Abstract
The first adult-repopulating hematopoietic stem cells (HSCs) are detected starting at day 10.5 of gestation in the aorta-gonads-mesonephros (AGM) region of the mouse embryo. Despite the importance of the AGM in initiating HSC production, very little is currently known about the regulators that control HSC emergence in this region. We have therefore further defined the location of HSCs in the AGM and incorporated this information into a spatial and temporal comparative gene expression analysis of the AGM. The comparisons included gene expression profiling (1) in the newly identified HSC-containing region compared with the region devoid of HSCs, (2) before and after HSC emergence in the AGM microenvironment, and (3) on populations enriched for HSCs and their putative precursors. Two genes found to be up-regulated at the time and place where HSCs are first detected, the cyclin-dependent kinase inhibitor p57Kip2/Cdkn1c and the insulin-like growth factor 2, were chosen for further analysis. We demonstrate here that they play a novel role in AGM hematopoiesis. Interestingly, many genes involved in the development of the tissues surrounding the dorsal aorta are also up-regulated during HSC emergence, suggesting that the regulation of HSC generation occurs in coordination with the development of other organs.
Introduction
Hematopoietic stem cells (HSCs) are at the center of the hematopoietic system. Their proliferative and multilineage differentiation potential endows them with the capacity to regenerate every blood cell type throughout the entire life of a person. For this reason, they are subject to tight regulatory processes to ensure an adequate supply of blood cells without the risk of HSC depletion or the development of blood-related malignancies.
Insight into the basic mechanisms of HSC regulation can be gained from the study of how these cells are first generated and expanded during development. The first cells that display adult HSC characteristics in transplantation assays are detected at embryonic day (E) 10.5 in a region of the embryo that includes the developing dorsal aorta, gonads, and mesonephros (AGM region).1 Subdissections have localized this first HSC activity to the dorsal aorta and the associated vitelline and umbilical vessels.2 Subsequently, adult HSC-type cells are also found in the yolk sac and the placenta.1,3,4 It is currently unclear why HSCs are harbored in multiple sites during development and whether they have multiple origins. After E12, HSC numbers start declining in the AGM as the fetal liver (FL) becomes colonized and an adult-type hematopoietic system is further established (reviewed in Dzierzak and Speck5 ).
Little is known about how HSCs are first generated in the AGM. In particular, the identification of the direct precursors of HSCs, the definition of the components of the regulatory microenvironment, and the discovery of cell-intrinsic regulators are issues that have only recently started to be addressed. There is now good evidence to suggest that HSCs in the AGM are either of endothelial origin or transit through an endothelial-like state during their generation.6,7 These hemogenic endothelial cells are thought to give rise to intra-aortic clusters of cells, which appear to be the first sign of HSC emergence in the dorsal aorta (reviewed in Ottersbach and Dzierzak8 ). Other reports suggest that there are HSC precursors in the ventral mesenchyme underneath the aorta that can give rise to HSCs, possibly via an endothelial-like intermediate.9,10 Indeed, these suggestions of either a mesenchymal or endothelial origin for hematopoietic cells are not necessarily mutually exclusive. Rather, as supported from recent studies using the embryonic stem cell system, they represent sequential steps in the same pathway.11
Even less is known about the architecture of the HSC regulatory microenvironment in the AGM. Comparison of the gene repertoire expressed by AGM-derived supportive and nonsupportive stromal cell lines has led to the identification of novel HSC regulators.12-14 Although expression studies on these cell lines provide some information into the hematopoietic supportive microenvironment, the gene repertoire of primary AGM tissue has not been examined.
With the aim to identify novel in vivo regulators of AGM hematopoiesis, we analyzed and compared the gene expression profiles of 3 different spatially and temporally isolated AGM primary tissue/cell preparations: (1) regions within the aorta that do or do not contain HSCs, (2) whole dorsal aortae before and after HSC emergence, and (3) cell populations enriched for HSCs and their putative precursors. We present here the list of up- and down-regulated genes and functional data suggesting that the cell cycle regulator p57Kip2 (Cdkn1c) and the growth factor insulin-like growth factor 2 (Igf2) are important regulators of AGM hematopoiesis.
Methods
Mice and tissue preparations
To obtain embryos, timed matings were set up between male wild-type (C57BL/10 × CBA)F1, Ly-6A GFP,15 or line 72 human β-globin16 transgenic and wild-type (C57BL/10 × CBA)F1 females, and between female p57Kip2+/−17 and C57BL/6 males homozygous for the Ly5.1 isoform. The day of vaginal plug observation was considered E0. On the appropriate day of gestation, females were killed and the embryos removed for dissections or morphologic analyses. All animals were housed according to institutional guidelines and experiments complied with the animal welfare laws. Animal studies were approved by the United Kingdom Home Office.
Long-term repopulation assays
Embryonic tissues were dissociated by collagenase treatment as described previously.18 Single-cell suspensions were coinjected intravenously with 2 × 105 spleen cells into irradiated mice (split dose of 9 Gy). Donor cell contribution to the peripheral blood was determined at 1 and 4 months after transplantation and recipients were considered positive when the donor marker showed at least 10% contribution to the peripheral blood by semiquantitative polymerase chain reaction (PCR) or 5% by Ly5 isoform-specific fluorescence-activated cell sorting (FACS) analysis.
Gene expression analysis
Embryonic tissues were dissected, pooled, and dissociated directly in Trizol Reagent (Invitrogen), or Ly-6A green fluorescent protein–positive (GFP+) cells were sorted from single-cell suspensions on a FACSVantage (BD Biosciences) directly into Trizol Reagent (total of 17 000 GFP+ from E9 aortae and 48 000 GFP+ cells from E11 aortae). Total RNA was isolated according to the manufacturer's instructions, DNase treated, and mRNA amplified by either 1 round (for unsorted embryonic tissues) or 2 rounds (for sorted cell populations) of linear T7-mediated amplification as described previously.19 Amplified RNA probes were labeled with either cyanin 3 (Cy3)– or Cy5-conjugated deoxyuridine triphosphate nucleotides during the reverse-transcription reaction and labeled probes hybridized overnight at 42°C to 15K mouse cDNA arrays obtained from the Netherlands Cancer Institute. Dye-swap and self-self hybridization experiments were included as controls. Microarrays were scanned with a ScanArray Express Microarray Scanner (PerkinElmer) and spot signals quantified using the Imagene 5 software (Biodiscovery). Normalization, analysis of variance, and K means clustering were performed using the Spotfire Decision Site software (TIBCO Spotfire). Detailed descriptions of the protocols and the raw and normalized data are deposited at the European Bioinformatics Institute's microarray data depository ArrayExpress (http://www.ebi.ac.uk/microarray-as/ae) under the accession number E-TABM-659.20 Biologic processes that are enriched in the total up-regulated or down-regulated population were determined using the freely accessible software DAVID (National Institute of Allergy and Infectious Diseases, http://david.abcc.ncifcrf.gov/summary.jsp) with all of the genes printed on the array serving as background.
RT-PCR, in situ hybridization, and immunohistochemistry
For the examination of the expression of Igf2 and its receptors in the different embryonic tissues by reverse-transcription–PCR, previously published primer sequences were used.21 Mouse β-actin (Actb) primer sequences are forward: CCTGAACCCTAAGGCCAACCG, reverse: GCTCATAGCTCTTCTCCAGGG. DNA fragments for the synthesis of riboprobes for in situ hybridization experiments were generated by RT-PCR from E11 AGM cDNA using the following primers: Th, forward: ATTGGAGGCTGTGGTATTCG, reverse: GGGTAGCATAGAGGCCCTTC; Cryab, forward: AGCTGCTGCTGAAGGAGTTG, reverse: CCAGACACCTGTTTCCTTGG; p57Kip2, forward: CTGACCTCAGACCCAATTCC, reverse: GATGCCCAGCAAGTTCTCTC. The Igf2 riboprobe was generated from a plasmid containing an Igf2 fragment encompassing nucleotides 1749-2169 (accession number NM 010514).22 Riboprobe synthesis, in situ hybridization experiments, and immunohistochemistry with antibodies against CD34 (BD Biosciences) and p57Kip2 (Santa Cruz Biotechnology) were carried out as previously described.4
AGM explant cultures and colony-forming assays
E11 AGMs were dissected and cultured at the air/liquid interface in M5300 myelocult (StemCell Technologies) supplemented with 10−5 M hydrocortisone (Sigma-Aldrich) in the presence or absence of recombinant mouse Igf2 (R&D Systems) at the indicated concentrations. After 3 days, AGMs were pooled, dissociated by collagenase treatment, and plated in triplicate as 10 000, 50 000, and 100 000 cells/mL in M3434 methylcellulose medium (StemCell Technologies). Colonies were scored after 7 days. Freshly dissected, uncultured E11 AGMs were also dissociated and plated in methylcellulose medium for comparison. As there was some variation in total colony numbers between experiments, we converted the colony composition into percentages for each individual experiment and calculated the average, standard deviation, and P values from those individual percentages.
Results
HSCs are enriched in the middle third of the E11 dorsal aorta
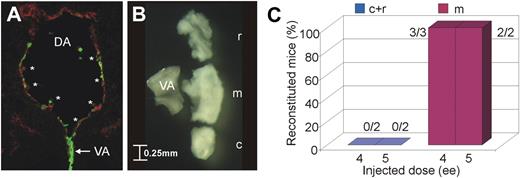
To facilitate the identification of new regulators, our initial aim was to further define the region within the AGM where HSCs are located. Upon examination of sections prepared from E11 Ly-6A GFP transgenic embryos, which express the green fluorescent protein (GFP) in all embryonic HSCs,4,15 it was noted that intra-aortic clusters were particularly abundant in the segment of the dorsal aorta adjacent to where the vitelline artery joins the dorsal aorta (Figure 1A clusters marked with asterisks; vitelline artery indicated by white arrow). Therefore, the aorta was cut into 3 equal parts: rostral (r), middle with the vitelline artery maintained for orientation (m) and caudal (c; Figure 1B). The caudal and rostral parts were pooled (c + r). The vitelline artery was removed from the middle region and the HSC activity in the c + r and m samples was determined by transplantation assay. As expected from the distribution of hematopoietic clusters, HSCs were found exclusively in the middle region of the aorta (Figure 1C). The expression profile of the caudal and rostral part was then compared with that of the HSC-rich middle region of the dorsal aorta.
HSC localization. (A) Transverse cryosection of an E11 Ly-6A GFP+ embryo (GFP in green) costained for the endothelial marker CD34 (red, Cy5; mounted with Vectashield; Vector Laboratories) showing the dorsal aorta with intra-aortic clusters marked by asterisks and the joining vitelline artery indicated by a white arrow. Ventral, down. DA indicates dorsal aorta; and VA, vitelline artery. Pictures were taken on a Zeiss LSM510NLO/FCS confocal microscope with a 40×/1.3 NA objective and images were analyzed with Zeiss LSM image software (both from Carl Zeiss BV). (B) Subdissection of an embryonic day (E) 11 dorsal aorta. Ventral, left; rostral, top. r indicates rostral; m, middle; and c, caudal. Aortae were dissected under a Nikon SMZ800 dissection microscope (Nikon), pictures were taken with a Pixera Pro 150ES camera (Pixera), and images were analyzed with the Pixera Viewfinder software (Pixera). (C) Percentage of mice reconstituted with the different parts of the E11 dorsal aorta (rostral and caudal parts pooled) after transplantation into irradiated mice. The number of pooled, transplanted tissues (ie, m or r + c) is indicated as embryo equivalent (ee) and the number of repopulated mice/number of mice injected is indicated for each bar.
HSC localization. (A) Transverse cryosection of an E11 Ly-6A GFP+ embryo (GFP in green) costained for the endothelial marker CD34 (red, Cy5; mounted with Vectashield; Vector Laboratories) showing the dorsal aorta with intra-aortic clusters marked by asterisks and the joining vitelline artery indicated by a white arrow. Ventral, down. DA indicates dorsal aorta; and VA, vitelline artery. Pictures were taken on a Zeiss LSM510NLO/FCS confocal microscope with a 40×/1.3 NA objective and images were analyzed with Zeiss LSM image software (both from Carl Zeiss BV). (B) Subdissection of an embryonic day (E) 11 dorsal aorta. Ventral, left; rostral, top. r indicates rostral; m, middle; and c, caudal. Aortae were dissected under a Nikon SMZ800 dissection microscope (Nikon), pictures were taken with a Pixera Pro 150ES camera (Pixera), and images were analyzed with the Pixera Viewfinder software (Pixera). (C) Percentage of mice reconstituted with the different parts of the E11 dorsal aorta (rostral and caudal parts pooled) after transplantation into irradiated mice. The number of pooled, transplanted tissues (ie, m or r + c) is indicated as embryo equivalent (ee) and the number of repopulated mice/number of mice injected is indicated for each bar.
Because of possible sensitivity issues and to add a temporal component to the expression analysis, 2 other sample sets were also prepared. The first sample set consisted of dorsal aortae from E9/E10 embryos (25-30 somite pairs [SPs]), when no HSCs can yet be detected by direct transplantation into adult recipients, and from E11 embryos (45-52 SPs), after HSCs have emerged in that tissue.2 Aortae were dissected with the immediate surrounding mesenchyme and their expression profiles compared. In this approach, gene expression changes in pre-HSCs/HSCs as well as their microenvironment can be monitored.
The second temporal sample set was designed to focus specifically on the expression changes that take place as pre-HSCs mature to become adult-repopulating HSCs. Enriched populations of HSCs and their putative precursors were isolated from Ly-6A GFP transgenic embryos. At E11, GFP+ cells can be detected in numerous endothelial cells of the dorsal aorta, where they are preferentially located on the ventral and lateral sides (Figure 1A). Individual GFP+ cells can also be found in the circulation and within the intra-aortic clusters. Endothelial cells of the vitelline artery are also highly GFP+. Although no HSC activity can be detected in the E9 aorta by direct transplantation into adult recipients, a limited number of Ly-6A GFP+ cells can already be observed. These are initially restricted to the ventral side (data not shown), where HSC activity is thought to originate. The number of GFP+ cells increases from 0.7% at E9 to 3% in the E11 aorta, suggesting a direct lineage relationship (pre-HSCs → HSCs) between the Ly-6A GFP+ cells at these 2 time points. Ly-6A GFP+ cells were sorted from E9 and E11 aortae for gene expression analysis.
The E11 aorta shows an up-regulation in genes involved in several different developmental processes
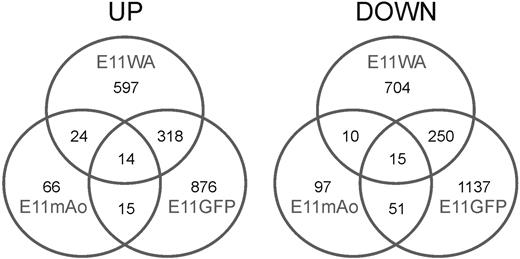
Probes from the 3 different sample sets were hybridized to 15K mouse cDNA arrays. The cDNA library that is spotted on these arrays was constructed from whole embryos and tissues from different stages and is therefore particularly enriched in developmentally relevant genes.23 Differentially expressed genes with a P value of .01 or less were identified through analysis of variance using the Spotfire Decision Site software package. The number of genes found to be “up-regulated” (ie, more highly expressed in E11 whole aortae [WA], E11 Ly-6A GFP+ cells, and in the middle region of the E11 aorta [mAo]) and “down-regulated” (ie, more highly expressed in the E9 aorta, E9 Ly-6A GFP+ cells, and in the rostral and caudal parts of the E11 aorta) is summarized in the 2 Venn diagrams in Figure 2, and the individual genes are listed in supplemental Table 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). We also analyzed the data with the limma software package (Walter and Eliza Hall Institute of Medical Research, http://bioinf.wehi.edu.au/limma)24 and identified a very similar set of genes (data not shown). More genes were found to be differentially expressed in the 2 temporal sample sets (E11WA and E11GFP) compared with the expression analysis of the different parts of the E11 aorta (E11mAo), and the overlap between the 2 temporal sample sets was approximately 30%. The genes found to be commonly up- or down-regulated in all 3 comparisons are listed in Table 1. The proteins encoded by the commonly up-regulated genes have a variety of different biologic functions and include, for example, 2 enzymes (Them4 and Rnaset2b), 2 ATPase subunits, a small heat shock protein (Cryab), a translation initiation factor (Eif3f), and 2 cell cycle regulators (Ccnd1 and p57Kip2/Cdkn1c). Approximately one-third of the commonly down-regulated genes code for proteins that are involved in gene regulation.
Number of differentially expressed genes. Venn diagrams showing (left) the number of genes more highly expressed in the E11 whole aorta (E11WA), E11 Ly-6A GFP+ cells (E11GFP), and the middle part of the E11 aorta (E11mAo), and (right) the number of genes showing the opposite expression pattern.
Number of differentially expressed genes. Venn diagrams showing (left) the number of genes more highly expressed in the E11 whole aorta (E11WA), E11 Ly-6A GFP+ cells (E11GFP), and the middle part of the E11 aorta (E11mAo), and (right) the number of genes showing the opposite expression pattern.
Genes differentially expressed in all 3 comparisons
| GenBank ID22 . | Gene name . | Gene symbol . | Accession no. . |
|---|---|---|---|
| Genes commonly up-regulated | |||
| BG062951 | Eukaryotic translation initiation factor 3, subunit F | Eif3f | NM 025344 |
| BG069380 | Thioesterase superfamily member 4 | Them4 | NM 029431 |
| BG071234 | Formin binding protein 1 | Fnbp1 | NM 019406 |
| BG071313 | Cyclin-dependent kinase inhibitor 1C (p57Kip2) | Cdkn1c | NM 009876 |
| BG075084 | Crystallin, alpha B | Cryab | NM 009964 |
| BG075879 | Proteolipid protein (myelin) 1 | Plp1 | NM 011123 |
| BG076602 | Cut-like homeobox 1 (Cux1), transcript variant 2 | Cux1 | NM 198602 |
| BG077799 | ATPase, H+ transporting, lysosomal V0 subunit C | Atp6v0c | NM 009729 |
| BG078776 | ATPase, H+ transporting, lysosomal V0 subunit E | Atp6v0e | NM 025272 |
| BG079781 | Ribonuclease T2B | Rnaset2b | NM 026611 |
| BG083088 | Cyclin D1 | Ccnd1 | NM 007631 |
| BG086406 | Angiotensin II receptor, type 2 | Agtr2 | NM 007429 |
| BG066003 | Unknown | ||
| C78810 | Unknown | ||
| Genes commonly down-regulated | |||
| BG063141 | NudC domain containing 1 | Nudcd1 | NM 026149 |
| BG064241 | Protein phosphatase 1, regulatory (inhibitor) subunit 15b | Ppp1r15b | NM 133819 |
| BG064278 | Nucleosome assembly protein 1-like 1 | Nap1l1 | NM 015781 |
| BG065281 | Glia maturation factor, beta | Gmfb | NM 022023 |
| BG067688 | Heterogeneous nuclear ribonucleoprotein A3 | Hnrnpa3 | NM 146130 |
| BG069073 | Transformer 2 alpha homolog (Drosophila) | Tra2a | NM 198102 |
| BG072727 | Bmi1 polycomb ring finger oncogene | Bmi1 | NM 007552 |
| BG077079 | A disintegrin and metallopeptidase domain 17 | Adam17 | NM 009615 |
| BG082860 | WD repeat domain 77 | Wdr77 | NM 027432 |
| BG085072 | WAP 4-disulfide core domain 2 | Wfdc2 | NM 026323 |
| BG086014 | High mobility group nucleosomal binding domain 1 | Hmgn1 | NM 008251 |
| AW542440 | Unknown | ||
| BG070468 | Unknown | ||
| BG069520 | Unknown | ||
| BG070685 | Unknown |
| GenBank ID22 . | Gene name . | Gene symbol . | Accession no. . |
|---|---|---|---|
| Genes commonly up-regulated | |||
| BG062951 | Eukaryotic translation initiation factor 3, subunit F | Eif3f | NM 025344 |
| BG069380 | Thioesterase superfamily member 4 | Them4 | NM 029431 |
| BG071234 | Formin binding protein 1 | Fnbp1 | NM 019406 |
| BG071313 | Cyclin-dependent kinase inhibitor 1C (p57Kip2) | Cdkn1c | NM 009876 |
| BG075084 | Crystallin, alpha B | Cryab | NM 009964 |
| BG075879 | Proteolipid protein (myelin) 1 | Plp1 | NM 011123 |
| BG076602 | Cut-like homeobox 1 (Cux1), transcript variant 2 | Cux1 | NM 198602 |
| BG077799 | ATPase, H+ transporting, lysosomal V0 subunit C | Atp6v0c | NM 009729 |
| BG078776 | ATPase, H+ transporting, lysosomal V0 subunit E | Atp6v0e | NM 025272 |
| BG079781 | Ribonuclease T2B | Rnaset2b | NM 026611 |
| BG083088 | Cyclin D1 | Ccnd1 | NM 007631 |
| BG086406 | Angiotensin II receptor, type 2 | Agtr2 | NM 007429 |
| BG066003 | Unknown | ||
| C78810 | Unknown | ||
| Genes commonly down-regulated | |||
| BG063141 | NudC domain containing 1 | Nudcd1 | NM 026149 |
| BG064241 | Protein phosphatase 1, regulatory (inhibitor) subunit 15b | Ppp1r15b | NM 133819 |
| BG064278 | Nucleosome assembly protein 1-like 1 | Nap1l1 | NM 015781 |
| BG065281 | Glia maturation factor, beta | Gmfb | NM 022023 |
| BG067688 | Heterogeneous nuclear ribonucleoprotein A3 | Hnrnpa3 | NM 146130 |
| BG069073 | Transformer 2 alpha homolog (Drosophila) | Tra2a | NM 198102 |
| BG072727 | Bmi1 polycomb ring finger oncogene | Bmi1 | NM 007552 |
| BG077079 | A disintegrin and metallopeptidase domain 17 | Adam17 | NM 009615 |
| BG082860 | WD repeat domain 77 | Wdr77 | NM 027432 |
| BG085072 | WAP 4-disulfide core domain 2 | Wfdc2 | NM 026323 |
| BG086014 | High mobility group nucleosomal binding domain 1 | Hmgn1 | NM 008251 |
| AW542440 | Unknown | ||
| BG070468 | Unknown | ||
| BG069520 | Unknown | ||
| BG070685 | Unknown |
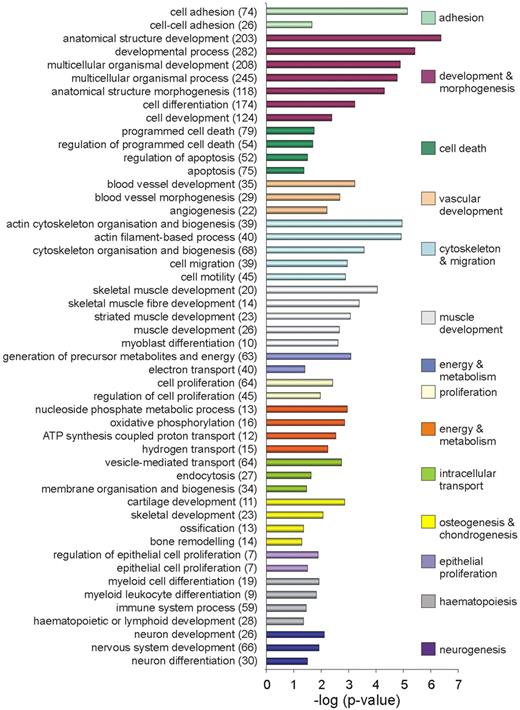
The functional annotation enrichment analysis tool from the freely accessible software DAVID was used to determine which biologic processes are enriched in the total up-regulated and down-regulated gene populations. Strikingly, the majority of biologic functions enriched in the down-regulated population are part of nucleic acid metabolism, DNA replication, transcription, translation, and further downstream steps of protein folding and posttranslational modifications (supplemental Figure 1). This may be due to many cells and tissues reaching their final differentiated state as development proceeds. Enriched in the up-regulated population are processes associated with development and morphogenesis such as cell death, adhesion, and migration (Figure 3). Apart from these more general developmental processes, an enrichment for genes specific to tissues known to develop in and around the dorsal aorta was found. This includes vascular development, muscle development, neurogenesis, and, most importantly, hematopoiesis.
Functional enrichment in the total up-regulated gene population. List of biologic processes significantly enriched (P < .05) in the up-regulated gene population. The number of differentially expressed genes representative of a particular process is given in brackets.
Functional enrichment in the total up-regulated gene population. List of biologic processes significantly enriched (P < .05) in the up-regulated gene population. The number of differentially expressed genes representative of a particular process is given in brackets.
Within the list of up-regulated genes, we looked for those previously implicated in hematopoiesis (supplemental Table 2). Included were genes known to be important for developmental hematopoiesis, such as Runx1, Gata1, and Gata3.25-28 Surprisingly, we also identified genes such as Foxo3,29 Id1,30,31 and Mll532,33 that have been implicated as yet only in adult hematopoietic processes. It will be interesting to determine whether these genes also play a role in AGM hematopoiesis. Among the genes commonly up-regulated in the 2 temporal sample sets were many globin-coding genes (supplemental Table 1).
p57Kip2 knockout mice display prolonged HSC activity in the AGM
To validate that regulators of AGM hematopoiesis were indeed being picked up by the expression screens, one of the genes commonly up-regulated in all 3 approaches (Table 1) was chosen for further analysis. The cyclin-dependent kinase inhibitor p57Kip2 was selected, as it is known to be highly expressed in adult HSCs, where it has been implicated in controlling cell proliferation.34,35 We first confirmed the expression pattern suggested by the microarray experiments, using in situ hybridization (Figure 4). There was almost no expression of p57Kip2 in the mesenchyme surrounding the E9 dorsal aorta (Figure 4A). More rostrally, p57Kip2 expression was detected in the nephrogenic chords (Figure 4B), but these would have been removed during the dissection of the aorta. In contrast, strong p57Kip2 expression was visible in the mesenchyme surrounding the E11 dorsal aorta with higher expression below the ventral aspect of the aorta (Figure 4C). In light of our finding that HSCs are enriched in the central part of the aorta, p57Kip2 expression was also analyzed along the length of the AGM. Compared with the middle region of the aorta (Figure 4E), p57Kip2 expression was markedly reduced in the caudal and rostral regions of the AGM (Figure 4D and F, respectively). Although most of the high expression is several cell layers removed from the aorta, some was observed just underneath the endothelial layer (Figure 4G asterisk) and weaker staining was occasionally detected in endothelial cells (Figure 4G arrows).
p57Kip2 expression and function in the AGM. Analysis of p57Kip2 expression by in situ hybridization in E9 wild-type (A-B), E11 wild-type (C-H), and E11 p57Kip2+/−m (I) embryos. (D) Caudal section; (F) rostral section. (G) Close-up of an E11 aorta with endothelial expression highlighted by arrows and subaortic expression with an asterisk. Stained sections were mounted with Hydromount. Ventral, down. DA indicates dorsal aorta; HG, hindgut; LB, lung bud; NC, nephrogenic chord; and VA, vitelline artery. (J) Graph showing percentage of mice repopulated with E11 and E12 AGMs (1 ee) from wild-type or p57Kip2 mutant embryos. The number of repopulated mice/total mice injected is indicated above each bar. (K) Cytospin of E11 AGM cells stained with an antibody for p57Kip2 (green, Alexa488) and mounted with Vectashield. Nuclear 4,6 diamidino-2-phenylindole staining in blue. White arrowheads indicate cells with cytoplasmic p57Kip2. Expression analysis by in situ hybridization for Cryab (L,Li) and Th (M,Mi) on wild-type (L-M) and p57Kip2+/−m (Li,Mi) E11 embryos. Stained sections were mounted with Hydromount (National Diagnostics). All pictures were taken with a Zeiss AxioSkop2 Wide-Field Microscope fitted with a Zeiss AxioCam MRc5, and images analyzed with the Zeiss AxioVision software (all from Carl Zeiss Ltd). Objectives used were 5×/0.15 NA (C,H-I), 10×/0.25 NA (A-B,D-F,L,Li,M,Mi), 20×/0.45 NA (G), and 40×/0.65 NA (K).
p57Kip2 expression and function in the AGM. Analysis of p57Kip2 expression by in situ hybridization in E9 wild-type (A-B), E11 wild-type (C-H), and E11 p57Kip2+/−m (I) embryos. (D) Caudal section; (F) rostral section. (G) Close-up of an E11 aorta with endothelial expression highlighted by arrows and subaortic expression with an asterisk. Stained sections were mounted with Hydromount. Ventral, down. DA indicates dorsal aorta; HG, hindgut; LB, lung bud; NC, nephrogenic chord; and VA, vitelline artery. (J) Graph showing percentage of mice repopulated with E11 and E12 AGMs (1 ee) from wild-type or p57Kip2 mutant embryos. The number of repopulated mice/total mice injected is indicated above each bar. (K) Cytospin of E11 AGM cells stained with an antibody for p57Kip2 (green, Alexa488) and mounted with Vectashield. Nuclear 4,6 diamidino-2-phenylindole staining in blue. White arrowheads indicate cells with cytoplasmic p57Kip2. Expression analysis by in situ hybridization for Cryab (L,Li) and Th (M,Mi) on wild-type (L-M) and p57Kip2+/−m (Li,Mi) E11 embryos. Stained sections were mounted with Hydromount (National Diagnostics). All pictures were taken with a Zeiss AxioSkop2 Wide-Field Microscope fitted with a Zeiss AxioCam MRc5, and images analyzed with the Zeiss AxioVision software (all from Carl Zeiss Ltd). Objectives used were 5×/0.15 NA (C,H-I), 10×/0.25 NA (A-B,D-F,L,Li,M,Mi), 20×/0.45 NA (G), and 40×/0.65 NA (K).
To determine a functional role for p57Kip2, knockout mice were analyzed for AGM HSC activity. p57Kip2 is an imprinted gene that is expressed solely from the maternal allele during mouse development. Heterozygous embryos in which the maternally derived allele is mutated (p57Kip2+/−m) are phenotypically indistinguishable from homozygous knockout embryos.17 We confirmed that there was no p57Kip2 expression in AGMs from p57Kip2+/−m embryos (compare Figure 4I with H), and used embryos of this genotype in all subsequent experiments. The 2 related cyclin-dependent kinase inhibitors p21Cip1 and p27Kip1 can, in some tissues, compensate for the absence of p57Kip2. We detected little or no expression of p21Cip1 or p27Kip1 around the p57Kip2+/+ E11 aorta, and did not observe an up-regulation in their expression when p57Kip2 was deleted (supplemental Figure 2).
When AGMs were dissected from E11 p57Kip2+/+ and p57Kip2+/−m embryos and directly transplanted, no difference in reconstitution of irradiated adult recipients was found (Figure 4J). However, whereas HSC activity starts declining in wild-type AGMs at E12.5, it remained high in p57Kip2+/−m AGMs. There are at least 3 possible explanations for this observation: (1) HSCs proliferate more in the absence of p57Kip2, (2) their migration from the AGM is delayed in mutant embryos, or (3) they are produced for a longer time in p57Kip2+/−m AGMs. As there was no increase in HSCs at E11, it is unlikely that p57Kip2+/−m HSCs proliferate more. Others have demonstrated that cytoplasmic p57Kip2 can interact with the actin filament-regulating LIM-domain–containing, protein kinase and sequester it in the nucleus.36 This interaction is thought to result in increased cell motility. We examined the subcellular localization of p57Kip2 protein in wild-type AGM cells and found several cells in which p57Kip2 was detectable in the cytoplasm (Figure 4K white arrowheads).
Although p57Kip2 is known to be highly expressed in adult HSCs34 and our microarray expression analysis also detected it in HSC-enriched populations, the widespread high expression seen in E11 subaortic mesenchymal tissue suggests that it may play a role in the regulatory microenvironment. Two cell lineages make up at least part of this region: (1) myogenic progenitors that migrate from the somites to the dorsal aorta to form the smooth muscle layer37 and (2) neural crest–derived sympathoadrenal precursors that form the sympathetic ganglia and the adrenal gland (reviewed in Huber38 ). αB crystallin (Cryab), a marker of myogenic progenitor cells, and tyrosine hydroxylase (Th), a marker of the sympathoadrenal precursor cells, were found to be up-regulated in our expression analysis (Table 1 and supplemental Table 1). Interestingly, expression analysis for these marker genes showed that both cell populations are expanded in p57Kip2+/−m AGMs (compare Figure 4Li,Mi with L,M). It remains to be determined how this may influence HSC behavior in the AGM.
Igf2 and its receptors are expressed in embryonic hematopoietic tissues
Recently, we observed that single growth factors, such as interleukin 3 and bone morphogenic protein 4, have a positive effect on AGM HSC expansion.12,18 Growth factor signaling pathways were highlighted in our functional enrichment analysis (data not shown), including the Igf2 pathway. Igf2 was up-regulated in the E11WA population, and 2 of its 3 receptors as well as several of its regulatory binding proteins were also differentially expressed (Figure 5A). Moreover, Igf2 and p57Kip2 are oppositely imprinted and suggested to act antagonistically in some tissues.39
Igf2 affects AGM hematopoiesis. (A) Venn diagrams showing the populations in which Igf2 and its receptors and binding proteins demonstrated differential expression. (Bi) Semiquantitative RT-PCR showing the expression of Igf2 and its receptors in several hematopoietic tissues of an E11 embryo and in the AGM-derived supportive stromal cell line UG26-1B6. (Bii) In situ hybridization on an E11 embryo section with a riboprobe specific to Igf2. Stained sections were mounted with Hydromount. DA indicates dorsal aorta; FL, fetal liver; M, myotome; and UGR, urogenital ridge. (Biii) Igf2 mRNA expression in the E11 dorsal aorta (ventral, down). Arrow highlights Igf2 expression in the intra-aortic cluster. Pictures were taken with a Zeiss AxioSkop2 Wide-Field Microscope fitted with a Zeiss AxioCam MRc5, and images analyzed with the Zeiss AxioVision software (all from Carl Zeiss Ltd). Objectives used were 5×/0.15 NA (ii) and 20×/0.45 NA (iii). (Ci) Outline of experiment to test the effect of Igf2 on colony formation from E11 AGM cells. (Cii) Bar graph showing the total number of colonies obtained from freshly isolated AGMs or from AGM cells after culture with or without Igf2 (n = 3). In the control cultures an equal volume of diluent was added. (Ciii) Bar graph showing the percentage of colony type obtained in each condition. Error bars indicate SD.
Igf2 affects AGM hematopoiesis. (A) Venn diagrams showing the populations in which Igf2 and its receptors and binding proteins demonstrated differential expression. (Bi) Semiquantitative RT-PCR showing the expression of Igf2 and its receptors in several hematopoietic tissues of an E11 embryo and in the AGM-derived supportive stromal cell line UG26-1B6. (Bii) In situ hybridization on an E11 embryo section with a riboprobe specific to Igf2. Stained sections were mounted with Hydromount. DA indicates dorsal aorta; FL, fetal liver; M, myotome; and UGR, urogenital ridge. (Biii) Igf2 mRNA expression in the E11 dorsal aorta (ventral, down). Arrow highlights Igf2 expression in the intra-aortic cluster. Pictures were taken with a Zeiss AxioSkop2 Wide-Field Microscope fitted with a Zeiss AxioCam MRc5, and images analyzed with the Zeiss AxioVision software (all from Carl Zeiss Ltd). Objectives used were 5×/0.15 NA (ii) and 20×/0.45 NA (iii). (Ci) Outline of experiment to test the effect of Igf2 on colony formation from E11 AGM cells. (Cii) Bar graph showing the total number of colonies obtained from freshly isolated AGMs or from AGM cells after culture with or without Igf2 (n = 3). In the control cultures an equal volume of diluent was added. (Ciii) Bar graph showing the percentage of colony type obtained in each condition. Error bars indicate SD.
RT-PCR was performed to confirm that Igf2 is expressed in the E11 AGM (Figure 5Bi). In addition to Igf2, we also detected transcripts for all of its 3 receptors in the E11 AGM. Igf2r seems to be expressed more highly than the other 2 receptors. Other tissues known to contain HSCs at E11, namely the yolk sac, the FL, and the placenta, were also examined. Igf2 and Igf2r transcripts were found at similarly high levels as in the AGM, whereas Ir and Igf1r showed different expression levels in these tissues. The E11 AGM stromal cell clone UG26-1B6, previously demonstrated to serve as a good in vitro model for the AGM HSC supportive microenvironment,40 showed high-level expression of all 3 Igf2 receptors, but no expression of Igf2 (Figure 5Bi). In situ hybridization performed for Igf2 transcripts on E11 embryo sections detected high levels of expression in several different tissues, including the myotome, the FL, and the AGM (Figure 5Bii). Within the region of the dorsal aorta, Igf2 was expressed in the endothelial and smooth muscle layer and in the intra-aortic clusters (Figure 5Biii arrow).
Igf2 affects hematopoietic stem/progenitor growth and/or maintenance in the AGM
To investigate whether Igf2 affects AGM hematopoiesis, AGM explants were cultured in the presence or absence of recombinant Igf2. After 3 days, AGM explants were dissociated and hematopoietic progenitor content was evaluated by in vitro colony-forming assays (Figure 5Ci). Although the total number of colonies obtained from E11 AGM cells was not affected by increasing Igf2 concentrations (Figure 5Cii), the analysis of colony types revealed differential effects of Igf2 on individual blood lineages (Figure 5Ciii). A striking effect was seen on colony-forming unit–Mix (CFU-Mix) progenitors, the most stem cell–like progenitor detectable in these in vitro colony-forming assays. Their percentage increased by 4.5-fold (4% to 18%; P = .01) upon the addition of 500 ng/mL of Igf2 and further to almost 7-fold (27%; P = .003) when the concentration of added Igf2 was doubled. This dose-dependent expansion of CFU-Mix colonies seemed to occur at the expense of CFU–granulocyte-macrophage (CFU-GM) and CFU-macrophage (CFU-M) colonies. The percentage of CFU-M progenitors was reduced from 76% to 51% (P = .03), whereas CFU-GM progenitor frequency decreased from 17% to 8% (P = .1), although the latter change failed to reach significance levels. Igf2 (both concentrations) also increased the percentage of erythroid burst-forming unit (BFU-E) progenitors by 7-fold (2% to 14%; P = .004). Taken together, these data seem to suggest that Igf2 has a positive effect on immature progenitors and the erythroid lineage, whereas it has a negative impact on more mature members of the granulocyte/monocyte lineage. To gain more insight into the mechanism by which Igf2 might exert these effects on progenitors, we also determined the CFU-C content in freshly isolated E11 AGMs. Remarkably, the total CFU-C content of fresh AGMs was more than double that of cultured AGMs (Figure 5Cii); however, this may at least in part be due to circulating progenitors still being present inside the aorta at the time of dissection, although these would get washed out during the culture step. The colony composition also differed from that of cultured AGMs (Figure 5Ciii). The most striking difference was seen in the percentage of CFU-GMs (57% fresh AGM vs 17% cultured control AGMs; P = .004) and CFU-Ms (20% vs 76%; P = .002). Although the percentages of CFU-Mix and BFU-E colonies are lower in fresh AGMs compared with Igf2-treated (1 μg/mL) AGMs, the absolute number of BFU-E colonies per 100 000 cells is the same (2.5 vs 2.7) and that of CFU-Mix colonies increased (9.2 vs 5.4). This suggests that hematopoietic progenitor cells in the AGM start to differentiate during the culture step and that Igf2 may act as a maintenance factor.
Discussion
Refinement of HSC location
Through subdissections of the E11 dorsal aorta, we have further refined the location of HSCs and shown that they are enriched in the middle region of the aorta near the junction of this vessel with the vitelline artery. The reason for this preferential location may be due to differences in the microenvironment along the length of the aorta. Alternatively, it is reasonable to suspect that the disturbance in blood flow at the junction of these 2 major vessels promotes HSC emergence. Two reports have recently highlighted the importance of blood flow and the associated biomechanical forces in HSC development.41,42 We have incorporated this new HSC localization information into the design of a comprehensive gene expression analysis of the dorsal aorta. Combined analyses of both spatial gene expression changes (in different parts of the aorta that do or do not support HSCs), as well as temporal gene expression changes (comparing tissues before and after HSC emergence) showed substantial overlap in the differentially expressed genes. Among the genes up-regulated at the time and location of HSC emergence are several implicated in various aspects of the adult hematopoietic system, including Foxo3, Id1, Mll5, and Sh2b3 (Lnk). It is expected that they play a role in the early stages of blood development. Some of these have also been linked to blood disorders and malignancies. Studying their precise function in the regulation of the initial stages of hematopoietic development may help to explain how their misregulation results in blood-related diseases.
Identification of 2 novel AGM hematopoietic regulators
To validate our approach and identification of novel candidate regulators of AGM hematopoiesis, we initiated functional studies on one of the genes commonly up-regulated in all 3 approaches. p57Kip2 is highly expressed in adult HSCs and is the only member of the Cip/Kip family to have an essential role during development. Although HSC development at E11 seems to proceed normally in the absence of p57Kip2, HSC activity remains high after E12 when it has already declined in wild-type embryos. The cause for this is presently unclear but may be due to prolonged production of HSCs in the AGM or a delay in their emigration. It is also not clear whether p57Kip2 influences HSC behavior cell-intrinsically or through modulation of the microenvironment. p57Kip2 seems to have an impact on the development of 2 cell types present in the mesenchyme, myogenic progenitors and sympathoadrenal precursors, both of which may influence AGM hematopoiesis (discussed in “Coordinate regulation in midgestation trunk tissues”). The prolonging of HSC activity in the AGM, in this case through the absence of p57Kip2, has, to our knowledge, never been observed and will be a useful tool for the study of the AGM as a temporary HSC niche and the migration of HSCs during development.
The growth factor Igf2 was chosen from the list of hematopoiesis-associated genes because it has previously been implicated in proliferation control of FL and adult BM HSCs.21 In addition, in contrast to the adult hematopoietic system, where several factors usually work in synergy, we recently discovered that single growth factors can have dramatic effects on AGM HSC development.18 Hence, by adding recombinant Igf2 to AGM explant cultures, we discovered that Igf2 affects early immature and committed hematopoietic progenitor cells. To our knowledge, this is the first time that a single factor has been demonstrated to have different effects on different AGM hematopoietic progenitor types.
The addition of Igf2 resulted in an increase of BFU-E colonies, which suggests that Igf2 may positively influence the proliferation and/or survival of erythroid progenitors. Considering that the absolute number of BFU-E colonies is the same in freshly isolated AGMs, we favor the latter interpretation.
We also detected an Igf2 dose-dependent increase in the earliest progenitors detectable in colony-forming assays (CFU-Mix) and an accompanying decrease in CFU-GM and CFU-M progenitors (compared with control cultured AGMs), suggesting that Igf2 may act as a survival factor or block their differentiation into more mature cell types. The latter seems plausible because a loss of CFU-Mix and CFU-GM with an accompanying increase in the proportion of more mature CFU-M progenitors was observed in control cultured AGMs compared with uncultured AGMs. We are currently investigating whether a similar increase of repopulating HSCs can also be achieved by the addition of Igf2. This would mean that Igf2 might act to maintain the HSC pool in the AGM. Other investigations are focusing on whether some of the Igf2-binding proteins we have identified are modulating the effects of Igf2 in the AGM. They can exert enhancing or inhibiting effects on Igf2 depending on cellular context, concentration, and type of binding protein (reviewed in Holly and Perks43 ), which may explain why they were detected among the up-regulated and down-regulated genes. In addition, they may also directly affect HSCs, as it has been reported for human IGFBP2.44 Interestingly, Igfbp3 and Igfbp4 have been shown to be more highly expressed in HSC-supportive AGM-derived stromal cell lines compared with nonsupportive cell lines.14
Igf2 is not produced by the HSC-supportive stromal cell line UG26-1B6. This is in contrast to the FL where Igf2 is secreted by CD3+ stromal cells and is responsible for their supportive capacity.21 Our detection of Igf2 expression within the intra-aortic clusters could indicate that Igf2 exerts its effects either in an autocrine fashion or via the microenvironment (because all 3 receptors are expressed on UG26-1B6 cells). In vivo, additional Igf2 may also be produced from the AGM microenvironment because we detected its expression also in the endothelial and the smooth muscle layer of the aorta. Signaling occurs mainly through Igf1r, which is up-regulated in the whole aorta as well as in the HSC-enriched population. Igf2 binding to the nonsignaling Igf2r regulates its bioavailability, targeting Igf2 for internalization and degradation, which may explain why Igf2r was among the down-regulated genes.
Coordinate regulation in midgestation trunk tissues
Using the bioinformatics package DAVID, we carried out an analysis of the biologic functions that are enriched in the total up-regulated gene population. It was intriguing to see that the expression of genes specific to other tissues that surround the aorta also increased at the time of HSC emergence. These included vascular genes, genes of the sympathetic nervous system, and genes expressed in (smooth) muscle cells. The fact that they are concomitantly up-regulated and that these other tissues develop in such close proximity to the aorta suggests that some regulators may be involved in the development of several different organs, as is certainly the case with some well-known developmental signaling pathways such as Wnt and Notch and, as shown here, p57Kip2.
However, this coregulation may also indicate that these tissues influence each other's development. There is an intimate connection between the vascular and the developing hematopoietic system, with specialized endothelial cells believed to be the direct precursors of HSCs in the AGM.6,7 Furthermore, it has also been demonstrated in avian embryos that at the peak of HSC production in the AGM, the hemogenic endothelium at the ventral side of the dorsal aorta is starting to be replaced by somite-derived endothelium, which may trigger the release of HSCs into the aortic lumen and is likely to cause the decline in HSCs in the AGM starting from E12.45,46 Similarly, lateral plate mesoderm-derived smooth muscle cells located ventrally to the dorsal aorta are also being replaced by smooth muscle precursors of somitic origin.37,47 How this may influence HSC production in the AGM is currently unclear. Wedged between the endothelial and smooth muscle cells is a single layer of pericytes. These share many regulators and markers with smooth muscle cells and have received a lot of attention recently as they have been shown to contain mesenchymal stromal cells (MSCs), which are important components of the adult HSC niche.48,49 Cells with MSC potential have also been detected in the AGM at the time of HSC emergence.50 However, whether these localize to the pericyte layer and form part of the AGM HSC supportive niche remains to be shown.
Neural crest cells migrate from the neural tube to the dorsal aorta at the time of HSC generation where they differentiate into cells of the sympathetic nervous system (reviewed in Huber38 ). Catecholamines, which are released from cells of the sympathetic nervous system, have recently been shown to induce adult HSC proliferation and migration.51,52 It will be interesting to determine whether they influence HSC production and migration during development.
In summary, we have carried out an extensive gene expression analysis of the AGM at the time of HSC emergence. In the up-regulated gene sets, we have identified several hematopoietic regulators as well as genes involved in the development of tissues surrounding the aorta. p57Kip2 and Igf2 were verified as regulators of AGM hematopoiesis and we are currently concentrating on several other candidate genes and their impact on HSC generation and/or expansion in the AGM.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Pumin Zhang for the p57Kip2 knockout mice; Karin van der Horn and Fredrik Wallberg for cell sorting services; Dr Natalia Ivanova for technical advice on RNA amplification; Richard Francis and especially Dr George Garinis for help and advice on bioinformatics; Suzanne van Nobelen and Dr Niels Galjart for the mouse Igf2 plasmid; Bahar Mirshekar-Syahkal, Gillian Kimber, and other members of the labs for technical assistance; and Dr Berthold Göttgens and Dr Anthony Green for helpful comments on the manuscript.
This work was supported by the Kay Kendall Leukemia Fund (KKL276, K.O.), the Medical Research Council (PhD studentship, M.I.M.), Dutch Organization for Scientific Research (916.36.601, E.D.), and the National Institutes of Health (R37DK054077, E.D.).
National Institutes of Health
Authorship
Contribution: M.I.M. performed research and analyzed data; A.P. performed research; E.D. designed experiments, analyzed data, and wrote the paper; and K.O. designed experiments, performed research, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Katrin Ottersbach, Department of Haematology, Cambridge Institute for Medical Research, University of Cambridge, Hills Rd, Cambridge CB2 0XY, United Kingdom; e-mail: ko268@cam.ac.uk.