA1 | ||||
| FAM50A | ||||
| 2 | 81/M | IgG κ | 1.5 | PSMC1 |
| IRF2BP2 | ||||
| GPATCH4 | ||||
| 3 | 75/F | IgG κ | 4 | OFD1 |
| ZNF292 | ||||
| HIPPI |
Gene products identified by SEREX in MGUS patients
| Gene product . | Gene ID . | Gene chromosomal locus . | Cellular localization . | Molecular function . | Cellular process . |
|---|---|---|---|---|---|
| OFD1 | 8481 | Xp22.2-p22.3 | Basal body/centrosome, cytosol | Unknown | Ciliogenesis |
| ZNF292 | 23036 | 6q15 | Nucleus | DNA binding | Transcription regulation DNA-dependent |
| AKAP11 | 11215 | 13q14.11 | Centrosome, cytosol | PKA binding, PP1-binding | Protein-kinase cascade |
| GPATCH4 | 54865 | 1q22 | Intracellular | Nucleic acid binding | Unknown |
| SON | 6651 | 21q22.1-22.2; 21q22.11 | Intracellular, nucleus | Nucleic acid binding, protein binding | Antiapoptosis |
| FAM50A | 9130 | Xq28 | Nucleus | Unknown | Spermatogenesis |
| SSSCA1 | 10534 | 11q13.1 | Unknown | Protein binding | Cell cycle, cell division, mitosis |
| IFT57/HIPPI | 55081 | 3q13.12 | Golgi, cilium IFT particles | DNA binding, protein binding | Ciliogenesis, proapoptosis, transcription |
| IRF2BP2 | 359948 | 1q42.3 | Nucleus | Corepressor of IRF-2BP2 | Transcription regulation DNA-dependent |
| PSMC1 | 5700 | 14q32.11 | Cytosol, nucleus, proteasome complex | ATP binding ATPase activity, nucleotide-protein binding | Regulation of ubiquitin-protein ligase activity |
| Gene product . | Gene ID . | Gene chromosomal locus . | Cellular localization . | Molecular function . | Cellular process . |
|---|---|---|---|---|---|
| OFD1 | 8481 | Xp22.2-p22.3 | Basal body/centrosome, cytosol | Unknown | Ciliogenesis |
| ZNF292 | 23036 | 6q15 | Nucleus | DNA binding | Transcription regulation DNA-dependent |
| AKAP11 | 11215 | 13q14.11 | Centrosome, cytosol | PKA binding, PP1-binding | Protein-kinase cascade |
| GPATCH4 | 54865 | 1q22 | Intracellular | Nucleic acid binding | Unknown |
| SON | 6651 | 21q22.1-22.2; 21q22.11 | Intracellular, nucleus | Nucleic acid binding, protein binding | Antiapoptosis |
| FAM50A | 9130 | Xq28 | Nucleus | Unknown | Spermatogenesis |
| SSSCA1 | 10534 | 11q13.1 | Unknown | Protein binding | Cell cycle, cell division, mitosis |
| IFT57/HIPPI | 55081 | 3q13.12 | Golgi, cilium IFT particles | DNA binding, protein binding | Ciliogenesis, proapoptosis, transcription |
| IRF2BP2 | 359948 | 1q42.3 | Nucleus | Corepressor of IRF-2BP2 | Transcription regulation DNA-dependent |
| PSMC1 | 5700 | 14q32.11 | Cytosol, nucleus, proteasome complex | ATP binding ATPase activity, nucleotide-protein binding | Regulation of ubiquitin-protein ligase activity |
Evaluating the prevalence of antibody responses against the identified antigens by testing healthy donors, as well as MGUS and MM patient sera
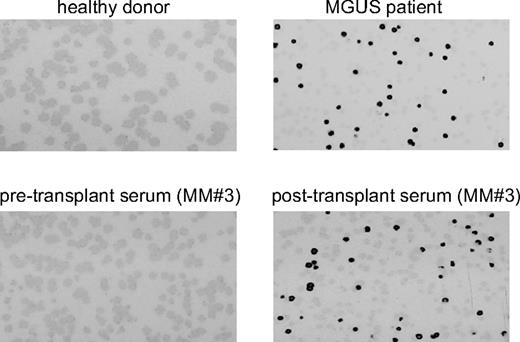
To verify the importance of these antigens in MGUS and MM, we have further analyzed, by phage plaque assay, the frequency of antibody response against these antigens in sera from additional 26 MGUS and 34 MM (11 newly diagnosed, 12 refractory, and 11 MM in remission after autotransplantation from 3 to 6 months) patients as well as 25 healthy donors. As shown in Table 3, antibody responses were observed in more than one MGUS patient against OFD1 (20.6%), ZNF292 (10.3%), AKAP11 (10.3%), and GPATCH4 (6.8%). To note, these 4 genes were detected by sera from additional samples besides the original sera used to identify these antigens, whereas the 6 other genes are identified only by the original sera used for screening the MM cDNA library by SEREX. Except for OFD1, no immune responses were observed against any of the identified antigens in MM patients. Importantly, 1 of 12 (8.3%) refractory MM patients and 3 of 11 (27.2%) MM patients in remission after transplantation had an antibody response to OFD1, with evidence of a significant increase in the antibody titer (from 1- to 4-fold) in one patient (Figure 1). No significant antibody responses were observed against any of these antigens in healthy donor sera. No difference was observed between peripheral blood and BM-derived serum samples.
Antibody response against the identified antigens in healthy persons, MGUS patients, and MM patients
| Gene product . | Healthy donors (n = 25) . | MGUS (n = 29) . | MM newly diagnosed (n = 11) . | MM post auto-SCT (n = 11) . | MM refractory (n = 12) . |
|---|---|---|---|---|---|
| OFD1 | 1/25 | 6/29 (20.6%) | 0/11 | 3/11 (27.2%) | 1/12 (8.3%) |
| ZNF292 | 1/25 | 3/29 (10.3%) | 0/11 | 0/10 | 0/12 |
| AKAP11 | 0/25 | 3/29 (10.3%) | 0/11 | 0/10 | 0/12 |
| GPATCH4 | 0/25 | 2/29 (6.8%) | 0/11 | 0/10 | 0/12 |
| SON | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| FAM50A | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| SSSCA1 | 1/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| IFT57/HIPPI | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| IRF2BP2 | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| PSMC1 | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| Gene product . | Healthy donors (n = 25) . | MGUS (n = 29) . | MM newly diagnosed (n = 11) . | MM post auto-SCT (n = 11) . | MM refractory (n = 12) . |
|---|---|---|---|---|---|
| OFD1 | 1/25 | 6/29 (20.6%) | 0/11 | 3/11 (27.2%) | 1/12 (8.3%) |
| ZNF292 | 1/25 | 3/29 (10.3%) | 0/11 | 0/10 | 0/12 |
| AKAP11 | 0/25 | 3/29 (10.3%) | 0/11 | 0/10 | 0/12 |
| GPATCH4 | 0/25 | 2/29 (6.8%) | 0/11 | 0/10 | 0/12 |
| SON | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| FAM50A | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| SSSCA1 | 1/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| IFT57/HIPPI | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| IRF2BP2 | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
| PSMC1 | 0/25 | 1/29 | 0/11 | 0/10 | 0/12 |
Phage plaque assay using multiple sera was performed. Screening results of multiple sera from healthy donors, MGUS, and MM patients.
OFD1 IgG-antibody response in MGUS and posttransplantation MM patients detected by phage plaque assay. Detection of antibody reactivity to the OFD1 protein determined by phage-plaque assay in healthy donors, as well as MGUS and posttransplantation MM patients.
OFD1 IgG-antibody response in MGUS and posttransplantation MM patients detected by phage plaque assay. Detection of antibody reactivity to the OFD1 protein determined by phage-plaque assay in healthy donors, as well as MGUS and posttransplantation MM patients.
Expression of the developmental antigen OFD1 in MM cell lines and primary MM cells
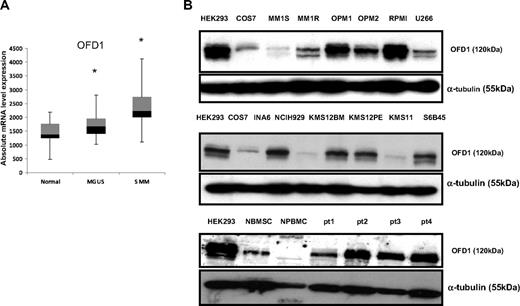
Because OFD1 had the highest frequency of immune response in MGUS and also had detectable immune response in MM, we have further focused our investigation on this gene. OFD1 is known to be a basal body/centrosome-related protein32 widely expressed during embryonic development, with an important role in organogenesis and fetal survival.19 Its expression decreases in adults and is restricted to some normal tissues and tumors. We evaluated the expression of OFD1 in CD138+ PCs from 22 healthy donors (N), as well as 44 MGUS and 12 SMM patients using a public database (National Institutes of Health Gene Expression Omnibus, accession no. GSE5900). As shown in Figure 2A, the range of OFD1 expression values was broader in PCs from MGUS and SMM patients compared with PCs from healthy persons (P < .05). We next analyzed the level of expression of OFD1 protein by Western blotting in 12 MM cell lines and in 4 primary MM cells. As seen in Figure 2B, OFD1 is overexpressed in the majority of the MM cell lines and in all 4 primary MM cells, compared with BMSCs and PBMCs from healthy persons.
OFD1 expression in MM cell lines and primary patient cells. (A) Quantitative mRNA expression level of OFD1 in plasma cells from healthy persons (n = 22), as well as MGUS (n = 44) and MM (n = 12) patients. Box plots define the median values, 25% to 75% of values around the median, and the range of values. *Statistically significant difference among the groups (P < .05). (B) Western blot analysis of OFD1 in MM cell lines and primary patient cells. OFD1 protein expression was analyzed in 12 MM cell lines (MM1s, MM1r, OPM1, OPM2, RPMI8226, U266, INA6, NCIH929, KMS12BM, KMS12PE, KMS11, S6B45), primary tumor cells from 4 MM patients, as well as BMSCs and PBMCs from healthy donors. Levels of OFD1 protein expression are compared with HEK293 and COS7 as positive controls for OFD1 expression.
OFD1 expression in MM cell lines and primary patient cells. (A) Quantitative mRNA expression level of OFD1 in plasma cells from healthy persons (n = 22), as well as MGUS (n = 44) and MM (n = 12) patients. Box plots define the median values, 25% to 75% of values around the median, and the range of values. *Statistically significant difference among the groups (P < .05). (B) Western blot analysis of OFD1 in MM cell lines and primary patient cells. OFD1 protein expression was analyzed in 12 MM cell lines (MM1s, MM1r, OPM1, OPM2, RPMI8226, U266, INA6, NCIH929, KMS12BM, KMS12PE, KMS11, S6B45), primary tumor cells from 4 MM patients, as well as BMSCs and PBMCs from healthy donors. Levels of OFD1 protein expression are compared with HEK293 and COS7 as positive controls for OFD1 expression.
OFD1 plays a role in the functional regulation of Hh and Wnt pathways in MM
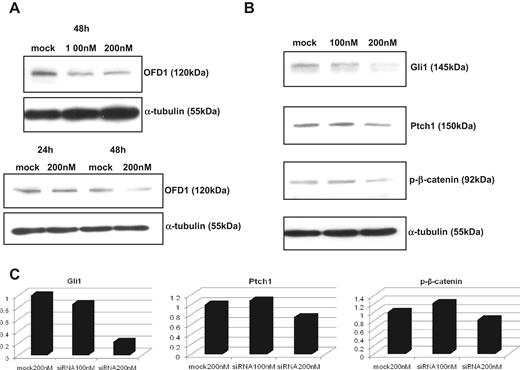
To understand the functional role of OFD1 in MM, we knocked it down in the MM1S cell line using a specific siRNA, as described in “Effect on Hh- and Wnt-related genes after OFD1 knockdown by specific siRNA” and shown in Figure 3A). Because Ofd1 is a developmental gene involved in functional regulation of Hh19 and Wnt pathway in mice,20 we next evaluated the effects of OFD1 inhibition on the expression of the Glioma transcription factor1 (GLI1)/Patched1 (PTCH1) and β-catenin genes, in MM. As seen in Figure 3B-C), Western blot analysis showed that specific knock-down of OFD1 led to a dose-dependent down-regulation of GLI1 and PTCH1, as well as p-β-catenin at 48 hours. GLI1 and PTCH1 are direct target genes of Hh pathway; therefore, their down-regulation suggests inhibition of Hh signaling. Unlike the results seen in mice, the inhibitory effect in MM cells is greater on GLI1 than PTCH1. β-Catenin is a key component of the Wnt pathway, cytoplasmatic levels of β-catenin are regulated by phosphorylation at serine residues, and p-β-catenin rapidly undergoes proteasomal degradation. Therefore, the observed down-regulation of p-β-catenin after OFD1 specific knock-down in MM cells suggests an increased β-catenin–dependent signaling. These findings suggests possible role of OFD1 in the functional regulation of Hh and Wnt pathways in MM.
OFD1-specific knock-down with siRNA in MM1S cells. (A) OFD1 knock-down with specific siRNA in MM1S cells reveals a time-dependent, but dose-independent, inhibition of OFD1. (B) OFD1-specific inhibition leads to a down-regulation of Hh- and Wnt-related genes at 48 hours. (C) The levels of expression of Gli1, Ptch1, and p-β-catenin proteins, after OFD1-specific knock-down, were quantitated by densitometry and normalized with tubulin.
OFD1-specific knock-down with siRNA in MM1S cells. (A) OFD1 knock-down with specific siRNA in MM1S cells reveals a time-dependent, but dose-independent, inhibition of OFD1. (B) OFD1-specific inhibition leads to a down-regulation of Hh- and Wnt-related genes at 48 hours. (C) The levels of expression of Gli1, Ptch1, and p-β-catenin proteins, after OFD1-specific knock-down, were quantitated by densitometry and normalized with tubulin.
CD8+ T cells against OFD1-derived peptides are detectable in both MGUS and MM patients
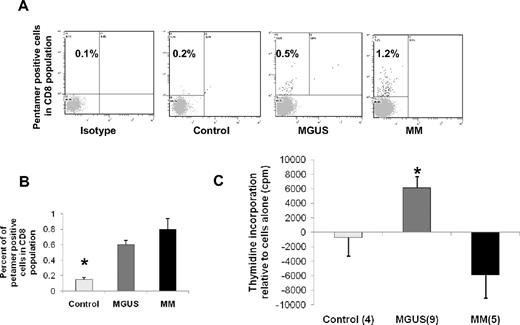
We further evaluated whether OFD1-derived peptides were able to stimulate CD8 T-cell responses in both MGUS and MM patients. To this aim, we used as an approach an HLA-A2–specific pentamer-based analysis. As shown in Figure 4A and B, we were able to detect pentamer-positive CD8 T cells against OFD1-derived peptide-3 in both MGUS and MM patients compared with isotype controls, although we did not observe the presence of pentamer-positive CD8 T cells in healthy donors. At least in part, these studies indicate that peptide-3 pentamer-positive CTLs against OFD1 were present in the T-cell population.
Increased frequency of OFD1-derived peptide-3-pentamer-positive CD8 cells and proliferation of PBMCs from MGUS patients in HLA-A2–restricted fashion. (A) When PBMCs from HLA-A2+ subjects were stained with OFD1-derived peptide-3 pentamers, the frequency of pentamer-positive cells in the CD8 population was increased in both MGUS and MM patients compared with isotype and healthy donor samples. (B) Composite results of pentamer staining in PBMCs were summarized from tested subjects (control = 4, MGUS = 6, and MM = 5). (C) PBMCs were isolated from HLA-A2+ subjects (control = 4, MGUS = 9, and MM = 5) and incubated with or without peptides (2 μg/mL) for 7 days. Proliferation was determined by thymidine incorporation. Results represent mean values with SEM compared with cells alone. *Statistical significance among the 3 groups (P < .05).
Increased frequency of OFD1-derived peptide-3-pentamer-positive CD8 cells and proliferation of PBMCs from MGUS patients in HLA-A2–restricted fashion. (A) When PBMCs from HLA-A2+ subjects were stained with OFD1-derived peptide-3 pentamers, the frequency of pentamer-positive cells in the CD8 population was increased in both MGUS and MM patients compared with isotype and healthy donor samples. (B) Composite results of pentamer staining in PBMCs were summarized from tested subjects (control = 4, MGUS = 6, and MM = 5). (C) PBMCs were isolated from HLA-A2+ subjects (control = 4, MGUS = 9, and MM = 5) and incubated with or without peptides (2 μg/mL) for 7 days. Proliferation was determined by thymidine incorporation. Results represent mean values with SEM compared with cells alone. *Statistical significance among the 3 groups (P < .05).
OFD1-derived peptide-3 is able to induce T-cell proliferation in MGUS patients compared with MM patients and healthy donors
Freshly isolated PBMCs collected from both MGUS and MM patients in addition to healthy donors were incubated in the presence or absence of OFD1-derived peptide-3 for 7 days. Proliferation was evaluated by thymidine incorporation. Even though we were able to detect pentamer-positive CD8 T cells against OFD1-derived peptide-3 in both MGUS and MM patients, we only observed T-cell proliferative responses (as seen in Figure 4C) against OFD1-derived peptide-3 in MGUS patients (n = 9) compared with MM (n = 5) and healthy donors (n = 4). We observed that PBMCs from MGUS and MM patients in addition to healthy donors were able to respond to common viral/fungal antigens used as positive controls (data not shown). These results suggest that OFD1-derived peptides are capable of inducing proliferative responses in T cells isolated from MGUS patients in vitro.
Discussion
MGUS is a premalignant PC disorder with increasing frequency in older persons and an age-adjusted prevalence of approximately 1% to 5%, and only 1% of patients per year develop symptomatic MM.34 Importantly, the majority of MGUS patients remain in stable disease for many years.6 The events leading to the transformation from MGUS to MM are still not clear, although mounting evidence supports the idea that the immune system in MGUS patients may control the progression to MM.10-13 Therefore, our objective here was to identify and characterize immunogenic antigens associated with this transformation. With this aim, we applied SEREX, an established method to identify immunogenic antigens in cancer patients. As previously described,30 we constructed an MM cDNA library using RNA isolated from CD138+ MM cells purified from the BM of a patient with an IgG-secreting MM, and screened it using sera from 3 MGUS patients with stable disease from 1 to 4 years. A panel of 10 novel antigenic targets that elicited an immune response was identified. These antibody responses were directed against intracellular proteins involved in apoptosis (SON, IFT57/HIPPI), DNA and RNA binding proteins (ZNF292, GPATCH4), signal transduction regulators (AKAP11), ciliary proteins (OFD1, IFT57/HIPPI), transcriptional corepressors (IRF2BP2), and proteins of the ubiquitin-proteasome pathways (PSMC1). Interestingly, the ubiquitin-proteasome pathway gene has already emerged as a crucial target for antimyeloma therapies.35-37 We further observed the frequent antibody response against these antigens in sera from patients with MGUS as well as MM. In patients with MGUS (Table 3), antibody response was most frequently detected against OFD1 (20.6%), followed by ZNF292 (10.3%), AKAP11 (10.3%), and GPATCH4 (6.8%). Interestingly, patients with refractory MM (1 of 12) and in remission after autotransplantation (3 of 11) had an antibody response against OFD1; evidence of a significant increase in antibody titer after transplantation suggests its importance as a potential therapeutic target (Figure 1). In contrast, no significant antibody responses were observed against any of these antigens in the sera of 25 healthy donors.
The relevance of the antigens detected here in MGUS is further supported by the fact that specific antibody responses to 6 of these antigens have been previously identified by SEREX (http://www2.licr.org/CancerImmunomeDB) in patients with other tumors or have been shown to be related in some ways with the tumorigenesis. For example, antibodies against OFD1 have been detected by SEREX in breast, ovarian, stomach, and lung cancer patient sera; and against ZNF292 in patients with breast, colorectal, testicular, and head and neck cancers.38 AKAP11 (A-kinase anchor protein), located on 13q14.11, a genetic locus whose deletion is implicated in MM disease initiation and progression,39,40 has been detected in patients with prostate cancer and is implicated in oral carcinogenesis.41 Family members of AKAP11, AKAP3, and AKAP4 have been previously identified as cancer testis antigens in ovarian cancer and MM, respectively.42,43 Immune responses detected in MGUS patients against GPATCH4 and IRFPB2, both on chromosome 1q, further highlight the importance of this genetic locus, whose amplification has been deemed a poor prognostic factor in MM.44 Indeed, GPATCH4 has been also identified by SEREX in melanoma patient sera, and its up-regulation has been found in hepatocellular carcinoma.45 Finally, antibodies against SON have been detected by SEREX in head and neck cancer,38 prostate cancer, in donor lymphocyte infusion responder MM patients,30 and against FAM50 in breast cancer patient sera. Importantly, both these genes have been involved in leukemogenesis.46,47 The immunogenicity of these antigens found in other cancers suggests that they might be appropriate targets for diagnostic and/or therapeutic approaches. The specific antibody response detected in a significant number of MGUS patients demonstrates that these patients are immune competent and able to up-regulate specific antitumor immune responses. Titers of OFD1 antibodies are absent in newly diagnosed MM patients, probably because of immune defects often associated with the disease initiation or progression. Interestingly, immune response reappears in posttransplantation MM patients in remission, an indication that transplantation can reconstitute immune competency. Importantly, our findings suggest that the presence of specific antibody titer in the preneoplastic condition of MGUS might control the progression, and, conversely, its disappearance might provide an early indication of progression to MM. Moreover, the presence of OFD1-antibody titers in a posttransplantation setting might reflect an immune response against tumor-specific antigens and a favorable outcome. The presence of OFD1-specific T cells, detected by pentamer-based analysis in MGUS and MM, but lack of specific T-cell activity in MM compared with MGUS, may further confirm an inhibitory mechanism operative in myeloma.
Among these 10 novel antigens, we focused our attention on OFD1 resulting from a specific antibody response in MGUS (20.6%) as well as posttransplantation MM patients (27.2%). OFD1 is a developmental antigen widely expressed during embryonic development, including in embryonic stem cells, with an important role in organogenesis and fetal survival.19 In adults, its expression remains significantly high in lymphoid-derived organs and cells (http://symatlas.gnf.org/deprecated/). The pattern of inheritance is X-linked dominant, being lethal in males, and the related genetic syndrome is characterized by craniofacial and digital abnormalities, as well as polycystic kidneys.48 Although the function of OFD1 is still not fully elucidated, it is a basal body/centrosome-related protein, localized at the base of primary cilia in fully differentiated cells.32,33 The cilium is a single microtubule-based structure that emanates from the surface of most cell types in the human body.49,50 It is anchored to the cell by the basal body, which develops from the mother centriole of the centrosome in a cell cycle-dependent fashion, thereby passively influencing the cell cycle. As indicated by our findings here and suggested by the work of others, signaling pathways that play a critical role in embryonic development and in tissue homeostasis, such as those of Hh and Wnt, appear to localize on the cilium. As a ciliary protein, Ofd1 is involved in the functional regulation of the Hh 19 and Wnt pathways20 in mouse models and recently has been implicated in promoting switch from canonical to noncanonical Wnt signaling in Zebrafish models.21 Mice mutant for Ofd1 fail to form cilia and show defects in left-right axis specification, reduced expression of the direct Hh target gene Ptch1, and impaired expression of Gli1 in some tissues, indicating that Hh signaling is defective in these embryos.19 Thus, loss of this centrosome/basal body protein blocks cilia formation, which disrupts Hh signaling. In addition, mouse embryonic stem cells lacking Ofd1 also show increased canonical Wnt responsiveness, indicating that primary cilium modulates canonical Wnt signal transduction.20 Based on these findings, we sought to get preliminary evidence whether OFD1 might also play a role in the Hh and Wnt pathways in MM. We observed that specific knock-down of OFD1 led to a down-regulation of Gli1, Ptch1, and p-β-catenin. This is an interesting finding that may need further evaluation in the future to understand the role of these pathways in MM and also the role of ciliary function in malignant transformation and progression.
Much evidence has shown that the immune responses are reasonably effective in MGUS patients compared with MM patients.10-13 In support of that viewpoint, we have further evaluated the ability of OFD1-derived peptides to induce, in vitro, specific immune responses in MGUS and MM patients. Using pentamer-based analysis, we demonstrated the presence of pentamer-positive CD8 T cells specific to OFD1-derived peptide-3 in both MGUS and MM patients but not in healthy donors, with a slightly higher percentage detected in MM than in MGUS patients. Peptide-3-specific proliferative in vitro assays showed PBMC proliferative responses, suggestive of functionally active T cells, only in MGUS compared with MM confirming intact immune function in MGUS patients. Studies by Spisek et al,12 showing the presence of antitumor-specific T-cell responses against SOX2 antigen in MGUS rather than in MM and healthy donors, are in agreement with our findings demonstrating OFD1-specific T-cell responses in MGUS. We have also observed that PBMCs from MGUS and MM patients, in addition to healthy donors, were able to respond to common antigens. The responses to tumor antigens, in general, are different from those observed for viral/fungal antigens and may explain some of our observations as well as results presented by others.12,51 Because of the self-nature of tumor antigens, T cells, in particular, are in a precarious situation: (1) they must maintain homeostasis and/or tolerance to tissue antigens that are present on tumor cells; and (2) they must also control the generation of autoimmunity that may be triggered by these antigens. Therefore, the lack of or inhibitory tumor-antigen responses in MM compared with MGUS may be the result of a possibility that MM has an impaired capacity to mount the immune responses and/or has more regulatory T cells that inhibit effector T cells and production of antibodies.
In conclusion, our results demonstrate the presence of a specific immune response in MGUS, suggesting the possibility of immune surveillance that may also have a role in the natural history of MGUS. On the other hand, the observed lack of significant immune response against these antigens in MM may be either reflective of the progression from MGUS to MM or possibly may represent suppressed immune function in MM. The causality of this observation remains difficult to determine, but identification of this panel of antigens may provide tools in the future to investigate this interesting question. Finally, the presence of specific T-cell proliferation directed against OFD1 in MGUS compared with MM is an indication to target this tumor-specific antigen to further assess whether cell-mediated immunity in response to OFD1 can induce regression in MGUS and tumor control in MM. We also demonstrated, for the first time, in human cells and in tumors of hematopoietic origin a role of OFD1 in Hh- and Wnt-related genes, suggesting that ciliary proteins may take part in the functional regulation of these tumorigenic pathways in MM. Altogether, these findings demonstrate OFD1 as an important immune target as well as highlight its possible role in signal transduction and tumorigenesis in MGUS and MM.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The paper is dedicated to the memory of Prof Salvatore Venuta, Chancellor of Magna Græcia University and Professor of Medical Oncology. Passionate scientist, meticulous clinician, and skilled manager, he had worked indefatigably to build up the Magna Græcia University and the T. Campanella Cancer Center of Catanzaro (Italy) for the love of knowledge, the future of young people, and the care of and dedication to patients.
The authors thank Dr P. G. Richardson and Dr R. Schlossman, patients, nursing staff, and clinical research coordinators of the Jerome Lipper Multiple Myeloma Center/Dana-Farber Cancer Institute and Veterans Administration Boston Healthcare System, for their help in providing primary tumor speciments and sera samples for this study.
This work was supported by the Specialized Program of Research Excellence in Myeloma, National Institutes of Health (NIH; grant P50CA-100707), a Career Development Award (S.B.), the Italian Telethon Foundation (B.F.), the Italian Association for Cancer Research, Milan, Italy (P. Tassone), the Department of Veterans Affairs merit review award, and Leukemia & Lymphoma Society Scholar in Translational Research Award and NIH (grant RO1-129494; N.C.M.), NIH grants P50CA-100707 and PO1-78378 (N.C.M., K.C.A.), and the Lebow Fund to Cure Myeloma (K.C.A.).
National Institutes of Health
Authorship
Contribution: S.B. designed and performed research, analyzed and interpreted the data, and wrote the manuscript; N.C.M. and K.C.A. contributed in the design and supervision of the research and performed the critical review of the manuscript; B.F., P. Tassone, and P. Tagliaferri analyzed the research, B.F. also provided the OFD1 antibody; Y.-T.T. and N.C.M. provided primary tumor specimens and sera samples; R.H.P. performed T-cell response studies; and D.C., S.A., J.J., K.P., C.S.M., and A.Z. contributed to the research. All the authors reviewed and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nikhil C. Munshi, Dana-Farber Cancer Institute, Harvard Medical School, 44 Binney St, M1B28, Boston, MA 02115; e-mail: Nikhil_Munshi@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal