Abstract
Alternatively activated macrophages (AAMs), triggered by interleukin-4 (IL-4) and IL-13, play a modulating role during Th2 cytokine-driven pathologies, but their molecular armament remains poorly characterized. Here, we established E-cadherin (Cdh1) as a selective marker for IL-4/IL-13–exposed mouse and human macrophages, which is STAT6-dependently induced during polarized Th2 responses associated with Taenia crassiceps helminth infections or allergic airway inflammation. The IL-4–dependent, arginase-1/ornithine decarboxylase–mediated production of polyamines is important for maximal Cdh1 induction, unveiling a novel mechanism for IL-4–dependent gene transcription. At the macrophage surface, E-cadherin forms a functional complex with the catenins that accumulates at sites of cell contact. Macrophage-specific deletion of the Cdh1 gene illustrates the implication of E-cadherin in IL-4–driven macrophage fusion and heterotypic interactions with CD103+ and KLRG1+ T cells. This study identifies the E-cadherin/catenin complex as a discriminative, partly polyamine-regulated feature of IL-4/IL-13–exposed alternatively activated macrophages that contributes to homotypic and heterotypic cellular interactions.
Introduction
Macrophages are implicated in functions as diverse as inflammation, wound healing, and immunity. To accommodate for this, they are able to adopt diverse activation states. Classically activated macrophages (CAMs), induced by Th1 cytokines and Toll-like receptor (TLR) ligands, play a pivotal role in inflammation and pathogen clearance.
More recently, it became clear that macrophages are also significantly altered by diverse noninflammatory cues, including the prototypical Th2 cytokines interleukin-4 (IL-4) and IL-13 (inducing alternatively activated macrophages [AAMs]), IL-10, transforming growth factor-β (TGF-β), glucocorticoids, immune complexes, and apoptotic cells, which has led to different macrophage classification systems.1-3 AAMs dampen Th1 cytokine-driven inflammation, coordinate adaptive immune responses, contribute to wound healing, and are implicated in Th2-driven pathologies, such as helminth infections and asthma.2 However, the molecular repertoire accounting for these functions remains poorly characterized.
For signaling, IL-4 binds IL-4Rα, thereby triggering parallel signaling pathways, including Janus-kinase/signal-transducer-and-activator-of-transcription-6 (JAK/STAT6), phosphoinositide-3-kinase (PI3K), and p38 mitogen-activated protein kinase (p38 MAPK).2,4 Intriguingly, recent data propose peroxisome-proliferator–activated receptor γ (PPAR-γ),5 PPAR-δ,6 and galectin-3 (gal3)7 to be required for maximal IL-4–induced gene transcription.
Until now, CAMs and AAMs have been mainly discriminated based on distinct gene expression profiles and metabolic programs, whereas reliable surface markers are largely lacking. Moreover, recent data question the usefulness of some “prototypical” AAM markers, such as arginase-1, to distinguish between CAMs and AAMs. Indeed, intracellular pathogens induce macrophage Arg1 expression in CAMs through the TLR pathway in a STAT6-independent manner.8
We previously reported a common gene signature for in vivo induced AAMs, identifying E-cadherin (Cdh1) as a marker for AAMs.9 This finding remained enigmatic, as E-cadherin expression, regulation, and function have only been investigated in detail in epithelial cells (ECs)10,11 and to some extent in Langerhans cells (LCs).12
E-cadherin is a transmembrane protein, whose intracellular domain is associated with β-catenin, p120, and α-catenin, and this complex forms tight junctions between ECs through homophilic interactions. In addition, E-cadherin can heterophilically interact with αEβ7 integrin (CD103) and the inhibitory killer cell lectin-like receptor G1 (KLRG1), expressed on diverse subsets of immune cells.13-16
Here, we report that E-cadherin is induced in AAMs by IL-4 and IL-13 in a JAK/STAT6-dependent manner. Importantly, IL-4–induced polyamine production is necessary for maximal Cdh1 induction, revealing a new regulatory mechanism for IL-4–dependent gene transcription. The E-cadherin/catenin complex is only formed at the macrophage surface during polarized Th2 responses, allowing its use as a selective surface marker for AAMs. Finally, we demonstrate that E-cadherin contributes to IL-4–induced macrophage fusion and heterotypic interactions with CD103+ and KLRG1+ T cells.
Methods
Mouse models of helminth infection and lung inflammation
All experiments were approved by the Ethics Committee at Vrije Universiteit Brussel, Brussels, Belgium, and met the standards required by the Belgian Council for Laboratory Animal Science guidelines. Genetic background, supplier, International Mouse Strain Resource nomenclature, and Mouse Genome Informatics IDs for all mouse strains are listed in Table 1. To generate mice in which the E-cadherin gene was disrupted in macrophages, floxed Cdh1F/F CD1 mice17 were crossed with LysM-Cre C57BL/6 mice. Homozygous LysM-Cre+/+-Cdh1F/F conditional KO (hereafter referred to as Cdh1Δ) mice were compared with LysM-Cre−/−-Cdh1F/F wild-type (WT) littermate controls (hereafter referred to as Cdh1F/F). IL-4 fails to induce E-cadherin on more than 95% of the Cdh1Δ macrophages, proving the efficiency of deletion (supplemental Figure 1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
List of mouse strains used
| . | Background . | IMSR nomenclature . | MGI ID . | Supplier . |
|---|---|---|---|---|
| BALB/c | BALB/c | BALB/cOlaHsd | Harlan | |
| C57BL/6 | C57BL/6 | C57BL/6JOlaHsd | Harlan | |
| IL-4−/− | BALB/c | BALB/c-Il4tm2Nnt/J | MGI:83819 | Dr F. Brombacher (University of Cape Town, Cape Town, South Africa) |
| IL-4Rα−/− | BALB/c | BALB/c-Il4ratm1Sz/J | MGI:1098188 | Dr F. Brombacher |
| STAT6−/− | BALB/c | C.129S2-Stat6tm1Gru/J | MGI:79436 | The Jackson Laboratory |
| Galectin-3−/− | C57BL/6 | B6.Cg-Lgals3tm1Poi/J | MGI:1202812 | Dr D. K. Hsu (University of California, Sacramento, CA) |
| LysM-Cre | C57BL/6 | B6.129P2-Lyz2tm1(cre)Ifo/J | MGI:1931699 | The Jackson Laboratory |
| Cdh1F/F | CD1 | CD1.129P2-Cdh1tm1Jjon/JA | MGI:3693661 | Dr J. Jonkers (The Netherlands Cancer Institute, Amsterdam, The Netherlands) |
| . | Background . | IMSR nomenclature . | MGI ID . | Supplier . |
|---|---|---|---|---|
| BALB/c | BALB/c | BALB/cOlaHsd | Harlan | |
| C57BL/6 | C57BL/6 | C57BL/6JOlaHsd | Harlan | |
| IL-4−/− | BALB/c | BALB/c-Il4tm2Nnt/J | MGI:83819 | Dr F. Brombacher (University of Cape Town, Cape Town, South Africa) |
| IL-4Rα−/− | BALB/c | BALB/c-Il4ratm1Sz/J | MGI:1098188 | Dr F. Brombacher |
| STAT6−/− | BALB/c | C.129S2-Stat6tm1Gru/J | MGI:79436 | The Jackson Laboratory |
| Galectin-3−/− | C57BL/6 | B6.Cg-Lgals3tm1Poi/J | MGI:1202812 | Dr D. K. Hsu (University of California, Sacramento, CA) |
| LysM-Cre | C57BL/6 | B6.129P2-Lyz2tm1(cre)Ifo/J | MGI:1931699 | The Jackson Laboratory |
| Cdh1F/F | CD1 | CD1.129P2-Cdh1tm1Jjon/JA | MGI:3693661 | Dr J. Jonkers (The Netherlands Cancer Institute, Amsterdam, The Netherlands) |
IMSR indicates International Mouse Strain Resource; and MGI, Mouse Genome Informatics.
To study helminth infection, mice were inoculated intraperitoneally with 10 Taenia crassiceps metacestodes; peritoneal cells were collected 8 or 12 weeks after infection and macrophages obtained via 3 hours plastic adherence.18
To sensitize C57BL/6 mice for allergic asthma, mice were injected intraperitoneally at days 0, 7, and 14 with 10 μg of grade 5 chicken egg ovalbumin (OVA; Sigma-Aldrich) adsorbed on 1 mg of Alum (Pierce Chemical) in phosphate-buffered saline (PBS). To sensitize for allergic hypersensitivity, mice were injected subcutaneously at day 0 with 20 μg of grade 5 OVA in complete Freund adjuvant (Sigma-Aldrich). Next, mice were challenged at day 21 and 22 for 30 minutes with aerosols, consisting of grade 3 OVA in PBS.19,20 Twenty hours later, mice were killed and bronchoalveolar leukocytes (BALs) were collected by PBS rinsing of the lungs. IL-4 and interferon-γ (IFN-γ) concentrations in BAL fluid were measured using Bio-plex (Bio-Rad).
In vitro stimulation of macrophages
Isolated macrophages (supplemental Methods) were cultured in the presence of 100 U/mL recombinant mouse (BD Biosciences) or human (PeproTech) IL-13, IL-4, IL-10, IFN-γ, 5 ng/mL rhTGF-β1 (PeproTech), or 100 ng/mL Escherichia coli lipopolysaccharide (LPS). To inhibit JAK, PI3K, or p38 MAPK, macrophages were pretreated for 1 hour with 100nM JAK inhibitor I, 50nM wortmannin, or 6μM SB203580, respectively (Calbiochem), followed by 6 or 24 hours of IL-4 stimulation. To study PPAR-γ and PPAR-δ involvement, macrophages were pretreated with 10μM GW9662 (PPAR-γ inhibitor) or 10μM L-165,041 (PPAR-δ ligand), followed by 24-hour IL-4 stimulation. As positive control for this procedure, PPAR-γ inhibition reduced Arg1 expression on average 4.3-fold, and PPAR-δ ligation stimulated Mgl1 expression on average 2.32-fold.5,6 Polyamines were depleted by 24-hour 5 mM 2-(difluoromethyl)ornithine (DFMO, ornithine decarboxylase [ODC] inhibitor) or 10μM N1,N11-diethylnorspermine (DENSPM; Tocris) pretreatment, with or without 10μM spermine or putrescine, followed by 24 hours of IL-4 treatment. To inhibit protein synthesis or the transcription machinery, 10 μg/mL cycloheximide or 2 μg/mL actinomycin D was added 30 minutes before IL-4 stimulation, respectively. Importantly, cycloheximide did not influence basal or IL-4–induced Cdh1 mRNA stability (supplemental Figure 2). All reagents were purchased from Sigma-Aldrich unless otherwise marked.
Gene expression analysis
After RNA extraction with TRIzol and reverse transcription with Superscript II (Invitrogen), quantitative real-time PCR was performed in an iCycler, with iQ SYBR-Green Supermix (Bio-Rad). Primer sequences and PCR program were reported earlier.21 Gene expression was always normalized using ribosomal protein S12 as housekeeping gene.
Immunoblotting
Macrophages were lysed in radio immunoprecipitation assay buffer (RIPA) containing Complete Protease Inhibitor Cocktail (Roche Diagnostics). A total of 25 μg of protein was separated on 8% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride membranes (PVDF; Millipore). After 2 hours of blocking with 5% nonfat dry milk, membranes were incubated overnight at 4°C with primary antibodies. After washing, membranes were incubated for 1 hour with peroxidase-coupled secondary antibody, and Immobilon chemiluminescent horseradish peroxidase substrate (Millipore) was applied to visualize proteins after exposure to an autoradiography film (GE Healthcare). Antibodies are listed in Table 2.
List of antibodies used
| Marker . | Clone . | Isotype . | Supplier . | Application . |
|---|---|---|---|---|
| Rat IgG2a/PE isotype control | R35-95 | Rat IgG2a | BD Biosciences | FC |
| CD124 (IL-4Rα chain)/PE | mIL4R-M1 | Rat IgG2a | BD Biosciences | FC |
| STAT6 (M-20) | Polyclonal | Rabbit Ig | Santa Cruz Biotechnology | WB |
| p-STAT6 (Tyr641) | Polyclonal | Rabbit Ig | Santa Cruz Biotechnology | WB |
| Rat IgG2a/pure isotype control | NA/LE | Rat IgG2a | BD Biosciences | FC |
| Mouse IgG2a/pure isotype control | X39 | Mouse IgG2a | BD Biosciences | IP |
| Anti–rat Ig/PE/APC | Polyclonal | Goat Ig | BD Biosciences | FC |
| CD16/CD32/pure Fc-Block | 2.4G2 | Rat IgG2b | BD Biosciences | FC |
| E-cadherin/pure | 36 | Mouse IgG2a | BD Biosciences | WB, IF, IP |
| E-cadherin/pure | ECCD2 | Rat IgG2a | Dr. M. Takeichi (University of Kyoto, Kyoto, Japan) | FC |
| p120 Catenin/pure | 98 | Mouse IgG1 | BD Biosciences | WB, IF |
| β-Catenin/pure | Polyclonal | Rabbit Ig | Sigma-Aldrich | WB, IF |
| α-Catenin/pure | Polyclonal | Rabbit Ig | Sigma-Aldrich | WB, IF |
| β-Actin/pure | AC-15 | Mouse IgG1 | Abcam | WB |
| CD11c/PerCp-Cy5.5 | N418 | Hamster IgG | eBioscience | FC |
| CD115/bio | AFS98 | Rat IgG2a | eBioscience | FC |
| Ly6c/APC | ER-MP20 | Rat IgG2a | Serotec | FC |
| Ly6c/FITC | AL21 | Rat IgM | BD Biosciences | FC |
| F4/80/APC–Alexa Fluor 750 | BM8 | Rat IgG2a | eBioscience | FC |
| CCR3/FITC | 83101 | Rat IgG2a | R&D Systems | FC |
| Ly6G/PE | 1A8 | Rat IgG2a | BD Biosciences | FC |
| IA/IE/PE | M5/114.15.2 | Rat IgG2b | BD Biosciences | FC |
| IA/IE/FITC | M5/114.15.2 | Rat IgG2b | eBioscience | FC |
| Marker . | Clone . | Isotype . | Supplier . | Application . |
|---|---|---|---|---|
| Rat IgG2a/PE isotype control | R35-95 | Rat IgG2a | BD Biosciences | FC |
| CD124 (IL-4Rα chain)/PE | mIL4R-M1 | Rat IgG2a | BD Biosciences | FC |
| STAT6 (M-20) | Polyclonal | Rabbit Ig | Santa Cruz Biotechnology | WB |
| p-STAT6 (Tyr641) | Polyclonal | Rabbit Ig | Santa Cruz Biotechnology | WB |
| Rat IgG2a/pure isotype control | NA/LE | Rat IgG2a | BD Biosciences | FC |
| Mouse IgG2a/pure isotype control | X39 | Mouse IgG2a | BD Biosciences | IP |
| Anti–rat Ig/PE/APC | Polyclonal | Goat Ig | BD Biosciences | FC |
| CD16/CD32/pure Fc-Block | 2.4G2 | Rat IgG2b | BD Biosciences | FC |
| E-cadherin/pure | 36 | Mouse IgG2a | BD Biosciences | WB, IF, IP |
| E-cadherin/pure | ECCD2 | Rat IgG2a | Dr. M. Takeichi (University of Kyoto, Kyoto, Japan) | FC |
| p120 Catenin/pure | 98 | Mouse IgG1 | BD Biosciences | WB, IF |
| β-Catenin/pure | Polyclonal | Rabbit Ig | Sigma-Aldrich | WB, IF |
| α-Catenin/pure | Polyclonal | Rabbit Ig | Sigma-Aldrich | WB, IF |
| β-Actin/pure | AC-15 | Mouse IgG1 | Abcam | WB |
| CD11c/PerCp-Cy5.5 | N418 | Hamster IgG | eBioscience | FC |
| CD115/bio | AFS98 | Rat IgG2a | eBioscience | FC |
| Ly6c/APC | ER-MP20 | Rat IgG2a | Serotec | FC |
| Ly6c/FITC | AL21 | Rat IgM | BD Biosciences | FC |
| F4/80/APC–Alexa Fluor 750 | BM8 | Rat IgG2a | eBioscience | FC |
| CCR3/FITC | 83101 | Rat IgG2a | R&D Systems | FC |
| Ly6G/PE | 1A8 | Rat IgG2a | BD Biosciences | FC |
| IA/IE/PE | M5/114.15.2 | Rat IgG2b | BD Biosciences | FC |
| IA/IE/FITC | M5/114.15.2 | Rat IgG2b | eBioscience | FC |
FC indicates flow cytometric analysis; WB, immunoblotting; IP, immunoprecipitation; and IF, immunofluorescence microscopy.
Flow cytometry
Cells were blocked with 10% normal rabbit serum (Gentaur) for 30 minutes at 4°C, followed by 30 minutes of incubation with anti-E-cadherin ECCD2 or isotype control. After washing, cells were incubated with phycoerythrin (PE)– or allophycocyanin (APC) coupled anti–rat Ig for 30 minutes, washed, and incubated with additional antibodies for 30 minutes. Data were acquired with a FACSCanto II (BD Biosciences) and analyzed using FlowJo (TreeStar).
Microscopy
Cells were grown on glass coverslips and fixed with methanol. Fc receptors were blocked for 30 minutes with anti-CD16/32, followed by a 1-hour incubation at room temperature with primary antibodies diluted in PBS plus 0.4% gelatin. After washing, cells were incubated for 1 hour with Alexa 488-coupled anti–mouse and Alexa 594-coupled anti–rabbit Ig (Invitrogen). Finally, cells were embedded in Vectashield with DAPI (Vector Laboratories), and samples were acquired at 20°C on an Olympus BX61 fluorescence microscope using a F view camera, Cell M software, and a 60× (Plan APO 1.42) or 100× (U Plan FLN 1.3) objective lens (all from Olympus Corporation).
Coimmunoprecipitation
Cells were lysed in 50mM Tris-HCl, pH 8, plus 1% NP-40, 150mM NaCl, and Complete Protease Inhibitor Cocktail. Lysates (500 μg of protein) were cleared during 2 hours at 4°C with 50 μL protein G-Sepharose beads (GE Healthcare). After removal of the beads, lysates were incubated overnight at 4°C with 1 μg of anti-E-cadherin clone 36 or isotype control and then during 1 hour with 50 μL of protein G-Sepharose beads. Beads were washed, boiled in sample solution, and proteins were separated on SDS-PAGE.
Polyamine measurement
Intracellular polyamine concentrations were measured as described earlier.22
Fusion assay
Equal numbers of DiI(red)– and DiO(green)–stained (Invitrogen) macrophages (105 total) were added to 8-well Permanox slides (Nunc) and stimulated with IL-4 for 24 hours.23 After paraformaldehyde fixation, Hoechst staining, and mounting in N-propyl gallate, 5 fields per condition were obtained on a Leica SP5 confocal system DMI6000 microscope with Advanced Fluorescence software (Leica Microsystems) using an Olympus 40× (HCX Plan APO 1.25) objective. Volocity software (Improvision) was applied to create a colocalization channel by selecting all image voxels, which contained both red and green fluorescent signals. This channel was used for visualization, and the average volume of colocalized voxels and the amount of nuclei per fused cell were calculated.
Adhesion assay
A total of 105 naive or IL-4–steered Cdh1F/F or Cdh1Δ thioglycollate-elicited peritoneal macrophages (thio-PEMs) were preincubated for 30 minutes with or without 10 μg/mL ECCD2 blocking antibodies in 96-well plates. MTC-1 cells were steered for 24 hours with 5 ng/mL TGF-β1 to induce CD103 and were fluorescently labeled with 2.5μM carboxyfluorescein succinimidyl ester (CFSE; Invitrogen).24 A total of 105 MTC-1 cells were added to the confluent macrophage monolayers. After 45 minutes of coculture, nonadherent cells were removed, fluorescence intensity in each well was measured using a Cytofluor II fluorescence plate reader (PerSeptive Biosystems), and the corresponding number of cells was calculated based on a standard curve.
KLRG1-reporter assay
A total of 105 Cdh1Δ or Cdh1F/F thio-PEMs were first stimulated in 24-well plates during 24 hours with different cytokines before adding 105 A5 KLRG1-reporter cells.25 After overnight coculture, cells were stained with anti-CD11b to gate out macrophages, and green fluorescent protein (GFP) expression was analyzed by fluorescence-activated cell sorter (FACS).
Results
Cdh1 gene expression is induced in mouse and human primary macrophages by the AAM-stimulating cytokines IL-4 and IL-13
We previously identified E-cadherin as one of the signature genes for in vivo induced AAMs.9 In vitro treatment of BALB/c and C57BL/6 thio-PEM with a range of macrophage modulators revealed a strong IL-4Rα-dependent Cdh1 induction by the AAM-inducing cytokines IL-4 and IL-13 (Figure 1A; supplemental Figure 3).
In vitro modulation and kinetics of E-cadherin gene expression in mouse and human macrophages. (A) BALB/c thio-PEMs, PEMs, and bone marrow-derived macrophages were left untreated (N) or were treated for 24 hours with indicated stimuli. (B) BALB/c thio-PEMs, with or without 2 μg/mL actinomycin D pretreatment, were IL-4 steered for 1, 3, 6, 12, 24, or 48 hours. (C) Human monocyte-derived macrophages were left untreated (N) or were treated for 48 hours with indicated stimuli. The fold Cdh1 induction relative to the expression in untreated macrophages (= 1) is shown and is the mean ± SEM of 3 mice.
In vitro modulation and kinetics of E-cadherin gene expression in mouse and human macrophages. (A) BALB/c thio-PEMs, PEMs, and bone marrow-derived macrophages were left untreated (N) or were treated for 24 hours with indicated stimuli. (B) BALB/c thio-PEMs, with or without 2 μg/mL actinomycin D pretreatment, were IL-4 steered for 1, 3, 6, 12, 24, or 48 hours. (C) Human monocyte-derived macrophages were left untreated (N) or were treated for 48 hours with indicated stimuli. The fold Cdh1 induction relative to the expression in untreated macrophages (= 1) is shown and is the mean ± SEM of 3 mice.
A kinetic analysis revealed a rapid increase in E-cadherin mRNA upon IL-4 stimulation of thio-PEM, which reached maximal levels after 6 hours, followed by a drop at 12 hours, to finally reach a plateau at 24 hours (Figure 1B). Blocking the transcription machinery with actinomycin D abolishes the IL-4 effect, indicating that de novo gene transcription is required (Figure 1B). Importantly, IL-4/IL-13–induced Cdh1 up-regulation can be reproduced in nonelicited mouse macrophages, such as resting PEMs and bone marrow–derived macrophages (Figure 1A). Moreover, human peripheral blood monocyte-derived macrophages also significantly up-regulate CDH1 in response to IL-4 and IL-13, establishing E-cadherin as an AAM marker in mouse and human (Figure 1C). The association of E-cadherin with AAMs is further strengthened by the observation that LPS and IFN-γ are unable to induce Cdh1 and dampen the IL-4 effect in mouse and human macrophages (except for IL-4 + LPS in BALB/c PEM, Figure 1A,C).
Although TGF-β does not alter the basal Cdh1 expression level, this cytokine synergizes with IL-4 for a further increase in Cdh1 mRNA in all BALB/c macrophage types tested (Figure 1A). IL-4/TGF-β collaboration is only observed if both cytokines are present simultaneously (supplemental Figure 4A) and cannot be explained by enhanced Cdh1 mRNA stability (supplemental Figure 4B) or TGF-β-mediated up-regulation of the IL-4Rα (supplemental Figure 5A), suggesting that IL-4 and TGF-β signaling intersect to maximize Cdh1 gene transcription in macrophages. IL-4/IL-10 collaboration is also observed in Balb/c thio-PEM and PEM (Figure 1A) and may be explained by an IL-10–mediated induction of the IL-4Rα (supplemental Figure 5A), as reported earlier.26
Arginase-1–dependent synthesis of polyamines is important for IL-4–mediated Cdh1 induction in macrophages
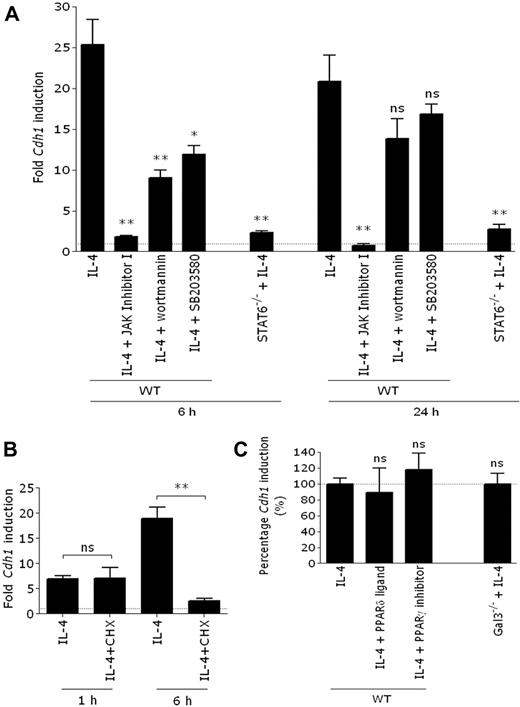
Having identified IL-4 and IL-13 as the most prominent Cdh1-inducing cytokines, we next investigated which IL-4Rα–initiated signaling pathways were responsible for this induction. Inhibition of JAK abolished the induction of Cdh1 gene transcription by IL-4, suggesting an essential role for JAK/STAT6 signaling (Figure 2A). In agreement, IL-4 was unable to induce high Cdh1 mRNA levels in STAT6−/− macrophages (Figure 2A). Nevertheless, both PI3K and p38 MAPK inhibition reduces IL-4–induced Cdh1 expression, illustrating that these pathways further enhance Cdh1 gene transcription (Figure 2A).
De novo–synthesized, STAT6-dependent factors are necessary for Cdh1 transcription. (A) WT BALB/c thio-PEMs, either pretreated or not with 100nM JAK inhibitor I, 50nM wortmannin (PI3K inhibitor), or 6μM SB203580 (p38 MAPK inhibitor), were IL-4 steered for 6 or 24 hours. STAT6−/− BALB/c thio-PEMs were IL-4 steered during 6 or 24 hours. (B) BALB/c thio-PEMs were pretreated with cycloheximide to block de novo protein synthesis, followed by 1 or 6 hours of IL-4 stimulation, and Cdh1 expression was determined. The fold induction of gene expression relative to the expression in the corresponding non–IL-4–treated thio-PEMs (n = 1) is shown. (C) WT BALB/c thio-PEMs, whether or not pretreated with 10μM GW9662 (PPAR-γ inhibitor) or 10μM L-165, 041 (PPAR-δ ligand), were IL-4 steered for 24 hours. Fold Cdh1 induction in IL-4–treated thio-PEM is set to 100%, and the effect of PPAR modifiers is shown relative to this level. WT or Gal3−/− C57BL/6 thio-PEMs were IL-4–stimulated for 24 hours, and the fold induction in IL-4–treated Gal3−/− thio-PEMs is shown relative to the induction level in WT thio-PEMs (100%). Values represent the mean ± SEM of 3 mice.*P < .05. **P < .01. ns indicates not significant.
De novo–synthesized, STAT6-dependent factors are necessary for Cdh1 transcription. (A) WT BALB/c thio-PEMs, either pretreated or not with 100nM JAK inhibitor I, 50nM wortmannin (PI3K inhibitor), or 6μM SB203580 (p38 MAPK inhibitor), were IL-4 steered for 6 or 24 hours. STAT6−/− BALB/c thio-PEMs were IL-4 steered during 6 or 24 hours. (B) BALB/c thio-PEMs were pretreated with cycloheximide to block de novo protein synthesis, followed by 1 or 6 hours of IL-4 stimulation, and Cdh1 expression was determined. The fold induction of gene expression relative to the expression in the corresponding non–IL-4–treated thio-PEMs (n = 1) is shown. (C) WT BALB/c thio-PEMs, whether or not pretreated with 10μM GW9662 (PPAR-γ inhibitor) or 10μM L-165, 041 (PPAR-δ ligand), were IL-4 steered for 24 hours. Fold Cdh1 induction in IL-4–treated thio-PEM is set to 100%, and the effect of PPAR modifiers is shown relative to this level. WT or Gal3−/− C57BL/6 thio-PEMs were IL-4–stimulated for 24 hours, and the fold induction in IL-4–treated Gal3−/− thio-PEMs is shown relative to the induction level in WT thio-PEMs (100%). Values represent the mean ± SEM of 3 mice.*P < .05. **P < .01. ns indicates not significant.
Although STAT6 appears to be crucial for the IL-4–mediated Cdh1 up-regulation in macrophages, no typical STAT6-binding sites (5′-TTC-(N)4-GAA-3′) are predicted in the E-cadherin promoter by the TFSEARCH algorithm.28 Although this does not exclude the binding of STAT6 to noncanonical promoter sequences, it might suggest the need for additional STAT6-regulated gene products in Cdh1 transcription. Indeed, although blocking de novo protein synthesis by cycloheximide did not influence the very early Cdh1 induction (1 hour), it inhibited a more sustained induction (6 hours; Figure 2B). Recently, IL-4–induced gal3, PPAR-γ, and PPAR-δ were proposed to regulate AAM markers. However, IL-4–mediated Cdh1 induction is unchanged following PPAR-γ inhibition, PPAR-δ ligation, or in gal3−/− macrophages (Figure 2C), arguing against an involvement of these proteins.
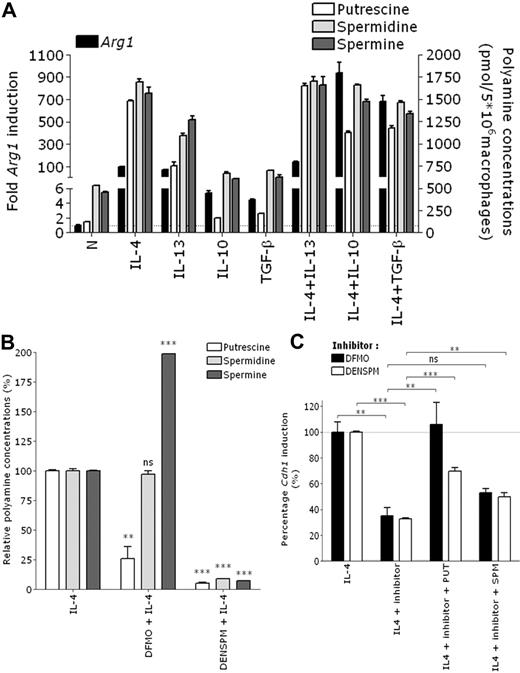
A known IL-4–inducible gene in macrophages is arginase-1, which converts L-arginine to L-ornithine. Next, ODC metabolizes L-ornithine to polyamines (putrescine → spermidine → spermine), which regulate E-cadherin expression in ECs.27 Confirming previous studies,28,29 treatment of BALB/c thio-PEM shows that, akin to Cdh1 regulation, Arg1 is strongly induced by IL-4 and IL-13, in synergy with IL-10 and TGF-β. IL-10 and TGF-β alone induce much lower levels of Arg1 (Figure 3A left y-axis). This results in an approximately 10-fold elevation of the intracellular putrescine concentration and 3-fold elevation of spermidine and spermine concentrations by IL-4, IL-4 plus IL-10, and IL-4 plus TGF-β (Figure 3A right y-axis).
Arginase-1–dependent synthesis of polyamines is important for IL-4–mediated Cdh1 induction in mouse macrophages. (A) BALB/c thio-PEMs were stimulated for 24 hours with indicated cytokines. The fold Arg1 induction relative to the expression in untreated macrophages (= 1) is shown (left y-axis). Intracellular polyamine concentrations were determined and shown as pmol/5 × 106 macrophages (right y-axis). (B) thio-PEMs were pretreated for 24 hours with DFMO or DENSPM, and after an additional 24 hours of IL-4 stimulation, intracellular polyamine concentrations were measured and plotted relative to the polyamine levels in nondepleted IL-4–treated macrophages (100%). (C) Same as panel B, with or without 10μM putrescine (PUT) or spermine (SPM) supplementation. The fold Cdh1 induction in polyamine-depleted IL-4–treated thio-PEM is shown relative to the induction level in IL-4–treated thio-PEMs (100%). Values represent the mean ± SEM of 3 mice. **P < .01. ***P < .001. ns indicates not significant.
Arginase-1–dependent synthesis of polyamines is important for IL-4–mediated Cdh1 induction in mouse macrophages. (A) BALB/c thio-PEMs were stimulated for 24 hours with indicated cytokines. The fold Arg1 induction relative to the expression in untreated macrophages (= 1) is shown (left y-axis). Intracellular polyamine concentrations were determined and shown as pmol/5 × 106 macrophages (right y-axis). (B) thio-PEMs were pretreated for 24 hours with DFMO or DENSPM, and after an additional 24 hours of IL-4 stimulation, intracellular polyamine concentrations were measured and plotted relative to the polyamine levels in nondepleted IL-4–treated macrophages (100%). (C) Same as panel B, with or without 10μM putrescine (PUT) or spermine (SPM) supplementation. The fold Cdh1 induction in polyamine-depleted IL-4–treated thio-PEM is shown relative to the induction level in IL-4–treated thio-PEMs (100%). Values represent the mean ± SEM of 3 mice. **P < .01. ***P < .001. ns indicates not significant.
To assess the potential contribution of polyamines to Cdh1 gene regulation, we pretreated macrophages for 24 hours with the ODC inhibitor DFMO, followed by a 24-hour treatment with IL-4. This short DFMO pretreatment only lowers putrescine concentrations 4-fold and even leads to a compensatory increase in spermine levels (Figure 3B), nevertheless resulting in a 65% reduction of IL-4–induced Cdh1 expression (Figure 3C). A full depletion of all polyamines by DENSPM pretreatment does not further lower Cdh1 mRNA levels, suggesting that part of the IL-4 effect is polyamine-independent. The importance of putrescine is further substantiated by a significant restoration of Cdh1 mRNA levels on supplementing putrescine to the DFMO plus IL-4 or DENSPM plus IL-4 cultures, which cannot be replicated to the same extent by spermine (Figure 3C). Culturing macrophages with polyamines alone has no impact on AAM gene expression (data not shown), illustrating that IL-4 signaling and polyamines need to collaborate. Importantly, altering the intracellular polyamine levels does not influence IL-4Rα expression (supplemental Figure 5A) or IL-4–induced STAT6 phosphorylation in macrophages (supplemental Figure 5B), excluding 2 potential levels of cooperation.
Together, these data reveal a novel role for arginase-1 and polyamines in IL-4–dependent gene regulation.
E-cadherin/catenin protein complexes are formed at the plasma membrane of AAMs
Full-length E-cadherin (120 kDa) and a shorter E-cadherin fragment (∼ 100 kDa), suggestive of proteolytic cleavage, are abundantly present in all conditions stimulating Cdh1 gene transcription (Figure 4A). Although E-cadherin is also present at a low basal level in unstimulated macrophages, no E-cadherin protein is detectable upon IFN-γ or LPS treatment (Figure 4A). Importantly, IL-4 and IL-13 also induce the E-cadherin protein in human peripheral blood monocyte–derived macrophages (Figure 4B).
The E-cadherin/catenin complex is formed at the plasma membrane of alternatively activated macrophages. (A) BALB/c thio-PEMs and (B) human monocyte–derived macrophages were treated for 24 hours with the indicated stimuli, followed by total cell lysate preparation and immunoblotting with antibodies against E-cadherin or β-, α-, or p120-catenin. β-Actin was probed as a loading control. (C) Naive (N) and IL-4–steered BALB/c thio-PEMs and NMe ECs were lysed, and immunoprecipitation was performed using anti–E-cadherin or isotype control antibodies. Immunoprecipitates (left) and total cell lysates (right) were immunoblotted with antibodies against E-cadherin, β-, α-, and p120-catenin. (D) NMe cells and BALB/c thio-PEMs, after 24 hours of treatment with the indicated stimuli, were stained with anti–mouse E-cadherin or isotype control and analyzed by FACS. The naive and NMe histograms show an overlay of isotype staining (dotted) with anti–E-cadherin staining (bold). For comparison, all other histograms show an overlay of anti–E-cadherin staining in stimulated macrophages (bold) with anti–E-cadherin staining in naive macrophages (dotted). Isotype stainings were similar in all conditions. (E) BALB/c thio-PEMs, whether or not pretreated for 24 hours with DFMO, were IL-4–steered for an additional 24 hours, stained with anti–mouse E-cadherin ECCD2 or isotype control and analyzed by FACS. E-cadherin surface expression values (ΔMFI = [median fluorescence intensity]anti-E-cad staining − [median fluorescence intensity]isotype staining) of a representative experiment were plotted.
The E-cadherin/catenin complex is formed at the plasma membrane of alternatively activated macrophages. (A) BALB/c thio-PEMs and (B) human monocyte–derived macrophages were treated for 24 hours with the indicated stimuli, followed by total cell lysate preparation and immunoblotting with antibodies against E-cadherin or β-, α-, or p120-catenin. β-Actin was probed as a loading control. (C) Naive (N) and IL-4–steered BALB/c thio-PEMs and NMe ECs were lysed, and immunoprecipitation was performed using anti–E-cadherin or isotype control antibodies. Immunoprecipitates (left) and total cell lysates (right) were immunoblotted with antibodies against E-cadherin, β-, α-, and p120-catenin. (D) NMe cells and BALB/c thio-PEMs, after 24 hours of treatment with the indicated stimuli, were stained with anti–mouse E-cadherin or isotype control and analyzed by FACS. The naive and NMe histograms show an overlay of isotype staining (dotted) with anti–E-cadherin staining (bold). For comparison, all other histograms show an overlay of anti–E-cadherin staining in stimulated macrophages (bold) with anti–E-cadherin staining in naive macrophages (dotted). Isotype stainings were similar in all conditions. (E) BALB/c thio-PEMs, whether or not pretreated for 24 hours with DFMO, were IL-4–steered for an additional 24 hours, stained with anti–mouse E-cadherin ECCD2 or isotype control and analyzed by FACS. E-cadherin surface expression values (ΔMFI = [median fluorescence intensity]anti-E-cad staining − [median fluorescence intensity]isotype staining) of a representative experiment were plotted.
In ECs, E-cadherin forms a complex with p120-, β-, and α-catenin, thereby linking the cell membrane to the cytoskeleton. In macrophages, the catenin mRNA levels are not modified by any of the stimuli tested (data not shown). α-Catenin protein and different p120-catenin isoforms are constitutively present in primary macrophages and the transformed Raw264.7 macrophage cell line. In contrast, β-catenin is only strongly detected in E-cadherin+ primary macrophages (Figure 4A) and E-cadherin-transfected30 Raw264.7 (supplemental Figure 6A), suggesting that E-cadherin protects β-catenin from degradation.
Immunoprecipitating E-cadherin from lysates of IL-4–steered thio-PEM pulls down all catenins, illustrating that a full E-cadherin/catenin complex is formed in AAMs. The amount of E-cadherin/catenin complexes in IL-4–treated macrophages is intermediate between the basal level found in naive macrophages and the high level in NMe ECs (Figure 4C).
Fluorescence microscopy on IL-4–steered thio-PEM (supplemental Figure 7) and E-cadherin–transfected Raw264.7 (supplemental Figure 6B) demonstrates the accumulation of E-cadherin/catenin complexes at the regions of cell contact, suggesting the potential involvement of E-cadherin in homotypic interactions between AAMs. To further confirm the surface availability of E-cadherin, FACS analysis was performed. E-cadherin surface expression is clearly detected on IL-4, IL-13, IL-4 + IL-13, IL-4 + IL-10, and IL-4 + TGF-β–treated thio-PEM, although the levels remain significantly below those in NMe ECs (Figure 4D). Finally, DFMO pretreatment reduces the E-cadherin surface expression in IL-4–steered macrophages by 70%, corroborating the important role of polyamines in this process (Figure 4E).
Induction of the E-cadherin/catenin complex in macrophages associated with T crassiceps helminth infection and allergic asthma
Prototypical Th2-associated pathologies, such as helminth infections and allergic asthma, strongly drive alternative activation of macrophages. We reported previously that T crassiceps helminths evoke alternatively activated peritoneal macrophages in BALB/c mice during the late stage of infection.18
Supporting our mRNA data (Figure 5A), the E-cadherin protein is only detected in PEM from WT, but not IL-4−/−, IL-4Rα−/−, or STAT6−/− 8- and 12-week-infected animals, illustrating the dominance of IL-4 signaling for in vivo E-cadherin induction. Besides β-catenin, also α-catenin is detected at a higher level in these in vivo induced AAMs compared with naive macrophages (Figure 5B). Similar to in vitro generated AAMs, E-cadherin is found at the cell surface, accumulating at sites of cell contact and colocalizing with β- and p120-catenin (Figure 5C). Finally, we wished to determine whether E-cadherin expression is restricted to IL-4–exposed macrophages or whether this molecule can be found in other hematopoietic cell types. FACS analysis on freshly isolated peritoneal cells from WT BALB/c- and C57BL/6-infected mice demonstrated the presence of E-cadherin at the surface of Ly6Chi inflammatory monocytes, immature differentiating macrophages, and mature macrophages, but not on any other cell type. No E-cadherin surface expression could be detected on any cell type in IL-4−/−, IL-4Rα−/−, and STAT6−/− mice (supplemental Figure 8).
The E-cadherin/catenin complex is induced in peritoneal alternatively activated macrophages elicited during T crassiceps infection. PEMs were isolated from BALB/c WT, IL-4−/−, IL-4Rα−/−, and STAT6−/− uninfected mice or 8 and 12 weeks after T crassiceps infection. (A) The fold Cdh1 induction relative to the expression in PEMs from the corresponding uninfected mice (= 1) is shown. (B) Peritoneal macrophages were analyzed by immunoblotting using antibodies against E-cadherin, β-, α- or p120-catenin. β-Actin was applied as a loading control. (C) Immunofluorescence microscopy on peritoneal macrophages isolated from T crassiceps infected WT BALB/c mice (8 weeks after infection). PEMs were grown on glass coverslips and after E-cadherin (i), β-catenin (ii,v), or p120-catenin (iv) labeling; images were obtained using an Olympus CellM fluorescence microscope. Subpanels iii and vi are merged pictures of the respective green and red images. Scale bars represent 20 μm.
The E-cadherin/catenin complex is induced in peritoneal alternatively activated macrophages elicited during T crassiceps infection. PEMs were isolated from BALB/c WT, IL-4−/−, IL-4Rα−/−, and STAT6−/− uninfected mice or 8 and 12 weeks after T crassiceps infection. (A) The fold Cdh1 induction relative to the expression in PEMs from the corresponding uninfected mice (= 1) is shown. (B) Peritoneal macrophages were analyzed by immunoblotting using antibodies against E-cadherin, β-, α- or p120-catenin. β-Actin was applied as a loading control. (C) Immunofluorescence microscopy on peritoneal macrophages isolated from T crassiceps infected WT BALB/c mice (8 weeks after infection). PEMs were grown on glass coverslips and after E-cadherin (i), β-catenin (ii,v), or p120-catenin (iv) labeling; images were obtained using an Olympus CellM fluorescence microscope. Subpanels iii and vi are merged pictures of the respective green and red images. Scale bars represent 20 μm.
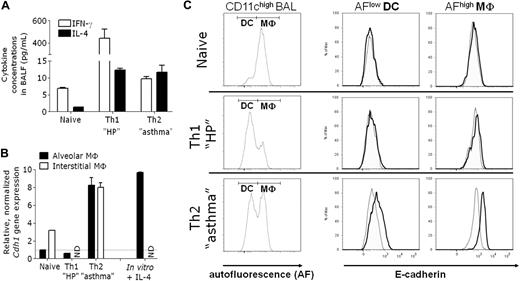
To further assess whether E-cadherin expression is a characteristic feature of macrophages in a polarized Th2 cytokine environment, we turned to mouse models of lung inflammation. Sensitization of C57BL/6 mice with OVA in Alum adjuvant, followed by OVA aerosols, triggers allergic airway inflammation,20 akin to moderate asthma associated with increased levels of IL-4, but not IFN-γ, in the bronchoalveolar lavage fluid (BALF; Figure 6A). In this model, both monocyte-derived macrophages and dendritic cells (DCs) were shown to regulate the lung pathology.31,32 First, we purified alveolar and interstitial lung macrophages from asthmatic or naive mice and compared the Cdh1 gene expression level in both conditions. Th2 inflammation significantly increases Cdh1 mRNA levels in both macrophage types (Figure 6B). Notably, in vitro stimulation of naive alveolar macrophages with IL-4 induces Cdh1 mRNA to the same extent, confirming allergic asthma as a strong AAM-driving pathology (Figure 6B). In agreement with the mRNA data, CD11chigh autofluorescence (AF)high alveolar macrophages33 from asthmatic mice express E-cadherin at the surface, whereas their naive counterparts do not (Figure 6C). Notably, 6-fold lower levels of E-cadherin are also induced on CD11chigh AFlow dendritic cells from asthmatic mice (ΔMFI = 163 ± 35 in DCs vs 945 ± 197 in macrophages; Figure 6C), whereas all CD11c− BAL cells score negative for E-cadherin (data not shown). Hence, although E-cadherin is associated with different types of monocyte-derived cells during Th2-driven diseases, mature macrophages are the highest expressers.
E-cadherin is induced in lung alternatively activated macrophages elicited during allergic asthma, but not hypersensitivity pneumonitis. Allergic asthma and hypersensitivity pneumonitis were induced in C57BL/6 mice. (A) IL-4 and IFN-γ concentrations were measured in the BALF. (B) Alveolar and interstitial lung macrophages were purified and Cdh1 expression in freshly isolated cells was determined. The fold induction of gene expression relative to the expression in naive alveolar macrophages (= 1) is shown. Naive alveolar macrophages were also cultured for 24 hours with or without IL-4 (in vitro + IL-4). The fold induction of Cdh1 gene expression in IL-4–treated relative to untreated alveolar macrophages (= 1) is shown. (C) BALs were stained with anti-CD11c and anti–E-cadherin or isotype control and analyzed via FACS. Within gated CD11chigh BAL, a distinction was made between AFlow DC and AFhigh alveolar macrophages and histogram overlays of isotype staining (dotted) and anti-E-cadherin staining (bold) are shown.
E-cadherin is induced in lung alternatively activated macrophages elicited during allergic asthma, but not hypersensitivity pneumonitis. Allergic asthma and hypersensitivity pneumonitis were induced in C57BL/6 mice. (A) IL-4 and IFN-γ concentrations were measured in the BALF. (B) Alveolar and interstitial lung macrophages were purified and Cdh1 expression in freshly isolated cells was determined. The fold induction of gene expression relative to the expression in naive alveolar macrophages (= 1) is shown. Naive alveolar macrophages were also cultured for 24 hours with or without IL-4 (in vitro + IL-4). The fold induction of Cdh1 gene expression in IL-4–treated relative to untreated alveolar macrophages (= 1) is shown. (C) BALs were stained with anti-CD11c and anti–E-cadherin or isotype control and analyzed via FACS. Within gated CD11chigh BAL, a distinction was made between AFlow DC and AFhigh alveolar macrophages and histogram overlays of isotype staining (dotted) and anti-E-cadherin staining (bold) are shown.
To assess whether E-cadherin expression can be induced in a mixed Th1/Th2 environment, we tested a mouse model of hypersensitivity pneumonitis, induced by the sensitization with OVA in complete Freund adjuvant and repeated OVA challenges.20 This approach results in an increase in IL-4 (similar to asthmatic mice) but an even higher increase in IFN-γ in BALF (Figure 6A). Recapitulating our in vitro findings (Figure 1A), IFN-γ counteracts the IL-4 effect in vivo, as no induction of Cdh1 mRNA and E-cadherin protein is detected during hypersensitivity pneumonitis (Figure 6B-C).
Together, these data propose E-cadherin as a surface marker that discriminates between AAMs and other macrophage activation states in vivo.
AAMs engage in homotypic and heterotypic interactions through E-cadherin
To gain insight into potential functions of E-cadherin in AAMs, we generated mice lacking E-cadherin expression in macrophages (Cdh1Δ). First, we evaluated whether E-cadherin on macrophages is able to interact heterotypically with KLRG1+ cells. Hereto, control Cdh1F/F or Cdh1Δ macrophages were pretreated with different stimuli and then cultured with KLRG1-reporter cells, which express a chimeric KLRG1/CD3ζ receptor driving nuclear factor of activated T cell (NFAT)–dependent GFP expression on ligation.25 Clearly, GFP is induced by Cdh1F/F macrophages stimulated with E-cadherin–inducing cytokine combinations, whereas stimulated Cdh1Δ macrophages do not induce GFP above background levels (Figure 7A).
IL-4–stimulated macrophages engage in homotypic and heterotypic interactions through E-cadherin. (A) A5 KLRG1-reporter cells were cocultured overnight with differentially stimulated Cdh1F/F control or Cdh1Δ thio-PEMs, and the percentage GFP+ reporter cells was determined by FACS. Data are mean ± SEM of 3 individual mice. (B) CD103+ CSFE-labeled MTC-1 cells were cocultured with untreated or IL-4–treated Cdh1F/F control or Cdh1Δ thio-PEM monolayers, which were either pretreated or not with ECCD2 antibodies. After 45 minutes, nonadherent cells were washed away, and the number of remaining MTC-1 cells was determined by fluorescence measurement. Data were plotted as the percentage change in the number of MTC-1 cells adhering to IL-4–treated thio-PEMs compared with untreated thio-PEMs. Mean ± SEM of 6 wells is shown for one representative experiment. (Ci-iii) Equal numbers of DiI (red) and DiO (green) labeled Cdh1F/F control (a-d) or Cdh1Δ (e-h) thio-PEMs were cultured for 24 hours with (b-d,f-h) or without (a,e) IL-4 on Permanox plastic and imaged by confocal microscopy. Volocity analysis software was applied to create a red/green colocalization channel (Ci). The average volume of colocalized voxels per object (Cii) and the amount of nuclei per fused cell (Ciii) were calculated. Data are mean ± SEM of 5 fields. **P < .01.
IL-4–stimulated macrophages engage in homotypic and heterotypic interactions through E-cadherin. (A) A5 KLRG1-reporter cells were cocultured overnight with differentially stimulated Cdh1F/F control or Cdh1Δ thio-PEMs, and the percentage GFP+ reporter cells was determined by FACS. Data are mean ± SEM of 3 individual mice. (B) CD103+ CSFE-labeled MTC-1 cells were cocultured with untreated or IL-4–treated Cdh1F/F control or Cdh1Δ thio-PEM monolayers, which were either pretreated or not with ECCD2 antibodies. After 45 minutes, nonadherent cells were washed away, and the number of remaining MTC-1 cells was determined by fluorescence measurement. Data were plotted as the percentage change in the number of MTC-1 cells adhering to IL-4–treated thio-PEMs compared with untreated thio-PEMs. Mean ± SEM of 6 wells is shown for one representative experiment. (Ci-iii) Equal numbers of DiI (red) and DiO (green) labeled Cdh1F/F control (a-d) or Cdh1Δ (e-h) thio-PEMs were cultured for 24 hours with (b-d,f-h) or without (a,e) IL-4 on Permanox plastic and imaged by confocal microscopy. Volocity analysis software was applied to create a red/green colocalization channel (Ci). The average volume of colocalized voxels per object (Cii) and the amount of nuclei per fused cell (Ciii) were calculated. Data are mean ± SEM of 5 fields. **P < .01.
A second unrelated KLRG1-reporter system34 confirms the potential of E-cadherin on AAMs to trigger KLRG1 signaling (supplemental Figure 9). Although macrophage E-cadherin levels are clearly adequate to provoke KLRG1 signaling, they are below a threshold level needed to trigger KLRG1-mediated T-cell inhibition (supplemental Figure 10).
To test whether E-cadherin+ macrophages could engage in heterotypic interactions with CD103+ T cells, Cdh1F/F and Cdh1Δ thio-PEMs were either pretreated for 24 hours with IL-4 or left untreated, before coculturing them with fluorescently labeled CD103+ MTC-1 T cells. IL-4 treatment of Cdh1F/F control macrophages leads to a significant increase in the number of adhering CD103+ cells relative to untreated Cdh1F/F macrophages, although this is not the case for Cdh1Δ macrophages. Blocking the E-cadherin–CD103 interaction with ECCD2 antibodies completely abolishes the increased MTC-1 adhesion to E-cadherin+ Cdh1F/F macrophages (Figure 7B), confirming the role of E-cadherin in this phenomenon.
E-cadherin was shown before to contribute to homotypic macrophage interactions and fusion,35 prompting us to study the fusion capacity of Cdh1Δ macrophages through a bifluorescent macrophage fusion assay.23 Although IL-4 increases the formation of multinucleated giant cells (MNG) in both Cdh1F/F WT and Cdh1Δ thio-PEMs (Figure 7Ci), the volume of each MNG (Figure 7Cii) and the average number of nuclei per MNG (Figure 7Ciii) are significantly lower for Cdh1Δ compared with Cdh1F/F macrophages. Remarkably, a partial reduction in E-cadherin expression after polyamine depletion is not sufficient to alter IL-4–stimulated macrophage fusion (supplemental Figure 11). We conclude that E-cadherin contributes to the fusion competence of AAMs.
Discussion
Helminths and allergens induce polarized Th2 responses, promoting alternative macrophage activation. Although AAMs are important regulators of these diseases,31,36 discriminative markers allowing their identification and contributing to their function are scarce. We identified E-cadherin as a molecule that associates with mouse and human macrophages exposed to IL-4 and IL-13 in the absence of Th1 stimuli. Consequently, E-cadherin expression is characteristic for macrophages associated with T crassiceps helminth infection and asthma, but not mixed Th1/Th2-cytokine–driven hypersensitivity pneumonitis.
In contrast to the wealth of information on E-cadherin in ECs, few data exist on its role in hematopoietic cells. LCs express high levels of E-cadherin, allowing them to form homotypic clusters and interact with ECs. Disruption of E-cadherin ligation leads to (partial) maturation of LCs37 and bone marrow–derived DCs.38 Contrary to the constitutive expression of E-cadherin in LCs and DCs, our data show that only minimal levels of E-cadherin are detected in mouse and human naive macrophages. Indeed, IL-4 or IL-13 is required to induce significant levels of E-cadherin in macrophages, both on cytokine stimulation in vitro or during Th2-driven pathologies in vivo. The fact that the concomitant presence of a Th1 cytokine or a TLR ligand abolishes E-cadherin expression in vitro and in vivo (during hypersensitivity pneumonitis) allows the use of this marker as a useful reporter of polarized Th2 responses and AAMs. This reporter function is further strengthened by our finding that also Ly6Chigh inflammatory monocytes and monocyte-derived inflammatory DCs up-regulate E-cadherin during polarized Th2 responses in vivo, although AAMs remain the highest expressers. In addition, the all-or-none difference in E-cadherin protein expression between AAMs and naive or differently activated macrophages is unique. Indeed, widely used AAM markers, such as MMR (CD206) or MGL-1/2, are already found at a significant level in naive macrophages, making their association with AAMs quantitative rather than discriminative.23,39
Similar to other IL-4–regulated macrophage markers, the induction of Cdh1 by IL-4 crucially depends on JAK/STAT6 activation. However, although the enhancers of highly up-regulated genes, such as arginase-1, contain at least one typical STAT6 response element,40 this is not the case for E-cadherin. The induction of Cdh1 by IL-4 within 1 hour suggests the rapid involvement of readily available IL-4–activated transcription factors. However, sustained Cdh1 transcription requires novel protein synthesis. Because STAT6 itself functions independently of protein synthesis,41 these data suggest the need for additional STAT6-dependent transcription factors or regulatory molecules. In this context, PPAR-δ, PPAR-γ, and gal3 are IL-4–regulated proteins that participate in a positive feedback loop to further enhance the IL-4–induced transcription of AAM genes.5-7 No PPAR response elements are found in the Cdh1 promoter, probably explaining the inability of PPAR-γ inhibition or PPAR-δ stimulation to alter Cdh1 transcription. Gal3 can induce gene expression via the stimulation of PI3K signaling. In our experiments, PI3K signaling indeed costimulates IL-4–induced Cdh1 transcription, although this happens independently of gal3. DAP12, another potential PI3K activator, was recently shown to enhance Cdh1 expression and might account for the observed PI3K effect in our results.23 Of note, p38 MAPK, a known inducer of a subset of IL-4–regulated genes,4 also costimulates the early IL-4 effect on Cdh1 transcription, but whether this is the result of the activation of p38-regulated transcription factors will need further investigation. As in LCs,12 a cross-talk is observed between IL-4 and TGF-β signaling, which synergistically enhance Cdh1 mRNA levels in all BALB/c macrophage populations tested. Our data show a requirement for simultaneous signaling and no effect on Cdh1 mRNA stability, favoring a model whereby IL-4- and TGF-β–induced transcription factors collaborate at the promoter site. However, these observations cannot be easily extrapolated to other cell types. Indeed, E-cadherin expression tends to be even inhibited by IL-4 in colon cancer and keratinocyte cell lines.42,43
Finally, we propose polyamines as novel agents involved in IL-4–dependent gene regulation. IL-4 potently induces Arg1 expression in macrophages, resulting in a strong elevation of intracellular polyamine levels that contribute to Cdh1 induction. Because Arg1 induction by IL-4 is STAT6-dependent,40 this mechanism may explain at least part of the STAT6 dependence of Cdh1 gene regulation. Although IL-4 and TGF-β also synergize to up-regulate Arg1 expression, this is unrelated to their synergy at the Cdh1 transcriptional level because combining both cytokines does not lead to higher polyamine levels compared with IL-4 alone. Among the polyamines, putrescine seems to be the most potent effector of Cdh1 gene transcription because (1) lowering the concentration of this polyamine alone suffices to significantly decrease the IL-4 effect, and (2) putrescine supplementation most efficiently restores E-cadherin levels in polyamine-depleted macrophages. High polyamine levels alone are not sufficient to induce expression of AAM markers, identifying these molecules as cofactors rather than as initiators of gene transcription. Polyamines do not induce IL-4Rα expression or STAT6 phosphorylation, so how they contribute to IL-4–induced gene expression remains unresolved. Possibly, polyamines modify DNA structures or induce polyamine-dependent transcription factors.44,45
Overall, the Cdh1 mRNA levels correlate well with E-cadherin protein content, suggesting the absence of major posttranscriptional regulation. One notable exception is the synergy between IL-4 and TGF-β, which was never observed at the protein level. p120 is usually required to stabilize E-cadherin at the plasma membrane, whereas β-catenin links this molecule to the actin cytoskeleton allowing cell-cell interactions.10 Similar to ECs, the β-catenin protein is almost undetectable in AAMs in the absence of E-cadherin, probably because of degradation by the proteasome, but rescued on association with that molecule. In contrast, p120- and α-catenin protein levels are significant in the absence of E-cadherin, suggesting the involvement of these proteins in macrophage housekeeping functions. Both catenins have reported effects on cell signaling when present in the cytoplasm or the nucleus, and it is tempting to speculate that these functions are altered because of the translocation of the catenins to the cadherin complex.10
The role of E-cadherin in mediating cell-cell interactions is shown by using IL-4–stimulated Cdh1Δ macrophages. Using KLRG1-reporter cells, we demonstrated that E-cadherin levels in AAMs are sufficient to ligate KLRG1 and induce downstream signaling. KLRG1 preferentially recruits SHIP-1 to its immunoreceptor tyrosine-based inhibitory motif (ITIM), thereby inhibiting suboptimal T-cell receptor signaling and natural killer cytotoxicity.25,34,46 However, we were unable to show any impact of E-cadherin+ AAMs on T-cell receptor signaling. This could reflect the need for high E-cadherin surface expression to reach threshold levels of KLRG1 signaling, as was suggested before.46
E-cadherin is also known to interact with αEβ7 integrin (CD103), promoting adhesion between cells expressing both proteins. We demonstrated that Cdh1F/F AAMs, but not their Cdh1Δ counterparts, trap CD103+ T-cell hybridomas in an E-cadherin–dependent fashion. Interestingly, CD103 is found on key orchestrators of the immune response, such as DCs and T-cell subsets.13-15,47 Hence, E-cadherin might serve to bring these cells in closer contact with anti-inflammatory AAMs, thereby potentially influencing their retention in tissues and phenotype during polarized Th2 responses.
Finally, Cdh1Δ macrophages were shown to fuse on IL-4 treatment, but the size and the number of nuclei in each giant cell are significantly lower compared with Cdh1F/F controls. These data provide the first genetic evidence for a model whereby different IL-4–induced molecules, including E-cadherin, need to collaborate to fully exploit the fusogenic capacity of macrophages, as has been suggested before.35 This finding could help to better understand the formation of MNG during the foreign body response or granuloma formation.
Overall, our data demonstrate that E-cadherin, although considered as an orchestrator of EC biology, could be important in the immune system as effector molecule of alternatively activated macrophages, increasing their fusion capacity and allowing these cells to interact with other hematopoietic cells (supplemental Figure 12).
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ella Omasta, Lea Brys, Hilde Verlinden, Petra D'Hooge, Marie-Thérèse Detobel, Nadia Abou, and Eddy Vercauteren for their technical aid, Marianthi Tatari for initiating the LysM-Cre x Cdh1F/F breeding, and Drs F. Brombacher, D. K. Hsu, and J. Jonkers for providing mice.
This work was supported by a doctoral grant from FWO-Vlaanderen and a postdoctoral grant from Stichting tegen Kanker.
Authorship
Contribution: J.V.d.B. designed and performed research, analyzed and interpreted results, made the figures, and wrote the paper; P.B. designed and performed research and analyzed and interpreted data; J.v.H. designed research and interpreted results; G.B., H.P., and P.D. provided materials; K.M., R.V.d.B., and A.P.-F. performed research; C.J.G. performed microscopy experiments; J.G., J.M.C.G., and P.D.B. designed research; and J.A.V.G. designed and performed research, interpreted results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jo A. Van Ginderachter, Laboratory of Cellular and Molecular Immunology, Department of Molecular and Cellular Interactions, VIB-Vrije Universiteit Brussel, Building E, Level 8, Pleinlaan 2, B-1050, Brussels, Belgium; e-mail: jo.vanginderachter@vib-vub.be.


![Figure 4. The E-cadherin/catenin complex is formed at the plasma membrane of alternatively activated macrophages. (A) BALB/c thio-PEMs and (B) human monocyte–derived macrophages were treated for 24 hours with the indicated stimuli, followed by total cell lysate preparation and immunoblotting with antibodies against E-cadherin or β-, α-, or p120-catenin. β-Actin was probed as a loading control. (C) Naive (N) and IL-4–steered BALB/c thio-PEMs and NMe ECs were lysed, and immunoprecipitation was performed using anti–E-cadherin or isotype control antibodies. Immunoprecipitates (left) and total cell lysates (right) were immunoblotted with antibodies against E-cadherin, β-, α-, and p120-catenin. (D) NMe cells and BALB/c thio-PEMs, after 24 hours of treatment with the indicated stimuli, were stained with anti–mouse E-cadherin or isotype control and analyzed by FACS. The naive and NMe histograms show an overlay of isotype staining (dotted) with anti–E-cadherin staining (bold). For comparison, all other histograms show an overlay of anti–E-cadherin staining in stimulated macrophages (bold) with anti–E-cadherin staining in naive macrophages (dotted). Isotype stainings were similar in all conditions. (E) BALB/c thio-PEMs, whether or not pretreated for 24 hours with DFMO, were IL-4–steered for an additional 24 hours, stained with anti–mouse E-cadherin ECCD2 or isotype control and analyzed by FACS. E-cadherin surface expression values (ΔMFI = [median fluorescence intensity]anti-E-cad staining − [median fluorescence intensity]isotype staining) of a representative experiment were plotted.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/114/21/10.1182_blood-2009-05-221598/4/m_zh89990944060004.jpeg?Expires=1769085234&Signature=CvsihIOw2intknylPI4pNWIPVuX-7SblYbm9hQPAgRawSh4DhGXmM4S~EFSdvchpmAIS3C~JJ~pJ3N8z0pAJ3xrUwmLqMg~PpbAVJoPk-xNHrigydkVPpIS0nGhL4iYLyzV1kS2AdE4wPiCcMjKKFPzMicyWaOjeW86MNRVzkBYioTmKW2Bc6wYW3K3WPFExVlzc6JKoBYsNMP2VvfhR99AXN5XwgDSsAp5isFiEOsoSUCiVG4ZNqkeexQGqCcZty3LFKcme5GJGS6dSOxhy~I4r7jL7I7~pnAPCUX5mnV7q6~jmx94OTauLeQ-lKpvPSti-33BLfMZqWvB22zmSaA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal