Abstract
Estimates of precursor frequency and assessment of functional characteristics of alloreactive CD4+ T cells are all biased by the need for long-term culture. In this study, direct visualization of human alloreactive CD4+ T cells on the single-cell level was achieved using cell surface expression of CD154 as a tool for identification. The average frequency of alloreactive CD154+CD4+ T cells among peripheral blood CD4+ T cells was 0.1%, with half of the cells displaying a naive phenotype. The proliferation capacity and expression of cytokines after allogeneic stimulation resided in these CD154+CD4+ T cells. The repertoire of alloreactive CD4+ T cells was biased to a Th17 response, and on average 24% of alloreactive CD154+CD4+ memory T cells produced interleukin-17 (IL-17) after polyclonal stimulation. Unexpectedly, mixed cell cultures from human leukocyte antigen (HLA)–identical donors also generated alloreactive CD154+CD4+ T cells and yielded the highest frequency compared with HLA-nonidentical combinations. Therefore, reactivity to minor histocompatibility antigens between HLA-identical subjects appears to be relatively common. Alloreactive HLA-identical T cells did not proliferate or express cytokines, but were driven to proliferation in the presence of exogenous IL-2.
Introduction
Expression of human leukocyte antigen (HLA) class I and II molecules on the cell surface is important for presentation of antigenic peptides to the immune system under normal circumstances. After organ transplantation, these molecules are critically involved in graft rejection as T-cell receptors of recipient T lymphocytes can interact with allogeneic HLA expressed by the donor organ. These alloreactive T cells can be either CD8+ T cells interacting with allogeneic HLA class I or CD4+ T lymphocytes interacting with allogeneic HLA class II.
The alloreactive CD4+ T cells are considered to be important cells in mediating rejection of transplanted organs. The mechanism of action may be by either the direct or indirect allogeneic antigen presentation pathway.1-4 For instance, immunization with allogeneic HLA peptides presented by acceptor HLA class II significantly increases both acute and chronic rejection after organ transplantation.5 In addition, CD4+ T-cell help is necessary to facilitate full differentiation of graft-infiltrating CD8+ T cells.6-8 However, other studies have pointed to a protective role of alloreactive CD4+ T cells, which are able to mediate immunologic tolerance to the graft.9,10
Visualization and characterization of alloreactive CD4+ T cells is necessary to elucidate the role of these cells in rejection or tolerance of the transplanted organ in humans. However, currently there are no assays available that can identify alloreactive CD4+ T cells on the single-cell level without prior long-term culture.
Cell surface expression of CD40 ligand (CD154) after antigenic stimulation is a key event in the bidirectional communication and activation of CD4+ T cells and the antigen-presenting cell.11,12 However, rapid internalization of CD154 after interaction with CD40 on the antigen-presenting cell largely prevents the detection of CD154 on the T-cell surface. Recently, live assays for flow cytometry have been developed that enable detection of antigen-specific CD4+ T cells, by stabilizing the expression of CD154 on their cell surface after antigenic stimulation.13,14 This new technique combines a high sensitivity with a low background staining. In addition, the antigen-specific T cells can now be directly visualized, analyzed by multicolor flow cytometry, and isolated for cell expansion and functional tests.
Using a modified CD154 live assay, we were able to identify alloreactive CD4+ T cells in different T-cell subsets with great specificity and sensitivity. In addition, expression of cytokines, typical for different types of T cells, could be shown to a variable extent. Most salient findings include an unexpected, high frequency of alloreactive T cells in HLA-identical combinations, indicative of minor histocompatibility antigen (mHag)–specific T cells and a bias to a Th17 response.
Methods
Blood samples
Buffy coats were purchased from Sanquin Blood Bank. These buffy coats were obtained from healthy human donors upon receiving written informed consent in accordance with the Declaration of Helsinki with regard to scientific use.
The current study did not require approval from an ethical committee according to the Dutch Medical Research Involving Human Subjects Act (WMO). Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation (Lymphoprep; Nycomed Pharma) and the cells were resuspended in RPMI-1640 (GibcoBRL) supplemented with 100 IU/mL penicillin, 100 μg/mL streptomycin, 2 mM l-glutamine, and 10% heat-inactivated AB+ pooled human serum (further referred to as standard culture medium). HLA typing was performed by serologic and DNA-based techniques according to international (American Society for Histocompatibility and Immunogenetics [ASHI]/European Federation of Immunogenetics [EFI]) standards. PBMCs obtained from siblings with similar HLA typing were considered to be HLA-identical combinations.
CD154 assays
A 2-way mixed leukocyte reaction (MLR) was performed by culture of PBMCs of 2 subjects in a round-bottom 96-well plate, at a density of 106 cells per well for each party. Prior to the MLR, one set of PBMCs was labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes; for protocol, see “CFSE labeling and cell proliferation”) or using an AmCyan-labeled anti-CD45 antibody (Becton Dickinson [BD]) to allow for identification of the 2 different parties during fluorescence-activated cell sorting (FACS) analysis. Preliminary experiments showed no influence of this labeling on CD154 expression on the alloreactive T cells. In a separate set of experiments, HLA-typed PBMCs were used to create MLRs with different numbers of mismatches on the level of HLA class I and II. As a control, cells were stimulated with autologous PBMCs stained with either CFSE or AmCyan-labeled anti-CD45 antibody. The live assay for detection of CD154+CD4+ T cells, as described by Frentsch et al,14 using the CD40 antibody (1 μg/mL, clone 14G7; Sanquin) was evaluated for detecting alloreactive T cells. In addition, CD154 was detected by intracellular staining after a 24-hour incubation of the PBMCs with addition of brefeldin (Golgiplug; BD Pharmingen) during the last 6 hours. In all assays, the APC-labeled CD154 antibody (clone TRAP1; BD Pharmingen) was used. Costimulation was given by addition of 1 μg/mL anti-CD28 (Serotec) and 1 μg/mL anti-CD49d (BD Pharmingen) to the MLR or autologous control, respectively. The net CD154 signal is determined by correcting for the autologous-induced CD154 signal.
The protocol for cytomegalovirus (CMV)–specific T-cell stimulation was described in detail previously and was used as a positive control for the experiments performed.15
Staining protocol for T-cell subsets
Antigen-specific T cells were characterized by staining the cell surface with allophycocyanin-Cy7 (APC-Cy7) or AmCyan-labeled anti-CD3, pacific blue–labeled anti-CD4, phycoerythrin (PE)–labeled CD45RO (all from BD), and fluorescein isothiocyanate (FITC; R&D Systems Europe Ltd)– or phycoerythrin-Cy7 (PECy7)–labeled CCR7 (BD) to dissect the different CD4+ T-cell subsets.16,17 Naive T lymphocytes are defined as CD45RO− and CCR7+, whereas memory T lymphocytes are CD45RO+. These can be further dissected into central memory and effector memory T lymphocytes, which are CCR7+ and CCR7−, respectively.
Intracellular cytokine expression
Intracellular cytokine staining was performed using costimulation (mixture of 1 μg/mL anti-CD28 and 1 μg/mL anti-CD49d) in the MLR. Cells were cultured for 6 hours in the presence of brefeldin and monensin (Golgistop; BD). Subsequently, the cell surface was stained to identify CD3+CD4+ T lymphocytes using antibodies directed against CD3 and CD4. In addition, we created a dump channel to exclude unwanted cells from the analysis using antibodies directed against CD8, CD14, CD16, and CD19 and by inclusion of a viability marker (Live/Dead Fixable Dead cell stain kits; Molecular Probes, Invitrogen Ltd). Next, after permeabilization, CD154 and cytokines were stained intracellularly with APC-labeled anti-CD154 and FITC-labeled anti–interleukin-2 (IL-2) combined with either PE-labeled anti–interferon-gamma (IFN-γ), PE-labeled anti–IL-17, or PE-labeled anti–IL-10. In addition, we also stained for IFN-γ and IL-17 using the following antibody combination: FITC-labeled anti–IFN-γ and PE-labeled anti–IL-17. At least 106 CD3+CD4+ T lymphocytes were analyzed to measure the cytokine-positive T cells. Percentages of cytokine-positive alloantigen-specific CD154+CD3+CD4+ T lymphocytes were determined by analyzing the samples on the fluorescence-activated cell sorter (FACS) Canto II (BD) using FACSDiva software Version 6.1.2 (BD Biosciences).
Isolation of alloantigen-specific CD154+CD4+ T cells
Alloantigen-specific CD154+CD4+ T cells were isolated using the live assay in the presence of costimulatory antibodies. For this purpose, 5 × 107 cells (2.5 × 107 of each fraction) were incubated in a 6-well plate for 24 hours in the presence of the CD40 antibody (1 μg/mL) and the combination of anti-CD28 and anti-CD49d (both at a final concentration of 1 μg/mL). Subsequently, isolation of CD154+CD4+ T cells was performed using the magnetic-activated cell sorting according to manufacturer's protocol (Miltenyi Biotec GmbH).
Two isolation strategies were chosen depending on the particular cell population of interest. For maximal enrichment of CD154+ cells, the “POSSELD2” mode was chosen, which resulted in high purity but low yields, and to obtain maximally CD154+-depleted T-cell fraction, the “DEPLETES” mode was chosen. Only CD154+-enriched fractions with more than 95% purity and CD154− T-cell fractions with less than 0.01% contaminating CD154+ T cells were used for further experiments.
CFSE labeling and cell proliferation
The different cell fractions (unseparated, CD154 enriched, and CD154 depleted) obtained as described in “Isolation of alloantigen-specific CD154+CD4+ T cells” from a 24-hour 2-way MLR (primary MLR) were labeled with CFSE by incubating the cells with 5 μM CFSE for 10 minutes at 37°C. Labeling was stopped by adding standard culture medium, and after washing twice the cells were seeded as responder cells in a 96-well plate at a density of 1 × 105 or 5 × 103 for restimulation. Restimulation was done using standard culture medium alone or supplemented with 200 U/mL recombinant human IL-2 (Alloga BV). As stimulators, we used irradiated (40 Gy) PBMCs of autologous, allogeneic, or third-party (2:2:2 mismatches [MMs] on HLA A:B:DR) origin at a ratio of 1:1. In addition, phytohemagglutinin (Murex Biotech Ltd; 1 μg/mL) was included as a positive control. After a 5- to 6-day culture, the cells were harvested and stained for CD3, CD4, and CD8, and by inclusion of the viability probe 7-amino-actinomycin D (BD Pharmingen) a distinction between viable and dead cells was enabled. The percentage of viable dividing T lymphocytes was determined by measuring the CFSE dilution using flow cytometry, and subsequent analysis was performed using FACSDiva software Version 6.1.2 and ModFit LT software (Verity Software House Inc).
Statistical analysis
For comparisons between groups, the t test, Mann-Whitney U test, 1-way analysis of variance, or Kruskal-Wallis test were used, as appropriate. Posthoc analysis was performed using Bonferroni test for multiple comparisons or Mann-Whitney U test. P values at α less than .05 for 2 sides were considered statistically significant.
Results
Expression of CD154 identifies alloreactive CD4+ T cells and is biphasic in time
The first set of experiments was performed to evaluate the optimal conditions for identification of CD154 expression on alloreactive CD4+ T cells. Initial experiments identified alloreactive CD4+ cells expressing CD154, using intracellular staining for CD154 in the presence of brefeldin. Costimulation by anti-CD49d and anti-CD28 increased the percentage of CD154+CD4+ cells, in both the naive and memory subset, by at least 2-fold without increasing the low background signal (< 0.01%), and was included in subsequent experiments (Figure 1A-B). Few alloreactive CD154+ T cells (< 0.005%) were observed among the CD8+ T-cell population.
Soluble costimulation enhances the alloantigen-specific CD154 signal in both intracellular staining protocol and live assay and kinetics of CD154-expressing CD4+ T-cell subsets. (A) Representative dot plot of the CD154 signal in CD4+ T cells using the CD154 live assay. Donor PBMCs were stimulated with CFSE-labeled donor PBMCs in medium only (unstimulated, i) or with costimulation (cos only, ii) provided by the combination of soluble anti-CD28 and anti-CD49d monoclonal antibodies for 24 hours. The MLR (inducing class II mismatches) was performed in the presence of costimulatory antibodies (alloantigen stimulated, iii). Next, the percentage of CD154+ cells within the different CD4+ T-cell subsets was measured, comparing the intracellular assay using brefeldin A (BFA) with the CD154 live assay using αCD40 (B). The closed and open bars represent the net CD154 signal (corrected for the background obtained for autologous stimulation) with and without costimulation, respectively. The results of 4 to 6 independent experiments for every condition are shown. The expression in time of CD154 on alloreactive (C) and CMV-specific (D) CD4+ T cells was monitored using the CD154 live assay with costimulation. Total CD4+ T cells (■), naive CD4+ T lymphocytes (▼; CD45RO−CCR7+; Tnaive), and memory CD4+ T lymphocytes (●; CD45RO+; Tmem) expressing CD154. A representative example of 4 separate experiments is shown.
Soluble costimulation enhances the alloantigen-specific CD154 signal in both intracellular staining protocol and live assay and kinetics of CD154-expressing CD4+ T-cell subsets. (A) Representative dot plot of the CD154 signal in CD4+ T cells using the CD154 live assay. Donor PBMCs were stimulated with CFSE-labeled donor PBMCs in medium only (unstimulated, i) or with costimulation (cos only, ii) provided by the combination of soluble anti-CD28 and anti-CD49d monoclonal antibodies for 24 hours. The MLR (inducing class II mismatches) was performed in the presence of costimulatory antibodies (alloantigen stimulated, iii). Next, the percentage of CD154+ cells within the different CD4+ T-cell subsets was measured, comparing the intracellular assay using brefeldin A (BFA) with the CD154 live assay using αCD40 (B). The closed and open bars represent the net CD154 signal (corrected for the background obtained for autologous stimulation) with and without costimulation, respectively. The results of 4 to 6 independent experiments for every condition are shown. The expression in time of CD154 on alloreactive (C) and CMV-specific (D) CD4+ T cells was monitored using the CD154 live assay with costimulation. Total CD4+ T cells (■), naive CD4+ T lymphocytes (▼; CD45RO−CCR7+; Tnaive), and memory CD4+ T lymphocytes (●; CD45RO+; Tmem) expressing CD154. A representative example of 4 separate experiments is shown.
Next, we tested the protocol for detecting CD154 on the cell surface, which uses anti-CD40 antibody in the culture medium to stabilize the expression of CD154 on the cell surface. CD154 expression on alloreactive CD4+ cells was rapid and biphasic. After an initial peak at 6 hours, the percentage of CD4 cells expressing CD154 decreased to nondetectable levels at 12 hours. CD154+CD4+ cells reappeared and peaked to a similar percentage at 24 hours, after which their percentage decreased again (Figure 1C). CMV antigen–specific CD4+ cells (and phorbol-12 myristate 13-acetate [PMA]/ionomycin-stimulated CD4+ cells; data not shown) showed a similar expression pattern for CD154 after antigen-specific stimulation (Figure 1D). The percentage of alloreactive CD154+CD4+ cells after 24 hours was on average 3 times higher using the anti-CD40 live assay compared with intracellular staining after incubation with brefeldin A (Figure 1B). The average percentage of alloreactive CD154+CD4+ cells was 0.1% (± 0.01%) in randomly performed MLRs of PBMCs mismatched for HLA class II (n = 10), with an average background signal of 0.01% in freshly isolated PBMCs. Cryopreserved cells showed an increased CD154 background staining (on average 0.03%) without an increase in specific signal.
Proliferating CD4+ alloreactive T cells are CD154+
Alloreactive T cells are historically defined as T cells proliferating in response to allogeneic antigen-presenting cells. We isolated the living alloreactive CD154+ T cells generated in the MLR of parties with a least 1 mismatch at HLA class II, and labeled them with CFSE. After restimulation with allogeneic PBMCs, the CD154+ T-cell fraction showed on average 90% of the labeled cells in the divided cell fraction at day 6 (Figure 2A top panel). Little proliferation of CD4+ T cells was observed in the CD154-depleted cell fraction. The presence of alloreactive CD154+CD4+ T cells was necessary for proliferation of allogeneic CD8+ T cells, as the latter was not observed anymore in the CD154-depleted cell fraction (Figure 2B bottom panel). From these experiments, it can be concluded that CD154+ T cells generated in the MLR comprise the CD4+ T-cell population that proliferates after allogeneic stimulation and that is necessary for in vitro proliferation of allogeneic CD8+ T cells.
Proliferating alloantigen-specific CD4+ T cells are exclusively CD154+. PBMCs of donor A were stimulated with PBMCs of donor B at a 1:1 ratio, mismatched for HLA class II, in the presence of the α-CD40 monoclonal antibody and the combination of soluble anti-CD28 and anti-CD49d for 24 hours. Alloreactive CD154+ T cells were then isolated using automated magnetic cell sorting; a typical flow cytometric example of cells before separation (i) as well as the CD154-depleted (ii) and -enriched (iii) fraction is depicted in panel A. Both fractions as well as the unseparated cells were labeled with CFSE and subsequently restimulated in absence or presence of irradiated allogeneic PBMCs for 6 days. On day 6, proliferation of CD3+CD4+ (B top panel) and CD3+CD8+ (B bottom panel) T lymphocytes was analyzed using flow cytometry. The dark blue peak at the far right of the x-axis represents the CFSE-labeled T-cell population that has not divided. With every cell division the CFSE-intensity is halved, and the different colors identify cell populations, which have undergone one or more rounds of cell division. The numbers within the figures indicate the percentage of cells that has divided. Shown is a representative example of 3 separate experiments performed.
Proliferating alloantigen-specific CD4+ T cells are exclusively CD154+. PBMCs of donor A were stimulated with PBMCs of donor B at a 1:1 ratio, mismatched for HLA class II, in the presence of the α-CD40 monoclonal antibody and the combination of soluble anti-CD28 and anti-CD49d for 24 hours. Alloreactive CD154+ T cells were then isolated using automated magnetic cell sorting; a typical flow cytometric example of cells before separation (i) as well as the CD154-depleted (ii) and -enriched (iii) fraction is depicted in panel A. Both fractions as well as the unseparated cells were labeled with CFSE and subsequently restimulated in absence or presence of irradiated allogeneic PBMCs for 6 days. On day 6, proliferation of CD3+CD4+ (B top panel) and CD3+CD8+ (B bottom panel) T lymphocytes was analyzed using flow cytometry. The dark blue peak at the far right of the x-axis represents the CFSE-labeled T-cell population that has not divided. With every cell division the CFSE-intensity is halved, and the different colors identify cell populations, which have undergone one or more rounds of cell division. The numbers within the figures indicate the percentage of cells that has divided. Shown is a representative example of 3 separate experiments performed.
The highest frequency of alloreactive CD154+CD4+ cells is found in HLA class II–matched MLRs and within the naive T-cell fraction
HLA-typed PBMCs were used to create 0, 1, or 2 mismatches on HLA-DR between the parties in the MLR. Surprisingly, an inverse relation was observed between the number of mismatches and the percentage of alloreactive CD154+CD4+ cells (Figure 3). The number of mismatches on HLA class I did not correlate with the frequency of alloreactive CD154+CD4+ cells (data not shown). Using HLA-identical combinations, a similar high frequency of alloreactive CD154+CD4+ cells was found, excluding the possibility of HLA mismatches not detected by routine analysis.
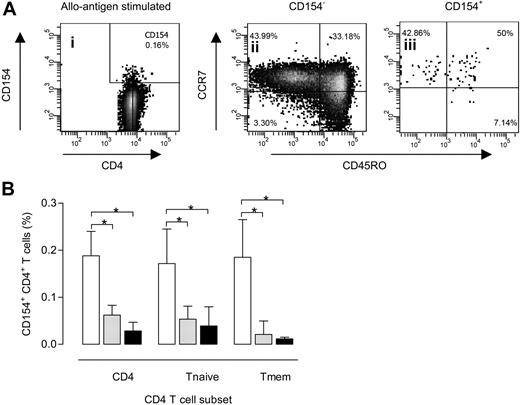
Distribution of alloantigen-specific CD154+CD4+ cells over naive and memory CD4+ T cells. A typical example of the distribution of CD154-expressing CD4+ T cells within the different T-cell subsets after a 24-hour allogeneic MLR (A). Briefly, viable lymphocytes are selected based on forward scatter (FSC)/side scatter (SSC) characteristics and 7-amino-actinomycin D expression. These lymphocytes are then plotted in a CD3 versus CD4 dot plot to select the CD3+CD4+ T (Th) lymphocytes. CD154 expression is than visualized using a dot plot of CD4 versus CD154 (i). Next, both CD154− (ii) and CD154+ (iii) CD4+ lymphocytes are displayed in a dot plot with CD45RO on the x-axis and CCR7 on the y-axis, allowing a dissection of the allogeneic response into the different CD4+ T-lymphocyte subsets. Naive CD4+ lymphocytes are defined as CD45RO−CCR7+; memory CD4+ lymphocytes, as CD45RO+CCR7+ (central memory cells) or CCR7− (effector memory cells). Subsequently, CD154+ expression for total, naive, and memory CD4+ lymphocytes at 24 hours is plotted against the number of MMs on HLA-DR where □,  , and ■ represent 0 (n = 6), 1 (n = 12), and 2 (n = 6) MMs on HLA-DR, respectively (B). *P < .05. Error bars represent SD.
, and ■ represent 0 (n = 6), 1 (n = 12), and 2 (n = 6) MMs on HLA-DR, respectively (B). *P < .05. Error bars represent SD.
Distribution of alloantigen-specific CD154+CD4+ cells over naive and memory CD4+ T cells. A typical example of the distribution of CD154-expressing CD4+ T cells within the different T-cell subsets after a 24-hour allogeneic MLR (A). Briefly, viable lymphocytes are selected based on forward scatter (FSC)/side scatter (SSC) characteristics and 7-amino-actinomycin D expression. These lymphocytes are then plotted in a CD3 versus CD4 dot plot to select the CD3+CD4+ T (Th) lymphocytes. CD154 expression is than visualized using a dot plot of CD4 versus CD154 (i). Next, both CD154− (ii) and CD154+ (iii) CD4+ lymphocytes are displayed in a dot plot with CD45RO on the x-axis and CCR7 on the y-axis, allowing a dissection of the allogeneic response into the different CD4+ T-lymphocyte subsets. Naive CD4+ lymphocytes are defined as CD45RO−CCR7+; memory CD4+ lymphocytes, as CD45RO+CCR7+ (central memory cells) or CCR7− (effector memory cells). Subsequently, CD154+ expression for total, naive, and memory CD4+ lymphocytes at 24 hours is plotted against the number of MMs on HLA-DR where □,  , and ■ represent 0 (n = 6), 1 (n = 12), and 2 (n = 6) MMs on HLA-DR, respectively (B). *P < .05. Error bars represent SD.
, and ■ represent 0 (n = 6), 1 (n = 12), and 2 (n = 6) MMs on HLA-DR, respectively (B). *P < .05. Error bars represent SD.
We found a similar percentage of naive T cells in the alloreactive CD154+CD4+ cells compared with the CD154−CD4+ cells (mean, 53% vs 47%; P = .11). The highest proportion of alloreactive CD154+CD4+ cells with a naive phenotype was observed for fully HLA class II–matched combinations compared with combinations of 1 or 2 mismatches (mean, 64% vs 49%; P = .14).
Time-dependent expression of CD25 and CD127 on alloreactive CD154+CD4+ cells: HLA-identical alloreactive CD154+CD4+ cells can be driven to proliferation with IL-2
Both IL-2 and IL-7 are important T-cell growth factors involved in cell proliferation and homeostasis. After 6 and 24 hours of allogeneic stimulation, the cell surface expression of the alpha-chain of the IL-2 receptor (CD25) and IL-7 receptor (CD127) on alloreactive CD154+CD4+ cells was determined and compared with CMV-specific T-cell stimulation, as a control antigen. At 24 hours, the majority of alloreactive CD154+CD4+ cells from fully HLA-DR–mismatched combinations were brightly CD25+ and part of the cells (on average 20%; n = 5 experiments) became negative for CD127 (Figure 4A-B), similar to the CMV-reactive CD4+ cells. This pattern of CD25 and CD127 expression in time was independent of the number of HLA-DR mismatches and also observed for alloreactive C154+CD4+ cells from HLA-identical combinations, although to a significantly lesser extent.
Kinetics of CD25 and CD127 expression of CD154+CD4+ T cells and proliferation of CD154+CD4+ T cells obtained from HLA-identical cocultures. PBMCs of one donor were cocultured with CFSE-labeled PBMCs of a different donor in the presence of costimulatory antibodies directed against CD28/CD49d and a monoclonal antibody against CD40. CFSE-labeled autologous PBMCs were used as a negative control and CMV-stimulated PBMCs, as a positive control. The cell surface expression of CD154 is plotted against CD25 (Ai-ii) or CD127 (Aiii-iv) at 6 (Ai,iii) and 24 (Aii,iv) hours (left panels). In the right panels of panel A, the expression of these markers is displayed for the different combinations of mismatches (MMs) for HLA-DR and CMV stimulation within the CD154+ fraction at 24 hours. Median fluorescence intensities (MFI) for CD25 and CD127 within the CD154+CD3+CD4+ T lymphocytes were related to the number of MMs on HLA-DR at 6 and 24 hours (B) and compared with the MFIs of CD25 (Bi) and CD127 (Bii) of CD154+ CMV-specific T cells and CD154−CD3+CD4+ T cells. *Significantly different compared with 6 hours. # and ^ indicate significantly different compared with CD154+ population with 0, 1, or 2 MMs on HLA-DR at 6 and 24 hours, respectively. Next, CD154+CD4+ T lymphocytes were isolated from combinations of PBMCs of 2 HLA-identical donors incubated at a 1:1 ratio for 24 hours. The CD154-enriched (Ci-iv top panel) as well as the CD154-depleted (Cv-viii bottom panel) fraction were CFSE-labeled and subsequently restimulated for 6 days using either irradiated PBMCs of the donors at a 1:1 ratio, 200 U/mL recombinant human (RHu) IL-2, or both. The dot plots show forward scatter (FSC) and side scatter (SSC) of the cells at day 6. Region 1 (R1) contains the apoptotic cells and region 2 (R2) signifies the viable CD154+ and CD154− T cells. The percentage of divided CD3+CD4+ T cells from R2, restimulated with irradiated PBMCs and RHu-IL-2, is calculated from the dilution of CFSE dye as a result of cell division (C histograms on right).
Kinetics of CD25 and CD127 expression of CD154+CD4+ T cells and proliferation of CD154+CD4+ T cells obtained from HLA-identical cocultures. PBMCs of one donor were cocultured with CFSE-labeled PBMCs of a different donor in the presence of costimulatory antibodies directed against CD28/CD49d and a monoclonal antibody against CD40. CFSE-labeled autologous PBMCs were used as a negative control and CMV-stimulated PBMCs, as a positive control. The cell surface expression of CD154 is plotted against CD25 (Ai-ii) or CD127 (Aiii-iv) at 6 (Ai,iii) and 24 (Aii,iv) hours (left panels). In the right panels of panel A, the expression of these markers is displayed for the different combinations of mismatches (MMs) for HLA-DR and CMV stimulation within the CD154+ fraction at 24 hours. Median fluorescence intensities (MFI) for CD25 and CD127 within the CD154+CD3+CD4+ T lymphocytes were related to the number of MMs on HLA-DR at 6 and 24 hours (B) and compared with the MFIs of CD25 (Bi) and CD127 (Bii) of CD154+ CMV-specific T cells and CD154−CD3+CD4+ T cells. *Significantly different compared with 6 hours. # and ^ indicate significantly different compared with CD154+ population with 0, 1, or 2 MMs on HLA-DR at 6 and 24 hours, respectively. Next, CD154+CD4+ T lymphocytes were isolated from combinations of PBMCs of 2 HLA-identical donors incubated at a 1:1 ratio for 24 hours. The CD154-enriched (Ci-iv top panel) as well as the CD154-depleted (Cv-viii bottom panel) fraction were CFSE-labeled and subsequently restimulated for 6 days using either irradiated PBMCs of the donors at a 1:1 ratio, 200 U/mL recombinant human (RHu) IL-2, or both. The dot plots show forward scatter (FSC) and side scatter (SSC) of the cells at day 6. Region 1 (R1) contains the apoptotic cells and region 2 (R2) signifies the viable CD154+ and CD154− T cells. The percentage of divided CD3+CD4+ T cells from R2, restimulated with irradiated PBMCs and RHu-IL-2, is calculated from the dilution of CFSE dye as a result of cell division (C histograms on right).
Irrespective of the HLA-DR mismatch, the alloreactive C154+CD4+ cells did not have characteristics of regulatory T cells as they were not CD25bright and CD127dim before culture (Figure 4) and were FoxP3 negative (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
To investigate the level of bystander activation, combinations of HLA-DR mismatches were made such that party A had 0 HLA-DR mismatches compared with CFSE-labeled party B (generating HLA-DR–matched alloreactive CD154+CD4+ cells in party B), whereas party B had 1 HLA-DR mismatch with party A (generating HLA-DR–matched and –mismatched alloreactive CD154+CD4+ cells in party A). These experiments did not show bystander activation of HLA-DR–matched CD154+CD4+ cells by the HLA-DR–mismatched CD154+CD4+ cells, as the median expression of CD25 and CD127 at 24 hours (median fluorescence intensity [MFI] CD25: 2640, MFI CD127: 3250) was similar to CD154+CD4+ cells from the initial experiments with 0-0 HLA-DR–matched combinations (MFI CD25: 2856, MFI CD127: 2587).
As expected, HLA-DR–matched alloreactive CD4+ cells (cells from party A) did not proliferate when a 1-way MLR was performed (stimulator cells from party B were irradiated to prevent proliferation; data not shown). However, proliferation of isolated alloreactive CD154+CD4+ cells from HLA-identical combinations could be induced by restimulation with both irradiated HLA-identical stimulator PBMCs (antigen-presenting cells) and exogenous IL-2 (Figure 4C top panel). The CD154−CD4+ cells from HLA-identical combinations did not show significant proliferation above background levels in the presence of stimulator PBMCs and exogenous IL-2 (Figure 4C bottom panel). Little proliferation was observed if CD154+CD4+ cells from HLA-identical combinations were restimulated with irradiated autologous PBMCs. In addition, restimulation with third-party (2:2:2 MMs on HLA-A:B:DR) and recombinant IL-2 showed no significant increased proliferation compared with the autologous control (supplemental Figure 2).
From these data, we conclude that alloreactive T cells generated from HLA-DR–matched combinations express the receptors for growth factors IL-2 and IL-7, but proliferate when only IL-2 is available, for example, from IL-2–producing HLA-mismatched alloreactive T cells.
Cytokine expression patterns of alloreactive CD154+CD4+ cells show a dominance of Th17 cells
The cytokines interferon-gamma (IFN-γ), interleukin-2 (IL-2), and IL-10 are best documented for their relation to acceptance and rejection of transplanted organs. In addition, we included expression of IL-17 in our analysis as this cytokine identifies a recently identified inflammatory T-cell subset (Th17 cells).
In general, cytokine expression in CD4+ cells after allogeneic stimulation gave no detectable responses above background level (on average 0.01%) in the total CD4+ T-cell population, confirming published data by others.18,19 Varying time points from 6 to 24 hours and increasing the number of CD4+ T cells for analysis to 0.5 to 1 million did not change these results. Only when we analyzed intracellular cytokine staining in the alloreactive CD154+CD4+ cells were we able to detect positive cells, albeit at low frequency (Figure 5). Remarkably, alloreactive CD4+ cells expressed IL-17 (median, 0.7%; range, 0%-17.1%), which normally does not exceed 1% of the total number of polyclonal stimulated CD4+ T cells (Figure 5A). Specifically, IL-2 expression showed a clear relation with the number of HLA-DR mismatches, with essentially no expression of IL-2 in HLA-DR–matched combinations. The percentages of alloreactive CD154+CD4+ cells expressing inflammatory cytokines such as IFN-γ and IL-17 were the highest within the HLA-DR–mismatched combination (Figure 6).
Alloantigen-specific cytokine-producing CD3+CD4+ T cells. A typical flow cytometric example of cytokine-producing cells is shown in panel A. For this purpose, labeled PBMCs were stimulated with unlabeled PBMCs of a different donor at a 1:1 ratio for 6 hours in the presence of soluble costimulation. As a negative control, these PBMCs were stimulated with unlabeled autologous PBMCs, whereas CMV and the combination of PMA and ionomycin were used as positive controls, the first serving as another antigen-specific stimulus and the latter as polyclonal stimulus. Percentages are of CD3+CD4+ T lymphocytes, whereas in brackets the percentage of cytokine-positive cells within CD154+CD4+ cells is given. (B) A summary of the results of 11 independent experiments, each using 2 different donors, where cytokine-producing cells are identified as CD3+CD4+ or subsequently using intracellular staining as positive for CD154, IL-2, IFN-γ, IL-10, or IL-17. (C) We determined the fraction of cytokine-positive cells within the CD154+ fraction.
Alloantigen-specific cytokine-producing CD3+CD4+ T cells. A typical flow cytometric example of cytokine-producing cells is shown in panel A. For this purpose, labeled PBMCs were stimulated with unlabeled PBMCs of a different donor at a 1:1 ratio for 6 hours in the presence of soluble costimulation. As a negative control, these PBMCs were stimulated with unlabeled autologous PBMCs, whereas CMV and the combination of PMA and ionomycin were used as positive controls, the first serving as another antigen-specific stimulus and the latter as polyclonal stimulus. Percentages are of CD3+CD4+ T lymphocytes, whereas in brackets the percentage of cytokine-positive cells within CD154+CD4+ cells is given. (B) A summary of the results of 11 independent experiments, each using 2 different donors, where cytokine-producing cells are identified as CD3+CD4+ or subsequently using intracellular staining as positive for CD154, IL-2, IFN-γ, IL-10, or IL-17. (C) We determined the fraction of cytokine-positive cells within the CD154+ fraction.
Alloantigen-specific cytokine-producing cells and relation with number of mismatches on HLA-DR. (A) Flow cytometric example of the IL-2/IFN-γ– and IFN-γ/IL-17–producing cells within the CD154+CD4+ cells for a couple with 0 (Ai and Aii) and 2 (Aiii and Aiv) mismatches (MMs) on HLA-DR, respectively. The results of n = 4 for 0 MM, n = 10 for 1 MM, on HLA-DR and n = 8 for 2 MMs on HLA-DR are displayed. The fraction of (B) IL-2–, (C) IFN-γ–, (D) IL-10–, and (E) IL-17–producing cells within the CD154+CD4+ lymphocytes is shown on the y-axis, whereas on the x-axis the number of MMs on HLA-DR is depicted. Each horizontal line represents the mean. *P < .05.
Alloantigen-specific cytokine-producing cells and relation with number of mismatches on HLA-DR. (A) Flow cytometric example of the IL-2/IFN-γ– and IFN-γ/IL-17–producing cells within the CD154+CD4+ cells for a couple with 0 (Ai and Aii) and 2 (Aiii and Aiv) mismatches (MMs) on HLA-DR, respectively. The results of n = 4 for 0 MM, n = 10 for 1 MM, on HLA-DR and n = 8 for 2 MMs on HLA-DR are displayed. The fraction of (B) IL-2–, (C) IFN-γ–, (D) IL-10–, and (E) IL-17–producing cells within the CD154+CD4+ lymphocytes is shown on the y-axis, whereas on the x-axis the number of MMs on HLA-DR is depicted. Each horizontal line represents the mean. *P < .05.
Next, we isolated and stimulated alloreactive CD154+CD4+ cells polyclonal with PMA-ionomycin. Compared with CD154−CD4+ cells, the alloreactive CD154+CD4+ cells showed a similar expression of IFN-γ and IL-10, but a higher expression of IL-2 (median, 51.5% vs 29.5% of the CD154−CD4+ cells; P < .05) and a strikingly higher expression of IL-17 (median, 13.4%; range, 7.2%-67.2% vs 0.8%; range, 0.2%-1.3% of the CD154−CD4+ cells; P < .001). Alloreactive CD154+CD4+ cells with a naive phenotype were predominantly single IL-2+ but some double-positive IL-2/IL-17 CD4+ cells were noted as well. The contribution of CD154+CD4+ cells coexpressing different cytokines increased with the differentiation from central-memory to effector-memory T cells (Figure 7).
Cytokine profiles of CD154+ and CD154− CD3+CD4+ T lymphocytes after polyclonal stimulation. A representative flow cytometric example of the cytokine profile of CD154+ and CD154− CD3+CD4+ T lymphocytes after a 6-hour stimulation with either costimulation (cos only) alone or combined with PMA and ionomycin (P/I) is shown in panel A. For this purpose, alloantigen-specific CD154+ T cells are isolated upon a 24-hour coculture of PBMCs of 2 different donors using magnetic cell separation. These fractions were stimulated with costimulation alone or a polyclonal stimulus. Cytokine-producing cells are enumerated using flow cytometry staining for IL-2 in combination with either IFN-γ (top panel), IL-17 (second panel), or IL-10 (third panel). In addition, IFN-γ was also combined with IL-17 (bottom panel). Averages (+ SEM) of 10 independent experiments of total cytokine-producing CD3+CD4+ T lymphocytes compared between the CD154+ (closed bars) and CD154− (open bars) fraction are depicted in panel B. Next, we analyzed the total percentages of cytokine-producing cells for each CD4+CD154+ T-cell subset. White bars represent Tnaive; gray bars, Tcm; and the black bars, Tem cytokine-producing cells (C). These were subsequently dissected to display single-positive and double-positive cytokine expression profiles of these CD3+CD4+ CD154+ T lymphocytes within the different T-cell subsets being naive (Di), Tcm (Dii), and Tem (Diii) using CCR7 and CD45RO monoclonal antibodies (D). *Significantly different (P < .05).
Cytokine profiles of CD154+ and CD154− CD3+CD4+ T lymphocytes after polyclonal stimulation. A representative flow cytometric example of the cytokine profile of CD154+ and CD154− CD3+CD4+ T lymphocytes after a 6-hour stimulation with either costimulation (cos only) alone or combined with PMA and ionomycin (P/I) is shown in panel A. For this purpose, alloantigen-specific CD154+ T cells are isolated upon a 24-hour coculture of PBMCs of 2 different donors using magnetic cell separation. These fractions were stimulated with costimulation alone or a polyclonal stimulus. Cytokine-producing cells are enumerated using flow cytometry staining for IL-2 in combination with either IFN-γ (top panel), IL-17 (second panel), or IL-10 (third panel). In addition, IFN-γ was also combined with IL-17 (bottom panel). Averages (+ SEM) of 10 independent experiments of total cytokine-producing CD3+CD4+ T lymphocytes compared between the CD154+ (closed bars) and CD154− (open bars) fraction are depicted in panel B. Next, we analyzed the total percentages of cytokine-producing cells for each CD4+CD154+ T-cell subset. White bars represent Tnaive; gray bars, Tcm; and the black bars, Tem cytokine-producing cells (C). These were subsequently dissected to display single-positive and double-positive cytokine expression profiles of these CD3+CD4+ CD154+ T lymphocytes within the different T-cell subsets being naive (Di), Tcm (Dii), and Tem (Diii) using CCR7 and CD45RO monoclonal antibodies (D). *Significantly different (P < .05).
The majority of IL-17+ CD4+ cells had a memory phenotype (on average 90%), coexpressed IL-2 (on average 50%), and did not express IFN-γ. Alloreactive IL-17+ CD4+ cells expressed the combination of chemokine receptors CCR4 and CCR6, in accordance with published data on Th17 cells (data not shown). Polyclonal-stimulated alloreactive CD154+CD4+ cells from HLA-identical combinations showed a normal frequency of IL-2+ CD4+ cells and a similar high frequency of IL-17–producing CD4+ cells (n = 4; median, 27%; range, 6%-58%).
Discussion
The lack of specific markers has prevented in-depth research of the role of human alloreactive CD4+ T cells in graft rejection or tolerance. In this study, we show that using a modified CD154 live assay, human alloreactive CD4+ T cells can be specifically identified and characterized after short-term stimulation. Using this approach, we observed several novel findings.
After engagement of the T-cell receptor, CD154 is rapidly synthesized and expressed on the cell within 1 to 2 hours, reaching a maximum expression at 6 hours. Similar kinetics was observed for alloreactive T cells. In a subset of human memory T cells, the CD154 is located in intracellular vesicles that translocate to the cell surface within minutes after stimulation, thereby ensuring a very fast CD40L-CD40 interaction.20,21 However, our results with alloreactive T cells do not indicate such a mechanism, as within 2 hours CD154 was not detectable.
CD154 expression on antigen or polyclonal stimulated CD4+ T cells was completely lost at 12 hours, but reappeared and once again became maximal at 24 hours. This pattern of CD154 expression was not mentioned in the initial study on the live CD154 assay, as the time points between 8 and 24 hours were apparently not monitored.14 However, the same typical biphasic pattern of CD154 expression after polyclonal stimulation of human CD4+ T cells was recently reported.22,23
After depletion of the alloreactive CD154+CD4+ cells, the proliferative capacity of the allogeneic PBMCs was almost totally abolished, including the proliferation of CD8+ T cells. The latter observation confirms previous results with fractionated allogeneic T cells showing the dependence of allogeneic CD8+ T-cell proliferation on the presence of allogeneic CD4+ T cells.24 Therefore, the CD154 positivity seems to identify the full allogeneic CD4+ T-cell repertoire classically defined as the capacity of T cells to proliferate in a mixed leukocyte cell culture. Of course, this does not exclude the possibility that some alloreactive CD4+ T cells are not recognized with this assay because they are negative for CD154. However, adding to the specificity of the CD154 assay, we were unable to identify alloreactive cytokine-positive CD4+ cells in the CD154−CD4+ cell fraction.
The percentages of alloreactive CD4+ cells within the PBMCs as observed with the CD154 live assay are within the presumed frequency (0.1%–0.01%).25 In addition, a significant part of the alloreactive CD154+CD4+ cells showed a naive T-cell phenotype, whereas CMV antigen–specific CD154+CD4+ cells have the expected memory phenotype. Surprisingly, the frequency of alloreactive CD154+CD4+ cells was inversely correlated with the number of HLA-DR mismatches. The percentage of alloreactive cells with fully matched HLA-DR combinations was on average 10-fold more than fully HLA-DR–mismatched combinations. A number of studies have suggested that HLA molecules themselves may provide antigenic peptides for allorecognition.2 However, HLA-identical combinations also yielded a relative high frequency of alloreactive CD154+CD4+ cells. Therefore, our finding indicates a much larger than expected frequency of alloreactive CD4+ cells that recognizes mHags. These cells do not proliferate in a MLR, which can be readily understood, as no intracellular IL-2 could be observed after direct allogeneic stimulation. The reason for the disparity between CD154 positivity and the lack of cytokine production is as yet unknown. As IL-2 production is dependent on the strength of T-cell receptor (TCR) stimulation, a plausible hypothesis could be that mHags are associated with weak TCR stimulation that allows for CD154 induction but not cytokine production. The importance of IL-2 for alloreactivity is well recognized and clinically exploited by the use of anti–IL-2 receptor antibodies to prevent organ graft rejection.26 However, the high frequency of IL-2, IFN-γ, and IL-17 positivity after polyclonal stimulation of HLA-DR–matched alloreactive CD154+CD4+ cells indicates the potential to become inflammatory effector cells. The lack of IL-2 production after allogeneic stimulation prohibits proliferation and has made isolation of mHag-specific CD4 T cells very difficult in the past. However, this problem can now be easily circumvented by isolation of allogeneic CD154+CD4+ cells with subsequent exogenous IL-2–induced proliferation. The latter may also be the mechanism that provides the route for these cells to become of pathologic importance after bone marrow or solid organ transplantation.
We were able to identify cytokine production of CD154+CD4+ cells on the single-cell level. The percentage of cytokine-positive cells was low and could be reliable detected only within the CD154+CD4+ cell fraction, again underscoring that expression of CD154 is the most comprehensive marker for CD4+ T-cell alloreactivity. The high percentage of cytokine-negative alloreactive CD154, even in fully HLA-DR–mismatched combinations, is remarkable and can only in part be explained by the high percentage of naive CD154+CD4+ cells. As polyclonal stimulation induced normal frequencies of cytokine-producing cells, one must assume that the interactions of allogeneic TCRs with HLA are strong enough to generate the costimulatory signal of CD154 and expression of IL-2 receptors at a later time point, but to a large extent not the pivotal cytokine IL-2. The cytokine-negative alloreactive CD154+CD4+ cells can probably be driven to proliferation in a paracrine fashion by proliferating, IL-2–producing alloreactive CD154+CD4+ cells from HLA-DR–mismatched combinations. Therefore, at least in vitro, a spark of IL-2 seems sufficient to light the fire of a complete allogeneic T-cell response.
Finally, we observed a Th17 response in the alloreactive memory CD154+CD4+ cells that has not been reported previously. After direct allogeneic stimulation, the IL-17–producing T cells are proportionally higher in the alloreactive CD4+ T-cell population than expected based on the frequency of IL-17–producing cells overall. In some donor combinations, we identified a more than 75% Th17 response among the alloreactive memory CD4+ cells after polyclonal stimulation. Present data indicate the importance of Th17 cells in the cellular host defense against bacteria and specifically Candida albicans, and Th17 cells may have an autoreactive capacity.27 Therefore, it is tempting to speculate that alloreactive Th17 cells represent antigenic cross-reactivity, suggesting that the alloantigens consist of epitopes that harbor similarities with those of certain pathogens known to induce Th17 cells. At present, there are no experimental data available to support such a hypothesis.
The functional role of Th17 cells in human organ transplantation is yet to be defined, but a remarkably high intragraft expression of IL-17 was recorded during acute cellular rejection of human kidney transplants.28,29 In addition, CD154 expression on effector T cells enhances the proinflammatory effects of IL-17 on proximal tubulus cells.30 Blocking the activity of IL-17 prevented early vascular graft rejection but not chronic vasculopathy in an animal model of transplantation.31 The presence of IL-17 was also shown to enhance early but not late graft-versus-host disease severity after allogeneic bone marrow transplantation.32
Combined with the finding that HLA class II mismatches are particularly important in early but not late human kidney allograft rejection,33 the present data indicate that IL-2–secreting alloreactive Th17 cells (as observed in HLA-DR–mismatched combinations) may be pivotal in early graft rejection. The emergence of Th17 cells specific for the minor antigen collagen type V, causing obliterative bronchiolitis in recipients of human lung transplants, provides a clear example of the potential pathogenic role of Th17 cells in HLA class II–matched transplantation.34 Therefore, targeting or depletion of alloreactive CD154+ Th17 cells could provide a new therapeutic strategy to prevent early allograft rejection and/or graft-versus-host disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: N.H.R.L. performed the experiments and wrote the paper; J.v.d.W. selected HLA-identical subject; N.M.v.B. helped with some statistical analysis; and M.G.H.B. designed the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michiel G. H. Betjes, Division of Nephrology, Department of Internal Medicine, Erasmus Medical Center, PO Box 2040, 3000 CA Rotterdam, The Netherlands; e-mail: m.g.h.betjes@erasmusmc.nl.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal