Abstract
Currently, limited molecular markers exist that can determine where in the spectrum of chronic myeloid leukemia (CML) progression an individual patient falls at diagnosis. Gene expression profiles can predict disease and prognosis, but most widely used microarray analytical methods yield lengthy gene candidate lists that are difficult to apply clinically. Consequently, we applied a probabilistic method called Bayesian model averaging (BMA) to a large CML microarray dataset. BMA, a supervised method, considers multiple genes simultaneously and identifies small gene sets. BMA identified 6 genes (NOB1, DDX47, IGSF2, LTB4R, SCARB1, and SLC25A3) that discriminated chronic phase (CP) from blast crisis (BC) CML. In CML, phase labels divide disease progression into discrete states. BMA, however, produces posterior probabilities between 0 and 1 and predicts patients in “intermediate” stages. In validation studies of 88 patients, the 6-gene signature discriminated early CP from late CP, accelerated phase, and BC. This distinction between early and late CP is not possible with current classifications, which are based on known duration of disease. BMA is a powerful tool for developing diagnostic tests from microarray data. Because therapeutic outcomes are so closely tied to disease phase, these probabilities can be used to determine a risk-based treatment strategy at diagnosis.
Introduction
Chronic myeloid leukemia (CML) is characterized by a reciprocal translocation between chromosomes 9 and 22 yielding the BCR-ABL fusion protein. It is this constitutively active tyrosine kinase that drives CML pathophysiology.1 CML usually presents in chronic phase (CP), but in the absence of effective therapy, CP CML invariably transforms to accelerated phase (AP) disease, and then to an acute leukemia, blast crisis (BC). BC is highly resistant to treatment, and all treatments are more successful when administered during CP.2 The current first-line therapy for early CML is the targeted tyrosine kinase inhibitor (TKI), imatinib mesylate (IM), which inhibits BCR-ABL and consequently its downstream targets.3 IM is most effective in early CP patients with a progressive increase in drug resistance, notably in the frequency of ABL tyrosine kinase domain (TKD) point mutations (a common cause of drug resistance), in late CP, AP, and BC patients.4-6 Responses in BC to TKI therapy are most often transient.5 Although CML has historically been divided into 3 phases, disease evolution is most likely a continuous process. Currently there are limited clinical markers,7,8 and no molecular tests that can predict the “clock” of CML progression for individual patients at the time of CP diagnosis, making it difficult to adapt therapy to the risk level of each patient.
Microarray-based expression analyses have been used extensively in the “discovery phase” for cancer-related biomarkers.9-14 According to the National Cancer Institute's Early Detection Research Network, the objective of exploratory studies in the discovery phase is to determine a short list of 1 to 10 high-priority candidates.15,16 A short list is critical as target validation is time, cost, and labor intensive; and a small set is highly desirable for the development of inexpensive diagnostic tests. However, choosing these “best” markers from thousands of genes is a daunting task. The merits of using a combination of biomarkers to predict outcome are well documented in the literature.15-19 However, most microarray-based analyses rely on univariate methods, such as the t test and the significance analysis of microarray statistic,20 which consider the expression profile of each gene individually. In contrast, multivariate gene selection methods consider multiple genes simultaneously, thereby accounting for the dependency between genes. These methods can be used systematically to identify signature genes that are predictive in combinations rather than individually.
We applied a probabilistic supervised method, called Bayesian modeling averaging (BMA),21 to a large gene expression microarray dataset of patients in various phases of CML14 to identify a small set of signature genes that predict CML disease progression. BMA is a multivariate method that takes the uncertainty of signature gene selection into consideration by averaging over multiple models (ie, sets of potentially overlapping relevant genes). BMA has many desirable features: is computationally efficient; systematically determines the number of predictive genes and models; yields posterior probabilities of the predictions, selected genes, and selected models; and each selected model typically consists of only a few genes.21,22 Six signature genes (NOB1, DDX47, IGSF2, LTB4R, SCARB1, and SLC25A3) were identified and validated that discriminated early CP from late CP, CP from AP, and CP from BC.
Methods
Patient samples
All patient samples used in these investigations were obtained from institutional review board–approved protocols from the Fred Hutchinson Cancer Research Center with written informed consent. All clinical investigations have been conducted according to the principles expressed in the Declaration of Helsinki. The 93 individual cases of CML profiled in the microarray studies have been previously described and are published.14 CP was defined as less than 10% blasts. AP was defined as 10% to 30% blasts or less than 10% blasts with clonal evolution, defined as cytogenetic abnormalities in addition to the Philadelphia chromosome (AP-cyto). BC was defined as more than 30% blasts. BC remission was defined as a return to CP from BC after therapy (BC-rem). For the array studies 42 CP, 9 AP, 8 AP-cyto, 30 BC, and 4 BC-rem individual patient samples were examined.14
In further validation studies in independent patient samples, we examined 45 early CP patients and 22 late CP patients by quantitative reverse transcription polymerase chain reaction (QPCR). The 45 early CP samples included de novo diagnostic samples from untreated patients analyzed before treatment with IM at 800 mg per day on the Novartis-sponsored RIGHT study. The 22 late CP patients had failed IM and were examined before treatment with nilotinib at 800 mg on the Novartis-sponsored AMN107 study.
Microarray studies
The procedures for RNA extraction, amplification, labeling, and hybridization to the Rosetta platform are previously published23,24 and the analytic methods used for analysis are as previously described.14 Briefly, analysis of variance (ANOVA) analysis identified 2612 differentially expressed genes (from a total of ≈ 24 000 genes) that discriminated CP from BC (P < .001).14 BMA was applied to these 2612 genes. Ingenuity Pathways Analysis was used to determine biologically enriched pathways and functions. The P values for the Ingenuity analyses are calculated using a right-tailed Fisher exact test and measure the statistical significance of a particular function or pathway in our data with respect to the reference set defined by Ingenuity Systems.
Quantitative PCR analysis
After cDNA synthesis, expression in patient samples was quantitated in duplicate by QPCR and normalized to GUSB expression using the Taqman Low Density Array platform (TLDA; Applied Biosystems).23 As PCR efficiencies were similar for all genes examined, relative gene expression was compared as: Δ Ct calculations: 2−ΔCt × 100, where Δ Ct = Ct gene of interest − Ct GUSB.
Classification and gene selection using microarray data
In classification, a classifier is built using a training set with the goal of predicting the classes of an independent test set. A major challenge of classification using microarray data is that the number of genes is usually much greater than the number of patient samples, and only a subset of these genes is relevant in distinguishing the different classes. Here, we report success with BMA from which we built classifiers consisting of a small number of genes to discriminate disease phases.
Bayesian model averaging
A full discussion of BMA is available in supplemental methods, available on the Blood website; see the Supplemental Materials link at the top of the online article. When classifying samples using microarray data, a primary goal is to identify a small set of predictive genes that can be validated easily. How to identify this “best set,” however, is unclear. In the BMA framework, sets of predictive genes are called “models.” In microarray analysis, there are typically many models (or sets of predictive genes) that fit the data well. BMA takes model uncertainty into consideration by computing the weighted average of the posterior probabilities that a test sample belongs to a given class over multiple “good” models. The weights are proportional to the goodness of fit of the model.22,25 We applied the iterative BMA algorithm26 to the 2612 differentially expressed genes derived from our prior ANOVA analysis14 to determine the best model of genes and phase of disease.
To briefly summarize our approach, we first ranked the genes associated with CML phase (chronic vs blast) using a univariate measure of the ratio of between-group to within-group sum of squares (BSS/WSS).27 Genes with large BSS/WSS ratios (ie, genes with relatively large variation between CML phases and relatively small variation within a phase) receive high rankings. We then applied the “leaps and bounds” algorithm28 and the Occam window method29 (supplemental Methods) to the top 30 ranked genes. Genes that were assigned low posterior probabilities (< 5%) are removed. Suppose m genes are removed. The next m genes from the rank ordered BSS/WSS ratios are added to the set of genes so that we maintain a window of 30 genes, and then we apply the leaps and bounds algorithm again. These steps of gene swaps and iterative applications of leaps and bounds were continued until all genes were subsequently considered.
Cross validation
Three-fold cross validation, in which a training set is randomly divided into 3 equal subsets, was used to assess the prediction accuracy of the iterative BMA method on the CML progression microarray data. Each of these 3 subsets is left out in turn for evaluation of classification accuracy, whereas the other 2 subsets are used as inputs to the classification algorithm. This process was repeated 100 times.
Computational assessment
BMA was used to predict probabilities that a given sample was CP or BC based on the gene expression of the genes populating the model. For example, say class 0 represents CP and class 1 represents BC. The true class labels of the test samples are unknown to the classification algorithm. Comparing the true class labels of the test samples to the labels predicted by the algorithm yields the number of classification errors. For a test sample assigned to BC, a predicted probability close to 1 is more desirable than a predicted probability slightly above 0.50, whereas the opposite is true for the predicted probability of a test sample assigned to CP. The Brier score30 is equal to the sum of squares of the difference between the true class and the predicted probability over all samples. When the predicted probabilities are constrained to 0 and 1, the Brier score is equal to the number of classification errors, and was used to compare the performance of the deterministic 0-1 classification methods with that of probabilistic methods such as BMA.
Results
BMA identifies genes associated with CML progression from microarray gene expression data
Although it is highly likely that more than a single set of genes is predictive of disease phase, most microarray analytic methods select a single set and ignore the uncertainty in the selection of this set. BMA takes this uncertainty into account by averaging over multiple models (ie, sets of potentially overlapping relevant genes)22,25 and it yields posterior probabilities of the inclusion of each gene in the model. In our previous work, we showed that BMA identified a small number of genes while achieving high prediction accuracy when applied to microarray data.21
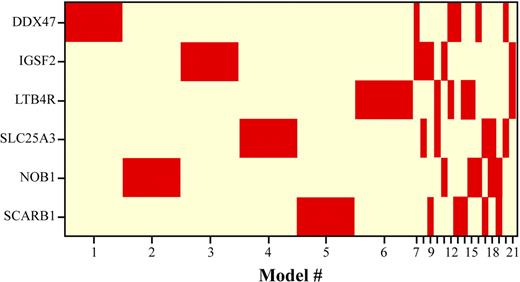
BMA was applied to a microarray dataset consisting of patients in various phases of CML14 to identify signature genes that predict CML disease progression (Figure 1). BMA identified 6 signature genes over 21 models from 2612 genes that discriminated 42 CP from 30 BC patients in our training set (Table 1). Using univariate methods, the top 3 ranked genes were DDX47 (rank = 1), IGSF2 (rank = 2), and LTB4R (rank = 3). Notably, these 3 genes were selected by the multivariate BMA method in addition to 3 others, NOB1, SCARB1, and SLC25A3, which were not as highly ranked by univariate methods (ranked 13, 69, and 77, respectively). The posterior probabilities reported for each gene (Table 1) are computed by summing the posterior probabilities of selected models in which each gene is included. Figure 2 illustrates the membership of the 6 signature genes in each of the 21 models selected by the iterative BMA algorithm. To summarize, each of the 6 signature genes is selected in a single-gene model with posterior probability of 12.87%, and in 5 different 2-gene models with posterior probabilities of 1.52%. Because the posterior probability of a gene is equal to the sum of the posterior probabilities of all the selected models containing the gene of interest in the BMA paradigm, each of the 6 genes is selected with posterior probability = 12.87% + (5 × 1.52%) = 20.47%. Under the BMA framework, we computed the predicted probability (ie, of being CP or BC) for each patient sample by averaging over the predicted probabilities from these 21 selected models, weighted by the posterior probabilities of the models.
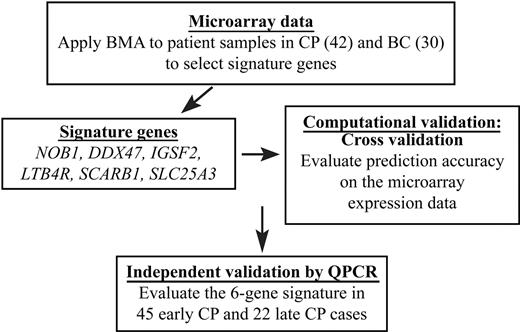
Experimental overview. A summary of our overall approach is shown. The merits of BMA include that the model is multivariate and considers multiple biomarkers simultaneously; that the uncertainty of gene selection is accounted for by averaging over multiple good models; that posterior probabilities are generated for all selected genes and models; and that it produces a high predictive accuracy with relatively few genes.
Experimental overview. A summary of our overall approach is shown. The merits of BMA include that the model is multivariate and considers multiple biomarkers simultaneously; that the uncertainty of gene selection is accounted for by averaging over multiple good models; that posterior probabilities are generated for all selected genes and models; and that it produces a high predictive accuracy with relatively few genes.
The 6-gene signature selected by BMA that discriminates chronic phase from blast crisis CML
| ID . | Gene . | Chromosome location . | Activity . | Cellular function . | Posterior probabilities, % . | Univariate (BSS/WSS) rank . |
|---|---|---|---|---|---|---|
| NM_016355 | DDX47 | 12p13.1 | RNA helicase | RNA processing, apoptosis | 20.5 | 1 |
| NM_004258 | IGSF2 | 1p13 | Signal transduction | Immune function, T-cell activation | 20.5 | 2 |
| NM_000752 | LTB4R | 14q11.2-q12 | 5-lipoxy-genase | Cell growth, apoptosis | 20.5 | 3 |
| NM_014062 | NOB1 | 16q22.1 | Unknown | Unknown | 20.5 | 13 |
| NM_005505 | SCARB1 | 12q24.31 | Cholesterol scavenger receptor | HDL and LDL metabolism | 20.5 | 69 |
| NM_005888 | SLC25A3 | 12q23 | Mitochondrial phosphate transport | ATP synthesis | 20.5 | 77 |
| ID . | Gene . | Chromosome location . | Activity . | Cellular function . | Posterior probabilities, % . | Univariate (BSS/WSS) rank . |
|---|---|---|---|---|---|---|
| NM_016355 | DDX47 | 12p13.1 | RNA helicase | RNA processing, apoptosis | 20.5 | 1 |
| NM_004258 | IGSF2 | 1p13 | Signal transduction | Immune function, T-cell activation | 20.5 | 2 |
| NM_000752 | LTB4R | 14q11.2-q12 | 5-lipoxy-genase | Cell growth, apoptosis | 20.5 | 3 |
| NM_014062 | NOB1 | 16q22.1 | Unknown | Unknown | 20.5 | 13 |
| NM_005505 | SCARB1 | 12q24.31 | Cholesterol scavenger receptor | HDL and LDL metabolism | 20.5 | 69 |
| NM_005888 | SLC25A3 | 12q23 | Mitochondrial phosphate transport | ATP synthesis | 20.5 | 77 |
The table indicates chromosomal location, function, the posterior probability of each gene in multivariate modeling, and its rank in univariate modeling (using the univariate measure of the ratio of between-group to within-group sum of squares (BSS/WSS).
BMA indicates Bayesain modeling averaging; and CML, chronic myeloid leukemia.
Model selection by BMA. This figure illustrates the membership of the 6 signature genes in the 21 models selected by the iterative BMA algorithm on the CML progression microarray data. The 6 signature genes are shown on the vertical axis, whereas the 21 models are shown in the horizontal axis. The widths of the columns are proportional to the posterior probabilities of the selected models. A red entry indicates that the corresponding gene is included in a given model.
Model selection by BMA. This figure illustrates the membership of the 6 signature genes in the 21 models selected by the iterative BMA algorithm on the CML progression microarray data. The 6 signature genes are shown on the vertical axis, whereas the 21 models are shown in the horizontal axis. The widths of the columns are proportional to the posterior probabilities of the selected models. A red entry indicates that the corresponding gene is included in a given model.
The prediction accuracy of the 6-gene signature was then validated using 3-fold cross validation. The 72 training samples were randomly divided into training and testing subsets in cross validation repeated 100 times: two-thirds of the samples were used to build the prediction model and the remaining one-third were used to evaluate the accuracy of the CP and BC prediction. The predicted probability that a patient sample was BC was computed using the BMA framework, and the number of classification errors was computed by thresholding the predicted probability at 0.5. In other words, if the predicted probability was less than 0.5, we classified a patient sample as CP; otherwise we would classify the sample as BC.
We compared the performance of different analytic methods by computing the average number of errors and average Brier scores, a relative probabilistic measure of the number of errors, over multiple cross validation runs (see “Computational assessment”). An analytic method with a relatively small number of errors and a relatively small Brier score thus achieves higher prediction accuracy. BMA produced an average number of errors of 0.20 and an average Brier score of 0.21 in the 24 test samples over 100 cross validation test sets, producing an average prediction accuracy of 99.17%. In addition, BMA selected fewer genes and achieved higher prediction accuracy than the simple method of predicting with the top 10 or 30 univariate genes (supplemental methods). In summary, our cross validation study demonstrated that BMA accurately predicts the phase of CML when applied to CML progression microarray data.
The 6-gene signature predicts advanced phase in 21 AP and BC remission samples
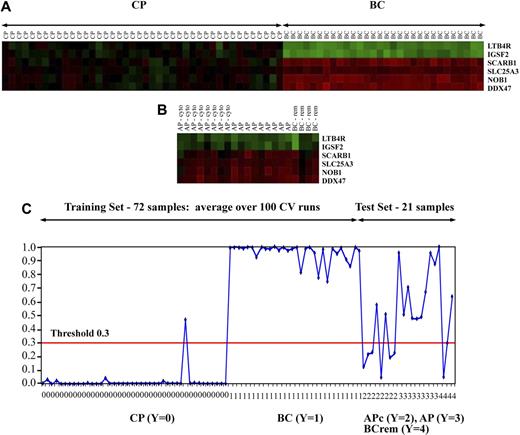
Our initial validation was performed on independent patient samples using microarrays and included 21 patients with disease between CP and BC. Figure 3A shows the expression of the 6 signature genes in the CP and BC training set. Figure 3B shows the expression of these 6 signature genes in 8 patients with AP solely by cytogenetic criteria (AP-cyto), 9 patients with AP disease by both cytogenetic and blast count criteria (AP), and 4 patients with a return to CP from BC after therapy (BC-remission or BC-rem). Notably, the expression pattern in these patients was variable, and in certain cases was more similar to that of BC patients than to CP patients. Therefore, we looked more closely at the predicted probabilities in these patients who subsequently underwent allogeneic transplantation.
Differential gene expression of the 6-gene signature in patient cases. Heatmaps of the 6-gene signature selected by the iterative BMA algorithm. (A) Heatmap of the gene expression of the 6-gene signature in the training set of CP and BC CML cases using microarray-based gene expression analyses. (B) Heatmap of the gene expression in the test set, also by microarray-based gene expression analyses. This group is made up of AP by cytogenetic criteria only (AP cyto); AP by cytogenetic and blast count criteria (AP); and BC after a return to second CP (BC-rem). Green indicates differentially decreased expression of each gene in each case compared with its expression in a pool of CP patient samples and red indicates differentially increased expression. The more saturated the color, the greater is the degree of differential expression. (C) A graphic depiction of the predicted posterior probability of all patient samples is shown. The samples are represented on the horizontal axis, whereas the predicted probabilities are represented on the vertical axis. For the training data consisting of 72 CP (group 0) and BC (group 1) cases, the average predicted probabilities over 100 cross validation runs are shown. For the test data, composed of 21 patients before allogeneic transplantation, the predicted probabilities of the 6 signature genes averaged over 21 models are shown. This “intermediate phase” test set is composed of group 2 AP by cytogenetic criteria only; group 3 AP by cytogenetic and blast count criteria; and group 4 BC after a return to second CP. For 17 of 21 patients, posttransplantation outcomes were available. Using a posterior probability threshold of 0.3, we found that 7 of 11 patients with predicted posterior probabilities ≥ 0.3 died of relapse or treatment-related mortality after transplantation versus only 1 of 6 with a predicted posterior probability < 0.3 (OR = 9).
Differential gene expression of the 6-gene signature in patient cases. Heatmaps of the 6-gene signature selected by the iterative BMA algorithm. (A) Heatmap of the gene expression of the 6-gene signature in the training set of CP and BC CML cases using microarray-based gene expression analyses. (B) Heatmap of the gene expression in the test set, also by microarray-based gene expression analyses. This group is made up of AP by cytogenetic criteria only (AP cyto); AP by cytogenetic and blast count criteria (AP); and BC after a return to second CP (BC-rem). Green indicates differentially decreased expression of each gene in each case compared with its expression in a pool of CP patient samples and red indicates differentially increased expression. The more saturated the color, the greater is the degree of differential expression. (C) A graphic depiction of the predicted posterior probability of all patient samples is shown. The samples are represented on the horizontal axis, whereas the predicted probabilities are represented on the vertical axis. For the training data consisting of 72 CP (group 0) and BC (group 1) cases, the average predicted probabilities over 100 cross validation runs are shown. For the test data, composed of 21 patients before allogeneic transplantation, the predicted probabilities of the 6 signature genes averaged over 21 models are shown. This “intermediate phase” test set is composed of group 2 AP by cytogenetic criteria only; group 3 AP by cytogenetic and blast count criteria; and group 4 BC after a return to second CP. For 17 of 21 patients, posttransplantation outcomes were available. Using a posterior probability threshold of 0.3, we found that 7 of 11 patients with predicted posterior probabilities ≥ 0.3 died of relapse or treatment-related mortality after transplantation versus only 1 of 6 with a predicted posterior probability < 0.3 (OR = 9).
As shown in Figure 3C, the predicted probabilities were more variable and intermediate between the CP and BC cases. Because BMA predicts the posterior probabilities of patient samples, the posterior probability of a patient sample can be taken as an indication of the strength of the prediction, that is, more CP-like or more BC-like. Because of the close relationship between disease phase and treatment outcomes, we then studied the relationship between these pretransplantation posterior probabilities and patient outcomes. Outcomes after transplantation were available for 17 of 21 patients. Using a posterior probability threshold of 0.3 in these patients, 7 of 11 patients with posterior probabilities 0.3 or higher ultimately died of relapse or treatment-related mortality after transplantation compared with only 1 of 6 deaths among patients with a posterior probability less than 0.3. These preliminary data suggest that AP patients with a higher predicted posterior probability using the 6-gene predictor behaved more like BC patients, who typically have the worst outcomes after transplantation.
The 6-gene predictor signature discriminates early from late CP by QPCR
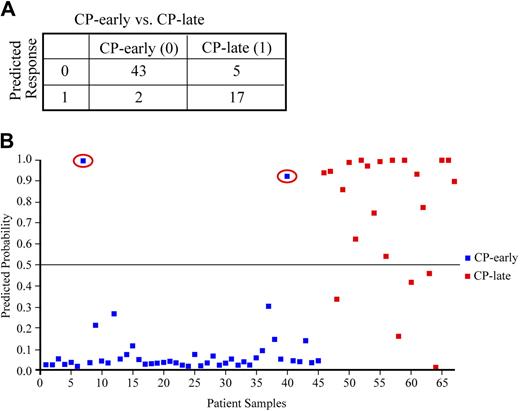
Based on our findings in AP patients, we hypothesized that the 6 signature genes could discriminate early from late CP, a distinction that is not possible with current clinical, pathologic, and molecular methods. The late CP designation is made based on the known duration of disease, which is unknown at the time of diagnosis. In addition, outcomes are also different in late CP patients who typically have poorer responses to TKI therapy than early CP patients.31 Early CP samples (45) were obtained from newly diagnosed and previously untreated patients who subsequently received IM. Late CP samples (22) were obtained from patients with an extended duration of CP disease who had also previously failed IM therapy. These samples were obtained before second TKI therapy with nilotinib. The expression of NOB1, DDX47, IGSF2, LTB4R, SCARB1, and SLC25A3 was examined by QPCR. As the QPCR expression data were on a different scale than the microarray expression data, the BMA model was refit for the 6 signature genes using leave-one-out cross validation. In leave-one-out cross validation, all but 1 patient sample (45 + 22 − 1 in our case) are used to compute the predictive models and the left-out patient sample is used to evaluate the prediction accuracy. Because the left-out sample was not used to derive the models, this analysis allows us to estimate the prediction accuracy for new diagnosis patients. As shown in Figure 4, using leave-one-out cross validation the 6-gene signature clearly discriminated early CP from late CP patients. Notably, there was no relationship between the predicted probabilities using the 6-gene signature and the white blood cell and peripheral blast counts at the time of the sample draw (correlation coefficient = −0.62 and −0.36, respectively). Among the 67 patients, 2 early CP patients and 5 late CP patients were misclassified (error rate = 7/66 = 10.6%). In terms of sensitivity and specificity analysis, our results achieved 77% sensitivity (17/22 = 77%) and 95% specificity (43/45 = 95%). Notably, both misclassified early CP patients subsequently failed IM therapy and among the misclassified late CP patients, 2 patients did well on subsequent nilotinib therapy, suggesting that therapeutic outcomes for these 4 patients were more like outcomes expected from their “predicted phase.”
The 6-gene signature discriminates early from late CP in independent patient samples by QPCR. Results of leave-one-out cross validation comparing diagnostic early CP (class 0, n = 45) and late CP (class 1, n = 22) patient samples (A) including graphic representation (B). Both early CP patients misclassified at diagnosis as late CP (indicated in the red circles) subsequently did poorly on imatinib mesylate therapy.
The 6-gene signature discriminates early from late CP in independent patient samples by QPCR. Results of leave-one-out cross validation comparing diagnostic early CP (class 0, n = 45) and late CP (class 1, n = 22) patient samples (A) including graphic representation (B). Both early CP patients misclassified at diagnosis as late CP (indicated in the red circles) subsequently did poorly on imatinib mesylate therapy.
BMA identifies potential biologic relationships associated with CML progression
Ingenuity Pathways Analysis (Ingenuity Systems, http://www.ingenuity.com) determines biologically enriched pathways and functions based on relationships in published literature. Using this database, networks around the 6 genes were created and merged. SCARB1, SLCA25, and IGSF2 made the largest contributions to these analyses as more is known about these 3 candidate genes. The merged network was enriched for the following functions: cellular movement (71 molecules, P << .001), cell death (84 molecules, P << .001), lipid metabolism (47, molecules, P << .001), and molecular transport (62 molecules, P << .001). The 2612 differentially expressed genes associated with CML progression (from which the 6-gene signature was derived) were then mapped onto this network with differential gene expression observed for NFKB, ERK, and heat shock proteins, and for the transcription factors CEBPA, CEBPB, MYB, MYC, SPI, SP2, and YY1 (supplemental Figure 1). These observations suggest that biologic relationships may exist between these genes because BMA identifies markers that work in combination.
Interestingly, 3 of the 6 signature genes identified by BMA (Table 1) are located on chromosome 12 (DDX47 on 12p13, SLC25A3 on 12q23, and SCARB1 on 12q24). Positional Gene Enrichment (PGE) analysis32 was used to investigate whether chromosome 12 is enriched with genes associated with CML progression. PGE (http://homes.esat.kuleuven.be/∼bioiuser/pge/index.php) is a web-based tool that identifies overrepresented chromosomal regions from a given gene list.32 We first identified 7017 differentially expressed genes associated with CML progression using significance analysis of microarray20 with a false discovery rate of 0.001. As shown in Table 2, the regions (p13 and q23) into which 2 of our signature genes (DDX47 and SLC25A3) fall are statistically enriched with differentially expressed genes (with raw P values for each well below .001 and adjusted P values equal to .046 and .019, respectively). Our findings were similar when restricted to the 2612 differentially expressed genes derived from the ANOVA analysis (supplemental Table 1). This finding suggests the possibility of gene amplification or chromatin modification in these regions leading to increased gene expression. Thus, BMA also identified an interesting chromosomal region or process for further investigation in future studies.
PGE analysis demonstrates enrichment of several chromosomal regions including 2 areas in which 2 of the 6 signature genes are located
| Chromosome . | Start position . | End position . | Chromosomal region . | No. of input genes in the region . | No. of genes in the region . | % enrichment . |
|---|---|---|---|---|---|---|
| 1 | 151596954 | 151789236 | q21.3 | 6 | 10 | 60.00 |
| 1 | 201403562 | 201587240 | q32.1 | 5 | 5 | 100.00 |
| 2 | 160664438 | 162639298 | q24.2 | 8 | 10 | 80.00 |
| 3 | 101566831 | 102795969 | q12.2-q12.3 | 6 | 10 | 60.00 |
| 3 | 137167257 | 137953935 | q22.2-q22.3 | 4 | 4 | 100.00 |
| 4 | 15313738 | 20229900 | p15.32-p15.31 | 8 | 16 | 50.00 |
| 4 | 77173975 | 80052363 | q21.1-q21.21 | 9 | 18 | 50.00 |
| 4 | 77173975 | 77450763 | q21.1 | 4 | 4 | 100.00 |
| 5 | 135392644 | 137503031 | q31.1-q31.2 | 7 | 14 | 50.00 |
| 8 | 6344580 | 6901669 | p23.1 | 7 | 10 | 70.00 |
| 9 | 4701155 | 5460547 | p24.1 | 6 | 10 | 60.00 |
| 11 | 59279108 | 59390594 | q12.1 | 4 | 4 | 100.00 |
| 12 | 2870294 | 6511381 | p13.33-p13.31 | 12 | 37 | 32.43 |
| 12 | 21175403 | 29378272 | p12.2-p11.22 | 16 | 55 | 29.09 |
| 12 | 32003620 | 32789750 | p11.21 | 5 | 7 | 71.43 |
| 12 | 91061030 | 97519906 | q21.33-q23.1 | 15 | 43 | 34.88 |
| 12 | 92385401 | 93377851 | q22 | 5 | 5 | 100.00 |
| 13 | 72227541 | 93857656 | q22.1-q32.1 | 12 | 30 | 40.00 |
| 19 | 56807156 | 56965572 | q13.33 | 4 | 4 | 100.00 |
| X | 55760897 | 56611026 | p11.21-p11.1 | 4 | 4 | 100.00 |
| Chromosome . | Start position . | End position . | Chromosomal region . | No. of input genes in the region . | No. of genes in the region . | % enrichment . |
|---|---|---|---|---|---|---|
| 1 | 151596954 | 151789236 | q21.3 | 6 | 10 | 60.00 |
| 1 | 201403562 | 201587240 | q32.1 | 5 | 5 | 100.00 |
| 2 | 160664438 | 162639298 | q24.2 | 8 | 10 | 80.00 |
| 3 | 101566831 | 102795969 | q12.2-q12.3 | 6 | 10 | 60.00 |
| 3 | 137167257 | 137953935 | q22.2-q22.3 | 4 | 4 | 100.00 |
| 4 | 15313738 | 20229900 | p15.32-p15.31 | 8 | 16 | 50.00 |
| 4 | 77173975 | 80052363 | q21.1-q21.21 | 9 | 18 | 50.00 |
| 4 | 77173975 | 77450763 | q21.1 | 4 | 4 | 100.00 |
| 5 | 135392644 | 137503031 | q31.1-q31.2 | 7 | 14 | 50.00 |
| 8 | 6344580 | 6901669 | p23.1 | 7 | 10 | 70.00 |
| 9 | 4701155 | 5460547 | p24.1 | 6 | 10 | 60.00 |
| 11 | 59279108 | 59390594 | q12.1 | 4 | 4 | 100.00 |
| 12 | 2870294 | 6511381 | p13.33-p13.31 | 12 | 37 | 32.43 |
| 12 | 21175403 | 29378272 | p12.2-p11.22 | 16 | 55 | 29.09 |
| 12 | 32003620 | 32789750 | p11.21 | 5 | 7 | 71.43 |
| 12 | 91061030 | 97519906 | q21.33-q23.1 | 15 | 43 | 34.88 |
| 12 | 92385401 | 93377851 | q22 | 5 | 5 | 100.00 |
| 13 | 72227541 | 93857656 | q22.1-q32.1 | 12 | 30 | 40.00 |
| 19 | 56807156 | 56965572 | q13.33 | 4 | 4 | 100.00 |
| X | 55760897 | 56611026 | p11.21-p11.1 | 4 | 4 | 100.00 |
To investigate the possibility of enrichment of differential expression on chromosome 12, we performed positional gene enrichment (PGE) analysis to detect overrepresented chromosomal regions in the CML progression-related data set. The table shows the chromosomal regions that were enriched in these analyses. Chromosome 12 is indicated in bold.
All raw P values were less than .001.
Discussion
We have identified and validated in independent patient samples, using the BMA algorithm, a 6-gene signature that discriminates CML disease phase. This algorithm calculates a posterior probability for each patient sample that ranges between 0 and 1; thus, values close to 0 represent CP patients and values close to 1 represent BC patients. The genes NOB1, DDX47, IGSF2, LTB4R, SCARB1, and SLC25A3 discriminated 42 CP from 30 BC patients with excellent predictive accuracy in cross validation studies. Notably, in a separate group of 21 patients with disease “intermediate” between CP and BC, either AP or BC disease in remission, BMA produced posterior probabilities that were also intermediate, suggesting some patients were more CP-like and others more BC-like. In an independent group of 45 early CP and 22 late CP patients, the expression of these 6 genes discriminated these groups with high accuracy, suggesting a biologic difference mapping with duration of chronic phase. Lastly, patients with 6-gene expression patterns more similar to blast crisis patients before allogeneic transplantation appeared to have more relapses and deaths compared with patients with 6-gene expression more similar to chronic phase patients.
These observations suggest that these 6 genes are particularly useful biomarkers as they can discriminate patients very early on the timeline of CML progression (early CP vs late CP). Currently, no clinical, pathologic, or molecular methods exist that can discriminate late from early CP; only time from diagnosis discriminates these 2 groups after a diagnosis has been made. Thus, the 6-gene signature could be used to classify early and late CP patients at diagnosis. The ability to classify patients as early and late CP at diagnosis is a useful prognostic marker as response rates for TKI therapies are lower in late CP patients and the incidence of ABL TKD point mutations, a common mechanism of acquired resistance, is also higher in late CP patients and more advanced disease patients.6
The 6 genes chosen by the BMA modeling approach are not common names in leukemia biology. This observation may simply reflect the limitations of our current knowledge, and does not discount them from being interesting candidates for disease biology investigations. SCARB1 plays a role in cancer cell proliferation33 and was also recently identified as differentially increased in expression in a panel of genes that discriminated TEL/AML1 acute lymphoblastic leukemia from other B-cell acute lymphoblastic leukemias in pediatric patients.34 The DDX family of RNA helicases are required for ATP binding, nucleic acid binding, and unwinding; and several family members have been characterized as being overexpressed in solid tumors.35 The SLC25 family of mitochondrial transporters is important in several physiologic and pathologic processes, although no direct role in cancer has, as of yet, been identified.36
BMA identifies small numbers of genes that work in combination to predict disease phase, and our analyses using Ingenuity Pathways Analysis also suggest that as disparate as each of the 6 genes appear individually, that they, and genes that they are related to, may share common biology. We found enrichment of cellular movement and cell death in addition to the expected lipid metabolism (SCARB1) and molecular transport (SLC25A3). Furthermore, when mapping the differential gene expression of the progression microarray data onto a merged network, hubs were identified that are known to play a role in CML and acute leukemia biology such as NFKB, ERK, and heat shock proteins. These biologic insights may have treatment implications as each of these can be targeted therapeutically.37-39 Lastly and most intriguingly, our 6-gene signature led us to identify areas of enrichment on chromosome 12, which will be examined in further studies.
In conclusion, the identification of this small set of phase-specific genes has several clinical implications. It is tempting to speculate that gene expression at diagnosis might give a better indication of response than pathologic diagnosis. A higher posterior probability in a newly diagnosed CP patient may indicate that a patient is at higher risk of failing first-line TKI therapy or developing ABL TKD point mutations. In this scenario, increased monitoring for early treatment failure with consideration for a more rapid switch to alternative therapy with either another TKI or even allogeneic transplantation may be indicated. The fact that this set of genes is so small makes this a testable hypothesis in a relatively inexpensive and efficient way. If we can indeed predict response at diagnosis by a simple QPCR assay, this will be a model for using molecular diagnostics to “tailor” therapy, a big step in the push to use genetic assays for “personalized” medicine.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the following grants: National Institutes of Health (NIH) K25CA106988 and R01GM084163-01A1 (K.Y.Y.); Leukemia & Lymphoma Society Translational Research Program grant and V Foundation for Cancer Research V Scholar Grant (V.G.O.); NIH R01GM084163-01A1, P50HL073996, U54AI057141, R24RR021863-01A1, R01DE012212-06, UL1RR025014-01, and a generous basic research grant from Merck (R.E.B.); NIH R01HDO54511-01A1 and R01GM084163-01A1, NSF llS0534094 and ATM0724721 (A.E.R.); and NCI CA18029 (J.P.R.).
National Institutes of Health
Authorship
Contribution: V.G.O. and K.Y.Y. conceived, designed, and performed the experiments, analyzed the data, and wrote the paper; Y.E.C. performed quantitative PCR; R.E.B. and A.E.R. provided guidance for statistical analyses; and J.P.R. conceived the experiments and helped organize the paper.
Conflict-of-interest disclosure: J.P.R. receives laboratory support and honoraria from Novartis Pharmaceuticals Corporation. The remaining authors declare no competing financial interests.
Correspondence: Vivian G. Oehler, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave North, D5-380, Seattle, WA, 98109; e-mail: voehler@u.washington.edu.
References
Author notes
*V.G.O. and K.Y.Y. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal