Abstract
The prognostic heterogeneity of the World Health Organization category of “systemic mastocytosis with associated clonal hematologic nonmast cell lineage disease” (SM-AHNMD) has not been systematically validated by primary data. Among 138 consecutive cases with SM-AHNMD, 123 (89%) had associated myeloid neoplasm: 55 (45%) myeloproliferative neoplasm (SM-MPN), 36 (29%) chronic myelomonocytic leukemia, 28 (23%) myelodysplastic syndrome (SM-MDS), and 4 (3%) acute leukemia. Of the myeloid subgroups, SM-MPN displayed a 2- to 3-fold better life expectancy (P = .003), whereas leukemic transformation was more frequent in SM-MDS (29%; P = .02). The presence of eosinophilia, although prevalent (34%), was prognostically neutral, and the overall results were not affected by exclusion of FIP1L1-PDGFRA-positive cases. We conclude that it is clinically more useful to consider specific entities, such as SM-MPN, systemic mastocytosis with chronic myelomonocytic leukemia, SM-MDS, and systemic mastocytosis with-acute leukemia, rather than their broad reference as SM-AHNMD.
Introduction
The 2008 World Health Organization (WHO) classification of systemic mastocytosis (SM) recognizes 4 major disease variants: (1) indolent SM, (2) SM with associated clonal hematologic nonmast cell lineage disease (SM-AHNMD), (3) aggressive SM, and (4) mast cell leukemia.1
In their study of 66 SM patients, Travis et al found SM-AHNMD to be the second most common subtype (after indolent SM), with 22 patients (33%).2 Eighteen patients (82%) had an associated clonal myeloid malignancy, with myelodysplastic syndrome (MDS) being observed most frequently (32%). In this series, the 5-year survival of SM-AHNMD patients was 28% compared with 61% for other SM patients. Horny et al similarly found SM-AHNMD to be the second most common subtype (n = 20) in their series of 64 SM patients.3 Eighteen patients (90%) had an associated myeloid neoplasm, with chronic myelomonocytic leukemia (CMML) representing the most frequent entity (39%). As in the series by Travis et al,2 the SM component was discovered incidentally in most cases, during evaluation of the associated hematologic disorder.
Although the pathophysiologic relationship between the SM and AHNMD components in SM-AHNMD patients remains to be precisely delineated, lineage distribution studies of the KITD816V mutation suggest that a common pluripotent hematopoietic stem cell is targeted in most patients.4-6 In some cases, however, the 2 clonal hematologic disorders may develop coincidentally.7
The current WHO category of SM-AHNMD does not clarify the prognostic impact of specific molecular and cytogenetic abnormalities, presence of eosinophilia, and specific therapies on life expectancy and risk of leukemic transformation within the context of individual disease subgroups. We recently reported on 342 adult SM patients and provided preliminary information for 138 SM-AHNMD patients.8 Here, we expand our description of SM-AHNMD patients, with the following aims: (1) to describe the clinical and laboratory characteristics of a large cohort of SM-AHNMD patients; (2) to evaluate the prevalence of molecular (KITD816V, FIP1L1-PDGFRA, and JAK2V617F mutations) and cytogenetic abnormalities in this cohort; (3) to evaluate whether SM-AHNMD patients can be stratified into prognostically relevant subgroups on the basis of life expectancy and risk of leukemic transformation; (4) to evaluate the prognostic impact of eosinophilia; and (5) to assess treatment outcome(s) of specific disease entities to cytoreductive therapy.
Methods
The current study was approved by the Mayo Clinic institutional review board. The study population was drawn from a previously reported larger study of 342 adult SM patients.8 Donor-informed consent was obtained in accordance with the Declaration of Helsinki. Clinical data and bone marrow (BM) histology were reviewed and SM-AHNMD confirmed per the 2008 WHO proposal1 ; for the purposes of consistency with other literature,9 SM associated with chronic eosinophilic leukemia (SM-CEL) was considered with or without the inclusion of FIP1L1-PDGFRA-positive cases. BM studies were performed as previously described.8 Mutation analysis for KITD816V and JAK2V617F was performed using DNA from archived cytogenetic pellets collected at the time of BM biopsy and according to previously published methods.8,10
Actuarial probability of survival was estimated using the Kaplan-Meier product limit method. Comparison between Kaplan-Meier curves was carried out by the log-rank test. The χ2 test and nonparametric tests were used to test for differences in proportions of nominal variables and medians of continuous variables, respectively. Statistical analyses of the data were carried out using the StatView software package (SAS Institute Inc, Version 9.1).
Results and discussion
Among 342 consecutive adult patients with SM, 138 (40%) had SM-AHNMD.8 Of the latter, 123 (89%) had an associated myeloid neoplasm, whereas the remainder had lymphoma (n = 7), myeloma (n = 5), chronic lymphocytic leukemia (n = 2), or primary amyloidosis (n = 1). The clinical and laboratory characteristics of the 123 SM patients with associated myeloid malignancies are presented in Table 1. (BM findings are summarized in supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article.) A total of 55 (45%) patients had SM-myeloproliferative neoplasm (SM-MPN), 36 (29%) SM-CMML, 28 (23%) SM-MDS, and 4 (3%) SM-acute leukemia (SM-AL).
Clinical and laboratory characteristics at diagnosis of SM patients with associated myeloid neoplasm
| Characteristic . | No (%) of patients . | Median (range) . | SM-MPN . | SM-CMML . | SM-MDS . | SM-AL . | P . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| SM-MPN vs SM-CMML . | SM-CMML vs SM-MDS . | SM-MPN vs SM-MDS . | Overall . | |||||||
| Total no. of SM | 342 | — | — | — | — | — | — | — | — | — |
| SM associated with myeloid neoplasm | 123 (36) | — | 55 (45) | 36 (29) | 28 (23) | 4 (3) | — | — | — | — |
| Male | 86 (70) | 41 (75) | 24 (67) | 17 (61) | 4 (100) | NS | NS | NS | NS | |
| Age, y | — | 67 (20-87) | 58 (26-83) | 70 (44-87) | 69 (50-80) | 50 (20-67) | < .001 | NS | < .001 | < .001 |
| Follow-up from SM diagnosis, mo | — | 14.5 (0-182.2) | 19.9 (0.9-182.2) | 12 (0.1-135.8) | 12.5 (0-136.3) | 13.3 (2.6-33.8) | NS | NS | .04 | NS |
| Clinical findings | ||||||||||
| Urticaria pigmentosa | 20 (16) | — | 11 (20) | 5 (14) | 4 (14) | 0 | NS | NS | NS | NS |
| Cutaneous symptoms* | 39 (32) | — | 19 (35) | 10 (28) | 10 (36) | 0 | NS | NS | NS | NS |
| Constitutional symptoms† | 80 (65) | — | 35 (64) | 27 (75) | 15 (54) | 3 (75) | NS | NS | NS | NS |
| Mediator-related symptoms‡ | 34 (28) | — | 17 (31) | 9 (25) | 6 (21) | 2 (50) | NS | NS | NS | NS |
| Musculoskeletal symptoms§ | 33 (27) | — | 21 (38) | 7 (19) | 3 (11) | 2 (50) | NS | NS | .01 | .02 |
| Splenomegaly (n = 119) | 67 (61) | — | 30 (56) | 28 (82) | 14 (52) | 1 (25) | .01 | .01 | NS | .02 |
| Leukemic transformation | 16 (13) | — | 6 (11) | 2 (6) | 8 (29) | NA | NS | .02 | NS | .02 |
| Transfusion-dependent | 32 (26) | — | 10 (18) | 8 (22) | 13 (46) | 1 (25) | NS | NS | .01 | .04 |
| Laboratory findings | ||||||||||
| Hemoglobin, g/dL | 123 (100) | 10.6 (6.4-17.4) | 11.8 (6.6-17.4) | 10.6 (7.7-15.4) | 9.5 (6.4-13.3) | 10.3 (7.5-13.4) | .02 | NS | < .001 | < .001 |
| White blood count, × 109/L | 123 (100) | 10.8 (1.2-87.2) | 12.5 (2.2-87.2) | 16.4 (2.8-51.1) | 5.6 (1.2-30.4) | 7.7 (4.6-30.1) | NS | < .001 | < .001 | < .001 |
| AEC ≥ 1.5 | 42 (34) | 31 (56) | 7 (19) | 3 (11) | 1 (25) | < .001 | NS | < .001 | < .001 | |
| Platelets, × 109/L | 123 (100) | 118 (2-1625) | 146 (7-1625) | 111 (12-723) | 122 (2-555) | 215 (90-469) | .02 | NS | NS | NS |
| Albumin (3.5-5.0 g/dL) | 94 (76) | 3.8 (2-4.9) | 3.8 (2-4.9) n = 38 | 3.8 (2.4-4.8) n = 31 | 3.8 (2.3-4.6) n = 22 | 4.1 (3.8-4.4) n = 3 | NS | NS | NS | NS |
| SAP (45-115 U/L) | 117 (95) | 219 (31-3680) | 212.5 (58-2586) n = 50 | 348 (63-3680) n = 35 | 179.5 (31-1546) n = 28 | 194 (67-265) n = 4 | NS | .02 | NS | NS |
| LDH (122-222 U/L) | 89 (72) | 168 (46-926) | 192 (74-912) n = 39 | 167 (91-926) n = 29 | 166 (46-432) n = 19 | 266 (244-288) n = 2 | NS | NS | NS | NS |
| Serum tryptase (< 11.5 ng/mL) | 50 (41) | 69.5 (3.7-1360) | 53.2 (3.7-1360) n = 26 | 94.6 (5.2-303) n = 16 | 56.2 (13.2-517) n = 8 | NA | NS | NS | NS | NS |
| BM, percentage MC | 107 (87) | 15 (1-70) | 10 (1-70) | 15 (5-70) | 15 (5-60) | 5 (5) | NS | NS | NS | NS |
| Abnormal karyotype (n = 102) | 29 (28) | — | 8 (18) n = 44 | 8 (25) n = 32 | 9 (41) n = 22 | 4 (100) n = 4 | NS | NS | NS | .003 |
| FIP1L1-PDFGRA mutation (n = 36) | 12 (33) | — | 12 (50) n = 24 | 8 (0) n = 8 | 4 (0) n = 4 | NA | .01 | NS | NS | .01 |
| KITD816V mutation (n = 80) | 50 (63) | — | 17 (52) n = 33 | 22 (76) n = 29 | 10 (59) n = 17 | 1 (100) n = 1 | NS | NS | NS | NS |
| JAK2V617F mutation (n = 80) | 6 (8) | — | 4 (12) n = 33 | 1 (3) n = 29 | 1 (6) n = 17 | 0 (0) n = 1 | NS | NS | NS | NS |
| Characteristic . | No (%) of patients . | Median (range) . | SM-MPN . | SM-CMML . | SM-MDS . | SM-AL . | P . | |||
|---|---|---|---|---|---|---|---|---|---|---|
| SM-MPN vs SM-CMML . | SM-CMML vs SM-MDS . | SM-MPN vs SM-MDS . | Overall . | |||||||
| Total no. of SM | 342 | — | — | — | — | — | — | — | — | — |
| SM associated with myeloid neoplasm | 123 (36) | — | 55 (45) | 36 (29) | 28 (23) | 4 (3) | — | — | — | — |
| Male | 86 (70) | 41 (75) | 24 (67) | 17 (61) | 4 (100) | NS | NS | NS | NS | |
| Age, y | — | 67 (20-87) | 58 (26-83) | 70 (44-87) | 69 (50-80) | 50 (20-67) | < .001 | NS | < .001 | < .001 |
| Follow-up from SM diagnosis, mo | — | 14.5 (0-182.2) | 19.9 (0.9-182.2) | 12 (0.1-135.8) | 12.5 (0-136.3) | 13.3 (2.6-33.8) | NS | NS | .04 | NS |
| Clinical findings | ||||||||||
| Urticaria pigmentosa | 20 (16) | — | 11 (20) | 5 (14) | 4 (14) | 0 | NS | NS | NS | NS |
| Cutaneous symptoms* | 39 (32) | — | 19 (35) | 10 (28) | 10 (36) | 0 | NS | NS | NS | NS |
| Constitutional symptoms† | 80 (65) | — | 35 (64) | 27 (75) | 15 (54) | 3 (75) | NS | NS | NS | NS |
| Mediator-related symptoms‡ | 34 (28) | — | 17 (31) | 9 (25) | 6 (21) | 2 (50) | NS | NS | NS | NS |
| Musculoskeletal symptoms§ | 33 (27) | — | 21 (38) | 7 (19) | 3 (11) | 2 (50) | NS | NS | .01 | .02 |
| Splenomegaly (n = 119) | 67 (61) | — | 30 (56) | 28 (82) | 14 (52) | 1 (25) | .01 | .01 | NS | .02 |
| Leukemic transformation | 16 (13) | — | 6 (11) | 2 (6) | 8 (29) | NA | NS | .02 | NS | .02 |
| Transfusion-dependent | 32 (26) | — | 10 (18) | 8 (22) | 13 (46) | 1 (25) | NS | NS | .01 | .04 |
| Laboratory findings | ||||||||||
| Hemoglobin, g/dL | 123 (100) | 10.6 (6.4-17.4) | 11.8 (6.6-17.4) | 10.6 (7.7-15.4) | 9.5 (6.4-13.3) | 10.3 (7.5-13.4) | .02 | NS | < .001 | < .001 |
| White blood count, × 109/L | 123 (100) | 10.8 (1.2-87.2) | 12.5 (2.2-87.2) | 16.4 (2.8-51.1) | 5.6 (1.2-30.4) | 7.7 (4.6-30.1) | NS | < .001 | < .001 | < .001 |
| AEC ≥ 1.5 | 42 (34) | 31 (56) | 7 (19) | 3 (11) | 1 (25) | < .001 | NS | < .001 | < .001 | |
| Platelets, × 109/L | 123 (100) | 118 (2-1625) | 146 (7-1625) | 111 (12-723) | 122 (2-555) | 215 (90-469) | .02 | NS | NS | NS |
| Albumin (3.5-5.0 g/dL) | 94 (76) | 3.8 (2-4.9) | 3.8 (2-4.9) n = 38 | 3.8 (2.4-4.8) n = 31 | 3.8 (2.3-4.6) n = 22 | 4.1 (3.8-4.4) n = 3 | NS | NS | NS | NS |
| SAP (45-115 U/L) | 117 (95) | 219 (31-3680) | 212.5 (58-2586) n = 50 | 348 (63-3680) n = 35 | 179.5 (31-1546) n = 28 | 194 (67-265) n = 4 | NS | .02 | NS | NS |
| LDH (122-222 U/L) | 89 (72) | 168 (46-926) | 192 (74-912) n = 39 | 167 (91-926) n = 29 | 166 (46-432) n = 19 | 266 (244-288) n = 2 | NS | NS | NS | NS |
| Serum tryptase (< 11.5 ng/mL) | 50 (41) | 69.5 (3.7-1360) | 53.2 (3.7-1360) n = 26 | 94.6 (5.2-303) n = 16 | 56.2 (13.2-517) n = 8 | NA | NS | NS | NS | NS |
| BM, percentage MC | 107 (87) | 15 (1-70) | 10 (1-70) | 15 (5-70) | 15 (5-60) | 5 (5) | NS | NS | NS | NS |
| Abnormal karyotype (n = 102) | 29 (28) | — | 8 (18) n = 44 | 8 (25) n = 32 | 9 (41) n = 22 | 4 (100) n = 4 | NS | NS | NS | .003 |
| FIP1L1-PDFGRA mutation (n = 36) | 12 (33) | — | 12 (50) n = 24 | 8 (0) n = 8 | 4 (0) n = 4 | NA | .01 | NS | NS | .01 |
| KITD816V mutation (n = 80) | 50 (63) | — | 17 (52) n = 33 | 22 (76) n = 29 | 10 (59) n = 17 | 1 (100) n = 1 | NS | NS | NS | NS |
| JAK2V617F mutation (n = 80) | 6 (8) | — | 4 (12) n = 33 | 1 (3) n = 29 | 1 (6) n = 17 | 0 (0) n = 1 | NS | NS | NS | NS |
Differences in proportions of nominal variables and medians of continuous variables were tested by using Fisher exact test or χ2 test, and Mann-Whitney test or Kruskal-Wallis test, respectively.
SM indicates systemic mastocytosis; AEC, absolute eosinophil count; SAP, serum alkaline phosphatase; LDH, lactate dehydrogenase; MC, mast cell; —, not applicable; NA, not applicable or not assessed; and NS, not significant.
Includes pruritus, flushing, urticaria, and angioedema.
Includes weight loss, fever, chills, and night sweats.
Includes headache, dizziness/lightheadedness, syncope/presyncope, hypotension, anaphylaxis, palpitation/tachycardia, bronchoconstriction/wheezing, and peptic ulcer disease.
Includes bone pain, arthralgias, and myalgias.
Among the 55 patients with SM-MPN, 30 had MPN-unclassifiable, 15 CEL, 6 essential thrombocythemia, 2 polycythemia vera, and 2 primary myelofibrosis. Patients classified as MPN-unclassifiable displayed clinical and laboratory features of MPN, including hepatosplenomegaly, leukocytosis, thrombocytosis, and a hypercellular bone marrow with prominent megakaryocytic proliferation, but failed to meet criteria for another specific MPN entity. The MDS component in the 28 patients with SM-MDS was refractory anemia with excess blasts-1/2 (n = 11), refractory anemia with ring sideroblasts (n = 6), refractory cytopenia with multilineage dysplasia (n = 4), refractory anemia (n = 3), and MDS associated with del(5q) (n = 1); 3 patients with MDS/MPN were also included in the SM-MDS category.
In contrast to previous reports, we observed a high prevalence of prominent eosinophilia (≥ 1.5 × 109/L) at presentation (n = 42; 34%), most commonly in the SM-MPN category (56%; P < .001; Table 1). Of the latter, 48% (n = 15) could be classified as SM-CEL (12 harbored FIP1L1-PDGFRA, 2 other clonal cytogenetic abnormalities, and 1 BM blasts > 5%). Clinical outcomes were similar for SM-MPN patients with and those without eosinophilia. However, SM-MPN patients with eosinophilia were more likely to be males (87% vs 58%; P = .03) and exhibit a “loose” pattern of mast cell infiltration in BM trephines (54% vs 5%; P < .001; supplemental Figure 1).
Results of cytogenetic and molecular testing are presented in Table 1 (individual cytogenetic abnormalities are listed in supplemental Table 2). Abnormal karyotype was most frequent in SM-AL (100%; n = 4); however, its distribution among the other subgroups was similar. KITD816V mutational frequency was similar among the different subgroups, and JAK2V617F occurrence was limited to SM-MPN. The frequency of abnormal karyotype in patients with prominent eosinophilia was 24%. Twenty-six SM patients with prominent eosinophilia underwent molecular testing for KITD816V and JAK2V617F; the respective mutational frequencies were 50% and 8%. Nineteen patients with prominent eosinophilia were evaluated for FIP1L1-PDGFRA, and 12 (63%) harbored the mutation; 8 of these 12 FIP1L1-PDGFRA-positve cases were also screened for KITD816V and JAK2V617F, and all tested negative.
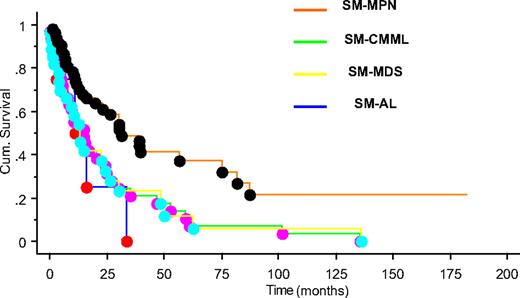
After a median follow-up of 15 months (range, 0-182 months), 90 (73%) deaths were recorded (100% with SM-AL, 89% SM-CMML, 82% SM-MDS, and 56% with SM-MPN). SM-MPN patients had a significantly longer median survival (∼ 31 months) compared with the SM-CMML (∼ 15 months), SM-MDS (∼ 13 months), and SM-AL (∼ 11 months) patients (P = .003; Figure 1). The superior survival seen in SM-MPN was not affected by the inclusion or exclusion of the 12 SM patients with FIP1L1-PDGFRA. Leukemic transformation (n = 16; 13% overall) was observed significantly more frequently in SM-MDS (29%), compared with SM-MPN (11%) or SM-CMML (6%; P = .02).
Overall survival of patients with systemic mastocytosis with associated myeloid neoplasm. Kaplan-Meier survival plot for patients classified by disease type: SM-MPN (orange), SM-CMML (green), SM-MDS (yellow), and SM-AL (blue) (P = .003).
Overall survival of patients with systemic mastocytosis with associated myeloid neoplasm. Kaplan-Meier survival plot for patients classified by disease type: SM-MPN (orange), SM-CMML (green), SM-MDS (yellow), and SM-AL (blue) (P = .003).
A summary of SM patients with associated myeloid neoplasia who received at least one cytoreductive therapy is provided in supplemental Table 3. In general, FIP1L1-PDGFRA-positive disease almost always responds to treatment with imatinib mesylate. Response is infrequent in the absence of this molecular marker. Approximately 50% of treated subjects responded to interferon-α or 2-chlorodeoxyadenosine. Response to hydroxyurea therapy was infrequent and modest.
We recently reported on the prognostic relevance of the WHO proposed classification for SM.8 In the current paper, we point out the limited clinical usefulness of the term “SM-AHNMD” and provide evidence to support its substitution by a prognostically more useful subcategorization that includes SM-MPN, SM-CMML, SM-MDS, and SM-AL. Finally, we demonstrate that FIP1L1-PDGFRA-negative eosinophilia is prevalent in SM associated with a myeloid malignancy but does not modify prognosis within a specific subcategory.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.P. designed the study, collected and analyzed the data, and wrote the paper; K.-H.L. collected and analyzed the data; T.L.L., C.F., and R.F.M. did sample preparation and/or molecular analysis; C.Y.L. reviewed the bone marrow histology; and A.T. designed the study, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Animesh Pardanani, Mayo Clinic, Division of Hematology, 200 First St SW, Rochester, MN 55905; e-mail: Pardanani.animesh@mayo.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal