Abstract
Hairy cell leukemia variant (HCLv) presents with high disease burden, lack of typical antigens like CD25, and poor response to standard treatments like cladribine. Occasionally, patients with classic HCL respond poorly. Clinical and molecular features of HCL and HCLv has not been compared. Rearrangements expressing immunoglobulin VH chain were sequenced, including 22 from 20 patients with HCLv and 63 from 62 patients with classic HCL. Most patients were seeking relapsed/refractory trials, representing a poor-prognosis population. VH4-34, a gene commonly used in autoimmune disorders, was observed in 8 (40%) HCLv and 6 (10%) classic (P = .004) HCL patients. Compared with 71 VH4-34− rearrangements, 14 VH4-34+ rearrangements were more frequently (P < .001) unmutated, defined as greater than 98% homologous to germline sequence. VH4-34+ patients had greater white blood cell counts at diagnosis (P = .002), lower response rate (P < .001) and progression-free survival (P = .007) after initial cladribine, and shorter overall survival from diagnosis (P < .001). Response and survival were more closely related to VH4-34 status than to whether or not patients had HCLv. VH4-34+ HCL is an important disorder that only partly overlaps with the previously described HCLv. Response to initial single-agent cladribine therapy is suboptimal; these patients should be considered for alternative approaches, including antibody-related therapy.
Introduction
Hairy cell leukemia variant (HCLv) is a B-cell disorder, recognized for nearly 30 years, which accounts for 10% of hairy-cell leukemia (HCL) cases. Morphology of variant cells was reported to be intermediate between that of classic HCL and prolymphocytic leukemias.1-3 Patients typically present with leukocytosis rather than leukopenia and often lack the neutropenia, anemia, and/or thrombocytopenia with which classic HCL patients present.1,2,4 By flow cytometry, B-cell antigens FMC7, CD11c, CD20, CD22, and surface immunoglobulin are strongly positive in both classic HCL and HCLv, whereas HCLv differs from classic HCL by lack of CD25, HC-2, and CD123 and by expression of CD27.3-7 CD103 is usually positive in both but can be negative in HCLv.2 HCLv lacking both CD25 and CD103 may be difficult to differentiate from splenic marginal zone lymphoma (SMZL)/splenic lymphoma with villous lymphocytes without also relying on morphologic differences between HCL and SMZL.2,6,7 In contrast to the high complete remission and overall response rates of classic HCL to the administration of purine analogs pentostatin and cladribine,8-10 response in patients with HCLv is limited to partial responses in approximately 50% of patients.2-4,11,12 Several complete responses of HCLv to monoclonal antibody-based therapy with and without chemotherapy have been reported.13-17
Like other mature B lymphocytes, the malignant cells in HCL patients have 1 or sometimes 2 different rearrangements for immunoglobulin heavy chain. Significant variability can be observed in the third complementarity determining region (CDR3), comprising the variable heavy (VH), Dbl homology, and junctional heavy domains. We previously reported a molecular characterization of 24 such rearrangements in 23 HCL patients18 and discussed previous studies in which 70 rearrangements in 69 patients were described.19-23 We reported that of 4 patients in our series with unmutated rearrangements, defined as greater than 98% homology to germline sequence, 3 presented with high tumor burden consistent clinically with variant disease. However, the CD25/VH status in these 3 patients was CD25−/VH4-34+, CD25−/VH4-34−, and CD25+/VH4-34+,18 suggesting that VH4-34 expression and adverse clinical behavior may not be confined to HCLv diagnosed by immunophenotype. A more recent VH study in HCL reported that 5 of 38 cases were HCLv, 2 of which were unmutated VH4-34.24 Another study reported that 5 of 83 cases were HCLv, and 5 classic cases were VH4-34+, 3 of which were unmutated.25 To better understand the association of VH4-34 and other VH genes with the variant immunophenotype and to determine whether molecular features could be prognostically important, independent of the diagnosis of classic HCL or HCLv, we studied immunoglobulin rearrangements and clinical factors in 82 HCL patients, 20 of whom had HCLv.
Methods
Patients and controls
Blood for DNA study was obtained as part of sample acquisition protocols with informed consent approved by the NCI Investigator's Review Board and in accordance with the Declaration of Helsinki. All samples were retrieved between 2001 and 2008. Of the 85 rearrangements in 82 patients examined, 24 rearrangements in 23 patients were published previously.18 Diagnoses of classic HCL and HCLv were rendered by a hematopathologist (M.S.-S.), based upon morphology and immunophenotype and according to World Health Organization classification.26
HCL was characterized by a proliferation of medium-sized lymphoid cells with mature chromatin; circumferential hairy-like cytoplasmic projections; reticulin fibrosis and interstitial infiltration in bone marrow; tartrate-resistant acid phosphatase (TRAP) positivity; bright coexpression of CD20, CD22, and CD11c; and expression of CD25, CD103, and CD123. HCLv was characterized by a proliferation of abnormal lymphoid cells varying from medium to large in size, often with prominent nucleoli and irregular nuclear contours, with hairy-like cytoplasmic projections. Like classic HCL, HCLv usually demonstrated bright coexpression of CD20, CD22, and CD11c and expression of CD103 but was negative for CD25 and frequently CD123.2,5,26 TRAP, when available, usually was positive. SMZL, which was excluded from this study, was diagnosed based upon morphology and immunophenotype, namely relatively small lymphoid cells with occasional cytoplasmic projections in polar orientation; moderate coexpression of CD20, CD22, and dim-to-moderate CD11c; and negativity for TRAP, CD103, and CD123.2,5,26 For each of the 82 patients assigned a diagnosis of HCL, the leukemic cells were examined in the same National Institutes of Health (NIH) laboratory for morphology and flow cytometry. In addition, bone marrow biopsy samples were evaluated at NIH for 63 patients, and bone marrow biopsy reports were obtained for 17 others. Of 32 asplenic patients, in 5 the spleen tissue was examined at NIH, and the pathology reports were obtained for 13 others.
cDNA synthesis and characterization of rearrangements
Total RNA was extracted from PAXgene Blood RNA tubes (PreAnalytiX GmbH) containing peripheral-blood mononuclear cells by use of the PAXgene Blood RNA kit (PreAnalytiX GmbH). Total RNA from CD11c-sorted cells was purified by use of the MagMax kit (Ambion). First-strand cDNA synthesis was performed by SuperScriptTM III RnaseH-Reverse Transcriptase (Invitrogen) from 1 to 3 μg of total RNA in reaction mix in the presence of Oligo(dT)20 primer (Invitrogen) and 10 mmol/L dNTP mix according to the manufacturer's protocol as described previously.18 cDNA was amplified in a single multiplexed polymerase chain reaction (PCR) consisting of 6 VH framework 1 primers combined with 1 JH consensus primer as designed for the BIOMED-2 study.27
All reactions were carried out in 100 μL containing 10 pmol of each primer (InVivoScribe Technologies), 200 μmol/L dNTPs, 0.5 U AmpliTaq Gold, and 10X PCR buffer II (Applied Biosystems). The DNA was amplified as follows: denaturation at 95°C for 7 minutes, 37 cycles of 94°C for 1 minute, 55°C for 30 seconds, 72°C for 1 minute, with a final extension of 10 minutes at 72°C. PCR products were analyzed on a 2% agarose gel and visualized with ethidium bromide staining, purified by the use of QIAquick PCR purification kit (QIAGEN), and cloned by the use of the Zero Blunt TOPO PCR Cloning kit (Invitrogen). Cloned PCR products were sequenced by the use of T3 consensus primers and Big-Dye terminators (Applied Biosystems). A monoclonal sequence was defined when at least 5 of 10 clones had a completely identical sequence. Intraclonal and ongoing somatic hypermutation were not observed. No 2 rearrangements from different patients were identical, which was evidence against any being a contaminant. The structure of each rearrangement was determined by sequence analysis by use of the IMGT database28 and the VBASE2 database.29
Cell purification with CD11c sorting
Peripheral-blood mononuclear cells were purified from 25 mL of heparin-containing peripheral blood of HCL patients by Ficoll-Paque Plus (GE Healthcare Bio-Science AB) by use of the manufacturer's protocol and then counted. The peripheral B-cell population was isolated by the Dynal B-cell–negative isolation kit (Invitrogen Dynal AS) and incubated with 20 μL of Fc receptor blocking reagent (Miltenyi Biotec) for 10 minutes at 4°C. One microgram of CD11c antibody (Santa Cruz Biotechnology) per 106 cells was added to the sample and incubated on ice for 1 hour. Cells were washed by adding 2 mL of 0.1% bovine serum albumin containing phosphate-buffered saline and centrifuged to 1200 rpm for 8 minutes at 4°C. The cells were then incubated with goat anti–mouse IgG Dynabeads (Invitrogen Dynal AS) for 15 minutes at 4°C with gentle tilting and rotation followed by magnetic capture of the CD11c-positive population, followed by addition of TRI Reagent (Ambion Inc) for future RNA purification.
Statistics
Comparisons between groups of patients were performed by the Fisher exact test or by Wilcoxon rank sum test as appropriate. Kaplan-Meier curves were compared by log-rank, and median survival of a population was considered 50% probability of survival. Calculations were performed using SAS, and P values presented were 2-sided.
Assessment of response to initial treatment with cladribine
Patients with sufficient medical records, including dates of treatment, complete blood counts, and bone marrow biopsy reports, were considered evaluable for retrospective response assessment, providing their first purine analog treatment was cladribine. Best response was considered after just the first course of cladribine and was determined on the basis of published criteria for HCL,9 except that hemoglobin had to reach 11 rather than 12 for a complete remission. Also, in patients who were red blood cell transfusion dependent, the 50% improvement in hemoglobin needed for partial response was assumed if the hemoglobin reached 9 g/dL without transfusions. Because bone marrow biopsy was not always used after cladribine, we also determined whether patients achieved hematologic remission, defined as meeting all criteria for complete remission except for the bone marrow biopsy. Date of progression was defined as the first documentation of relapse from partial response in responding patients or progressive disease in nonresponding patients.
Results
Patient characteristics
Of 82 patients with HCL, 62 (76%) were considered classic HCL, whereas 20 (24%) were considered HCLv. Immunophenotypic characteristics are shown in Table 1 for the 26 patients who had either HCLv or VH4-34+ rearrangements or both. The remaining 56 classic HCL patients are summarized in Table 2. HCL cells from all patients expressed the B-cell markers CD19, CD20 (bright in 100%), and CD22 (bright in 98%), as well as CD11c (bright in 96%). HCL cells from all classic patients expressed CD25, which was bright in 44 (71%), positive in 17 (27%), and in 1 (2%) patient varied from bright to negative. In contrast, the HCLv cells were dim to negative or negative for CD25. CD123 was expressed homogenously in 31 (100%) of 31 classic patients and in 3 (38%) of 8 HCLv patients evaluated, and 1 HCLv patient had partial CD123 expression. CD103 was expressed homogenously in 77 (94%) of the 82 patients. Two patients with classic HCL had partial CD103 expression, 1 of whom had previously documented homogenous CD103 expression. The 3 CD103− patients also were CD25− but had either TRAP+ or CD11c bright-positive cells consistent with HCLv rather than SMZL. Patient BL26 had partial positive CD11c expression with lack of CD25 and CD103, suggesting SMZL. However, the splenic pathology showed red pulp rather than a white pulp infiltration, and positivity for both TRAP and DBA.44 were more consistent with HCLv than with SMZL.2,5,30 Lymphoplasmacytic lymphoma was considered but the morphology, the bright expression of CD22 and CD20, and the lack of CD25 were against this diagnosis.31 One patient previously diagnosed as HCLv was diagnosed with SMZL on the basis of flowcytometry and morphology and was therefore not included in the 82 patients reported. The male/female ratio was 74:8 and not different with respect to variant status. Age from diagnosis was older for HCLv than for classic HCL (P = .02), with median age 55 versus 46 years.
Immunophenotypic and molecular characteristics of patients with VH4-34 or HCLv (n = 26)
| Patient no. . | Age . | Sex . | CD19 . | CD20 . | CD22 . | CD25 . | CD11C . | CD5 . | CD10 . | CD23 . | FMC7 . | CD103 . | CD123 . | LC . | TR . | Dx . | VH . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BH18 | 57 | M | ++ | +++ | +++ | +++ | +++ | − | − | − | ++ | ++ | NA | κ | + | HCL | VH4-34 |
| BH32 | 58 | M | ++ | +++ | +++ | +++ | +++ | NA | NA | NA | NA | ++ | ++ | λ | NA | HCL | VH4-34 |
| BH34 | 56 | M | ++ | +++ | +++ | P+ | +++ | − | − | − | +++ | + | NA | λ | + | HCL | VH4-34 |
| BN27 | 38 | M | ++ | +++ | +++ | +++ | +++ | − | − | − | ++ | ++ | NA | κ | + | HCL | VH4-34 |
| BN29 | 55 | M | ++ | +++ | +++ | +++ | +++ | − | − | − | ++ | ++ | NA | λ | NA | HCL | VH4-34 |
| HH14 | 50 | M | ++ | +++ | +++ | ++ | +++ | − | − | − | ++ | ++ | ++ | κ | − | HCL | VH4-34 |
| BH16 | 52 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | + | NA | κ | + | HCLv | VH4-34 |
| BH28 | 42 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | λ | + | HCLv | VH4-34 |
| BH29 | 55 | M | ++ | +++ | +++ | +/− | +++ | − | − | − | ++ | ++ | ++ | λ | + | HCLv | VH4-34 |
| BL18 | 69 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | κ | + | HCLv | VH4-34 |
| BL26 | 55 | M | ++ | +++ | ++ | − | P+ | − | − | − | ++ | − | NA | κ | + | HCLv | VH4-34 |
| BN15 | 61 | M | ++ | +++ | +++ | − | +++ | − | + | − | ++ | ++ | − | κ | NA | HCLv | VH4-34 |
| BN21 | 71 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | − | NA | κ | NA | HCLv | VH4-34 |
| BN35 | 55 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | − | κ | NA | HCLv | VH4-34 |
| HH10 | 40 | F | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | − | κ | + | HCLv | VH1-18 |
| BN17 | 72 | M | +++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | λ | NA | HCLv | VH3-23 |
| BN20 | 50 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | κ | + | HCLv | VH3-48 |
| BN23 | 35 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | P+ | λ | NA | HCLv | VH3-66 |
| BL14 | 38 | M | ++ | +++ | +++ | +/− | +++ | − | − | − | ++ | ++ | + | κ | + | HCLv | VH4-31 |
| BN16 | 63 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | λ | + | HCLv | VH4-31 |
| BN22 | 55 | F | ++ | +++ | ++ | +/− | ++ | − | − | − | ++ | ++ | NA | λ | + | HCLv | VH4-39 |
| BN39 | 77 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | + | κ | NA | HCLv | VH4-39 |
| BN24 | 73 | M | ++ | +++ | +++ | − | ++ | − | − | − | ++ | ++ | NA | − | NA | HCLv | VH4-61 |
| BN04 | 57 | M | ++ | +++ | +++ | − | +++ | P+ | − | − | ++ | − | NA | κ | + | HCLv | VH4-61 |
| HH19 | 44 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | κ | NA | HCLv | VH5-a |
| BN19 | 37 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | − | κ | NA | HCLv | VH6-1 |
| Patient no. . | Age . | Sex . | CD19 . | CD20 . | CD22 . | CD25 . | CD11C . | CD5 . | CD10 . | CD23 . | FMC7 . | CD103 . | CD123 . | LC . | TR . | Dx . | VH . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BH18 | 57 | M | ++ | +++ | +++ | +++ | +++ | − | − | − | ++ | ++ | NA | κ | + | HCL | VH4-34 |
| BH32 | 58 | M | ++ | +++ | +++ | +++ | +++ | NA | NA | NA | NA | ++ | ++ | λ | NA | HCL | VH4-34 |
| BH34 | 56 | M | ++ | +++ | +++ | P+ | +++ | − | − | − | +++ | + | NA | λ | + | HCL | VH4-34 |
| BN27 | 38 | M | ++ | +++ | +++ | +++ | +++ | − | − | − | ++ | ++ | NA | κ | + | HCL | VH4-34 |
| BN29 | 55 | M | ++ | +++ | +++ | +++ | +++ | − | − | − | ++ | ++ | NA | λ | NA | HCL | VH4-34 |
| HH14 | 50 | M | ++ | +++ | +++ | ++ | +++ | − | − | − | ++ | ++ | ++ | κ | − | HCL | VH4-34 |
| BH16 | 52 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | + | NA | κ | + | HCLv | VH4-34 |
| BH28 | 42 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | λ | + | HCLv | VH4-34 |
| BH29 | 55 | M | ++ | +++ | +++ | +/− | +++ | − | − | − | ++ | ++ | ++ | λ | + | HCLv | VH4-34 |
| BL18 | 69 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | κ | + | HCLv | VH4-34 |
| BL26 | 55 | M | ++ | +++ | ++ | − | P+ | − | − | − | ++ | − | NA | κ | + | HCLv | VH4-34 |
| BN15 | 61 | M | ++ | +++ | +++ | − | +++ | − | + | − | ++ | ++ | − | κ | NA | HCLv | VH4-34 |
| BN21 | 71 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | − | NA | κ | NA | HCLv | VH4-34 |
| BN35 | 55 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | − | κ | NA | HCLv | VH4-34 |
| HH10 | 40 | F | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | − | κ | + | HCLv | VH1-18 |
| BN17 | 72 | M | +++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | λ | NA | HCLv | VH3-23 |
| BN20 | 50 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | κ | + | HCLv | VH3-48 |
| BN23 | 35 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | P+ | λ | NA | HCLv | VH3-66 |
| BL14 | 38 | M | ++ | +++ | +++ | +/− | +++ | − | − | − | ++ | ++ | + | κ | + | HCLv | VH4-31 |
| BN16 | 63 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | λ | + | HCLv | VH4-31 |
| BN22 | 55 | F | ++ | +++ | ++ | +/− | ++ | − | − | − | ++ | ++ | NA | λ | + | HCLv | VH4-39 |
| BN39 | 77 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | + | κ | NA | HCLv | VH4-39 |
| BN24 | 73 | M | ++ | +++ | +++ | − | ++ | − | − | − | ++ | ++ | NA | − | NA | HCLv | VH4-61 |
| BN04 | 57 | M | ++ | +++ | +++ | − | +++ | P+ | − | − | ++ | − | NA | κ | + | HCLv | VH4-61 |
| HH19 | 44 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | NA | κ | NA | HCLv | VH5-a |
| BN19 | 37 | M | ++ | +++ | +++ | − | +++ | − | − | − | ++ | ++ | − | κ | NA | HCLv | VH6-1 |
Dx indicates diagnosis; HCL, classic hairy-cell leukemia; HCLv, variant HCL; LC, light chain; NA, not available; P+, part positive; and TR, tartrate-resistant acid phosphatase stain.
Immunophenotypic and molecular characteristics of patients with non–VH4-34 HCL (n = 56)
| . | CD19 . | CD20 . | CD22 . | CD25 . | CD11C . | CD5 . | CD10 . | CD23 . | FMC7 . | CD103 . | CD123 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Evaluable | 56 | 56 | 56 | 56 | 56 | 50 | 49 | 52 | 52 | 56 | 29 |
| Bright + (+++) | 1 | 56 | 56 | 40 | 56 | 2 | |||||
| Positive (++) | 54 | 16 | 2 | 2 | 50 | 54 | 27 | ||||
| Dim + (+) | 1 | 2 | 2 | ||||||||
| Dim to negative (+/−) | 1 | 1 | |||||||||
| Part positive (P+) | 5 | 1 | 3 | 2 | |||||||
| Negative (−) | 44 | 44 | 46 |
| . | CD19 . | CD20 . | CD22 . | CD25 . | CD11C . | CD5 . | CD10 . | CD23 . | FMC7 . | CD103 . | CD123 . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Evaluable | 56 | 56 | 56 | 56 | 56 | 50 | 49 | 52 | 52 | 56 | 29 |
| Bright + (+++) | 1 | 56 | 56 | 40 | 56 | 2 | |||||
| Positive (++) | 54 | 16 | 2 | 2 | 50 | 54 | 27 | ||||
| Dim + (+) | 1 | 2 | 2 | ||||||||
| Dim to negative (+/−) | 1 | 1 | |||||||||
| Part positive (P+) | 5 | 1 | 3 | 2 | |||||||
| Negative (−) | 44 | 44 | 46 |
One patient had monophasic light chain. Median patient age = 45 years; male/female = 50:6; κ/λ light chain = 25:31; TRAP +:− = 38:1.
HCL indicates classic hairy cell leukemia; and TRAP, tartrate-resistant acid phosphatase stain.
VH usage
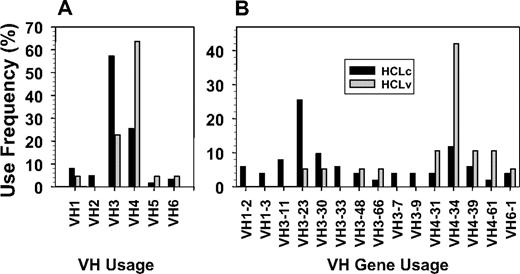
To investigate immunoglobulin VH usage in classic and variant HCL, a total of 63 rearrangements in 62 classic HCL and 22 rearrangements in 20 HCLv patients were amplified, sequenced, and analyzed. As shown in Figure 1A, VH3-family VH chains were more common in classic HCL than HCLv (57% vs 23%, P = .007), and VH4 was more common in HCLv than in classic disease (64% vs 25%, P = .002). The usage in classic HCL is similar to the reported32 usage of VH families 1 to 7 in 206 rearrangements from normal (CD5−/IgM+) B cells, namely 13.1%, 1.9%, 53.9%, 24.8%, 2.9%, 2.4%, and 1%, respectively. However, in HCLv, VH3 usage was lower (P = .007) and VH4 usage was greater (P < .001) compared with normal B cells.
VH use frequency in HCL. A, V families; B, VH gene usage. Patients had classic (back bars) or variant (shaded bars) HCL.
VH use frequency in HCL. A, V families; B, VH gene usage. Patients had classic (back bars) or variant (shaded bars) HCL.
VH gene usage
When further subdivided into individual genes within each family, a total of 30 different VH genes were cloned. Half (15/30) of these occurred in more than 1 patient, and their frequencies are shown in Figure 1B. The most frequently used classic VH genes were VH3-23 (n = 13, 21%), VH4-34 (n = 6, 10%), and VH3-30 (n = 5, 8%). Of the 3 patients with 2 rearrangements each, 1 with HCLv expressed both VH3-66 and VH3-30-3, 1 with HCLv expressed both VH4-39 and VH3-30, and 1 with classic HCL expressed both VH1-2 and VH3-74. The most frequently used variant VH gene was VH4-34 (n = 8, 36%), and the largest difference between classic and variant rearrangements was observed with incidence of VH4-34 (10% vs 36%, P = .007). Thus, VH4-34 was more common in but not confined to HCLv.
Somatic hypermutation
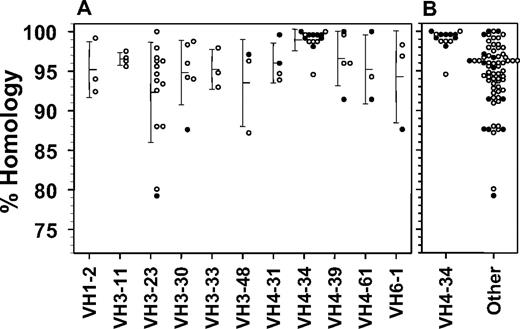
To determine whether the frequency of somatic mutations in the VH rearrangements differed with respect to variant disease or VH4-34 expression, they were cloned and compared with the IMGT database. Figure 2 shows homology to germline sequence for the 11 different rearrangements observed in at least 3 patients each. All but 1 VH4-34+ rearrangement was unmutated compared with 15% of the others (P < .001). Homology to germline sequences was greater for VH4-34+ compared with VH4-34− rearrangements (median homology 99.4% vs 95.6%, P < .001). In contrast, there was no relationship between homology and variant versus classic phenotype (97.6% vs 95.9%, P = .12), despite the relationship between variant phenotype and VH4-34 positivity. In fact, when considering the 22 variant and 63 classic rearrangements separately, VH4-34 positivity in each group is still associated with increased homology (Table 3, P = .006-.02). Thus, VH4-34 expression is associated with high homology to germline sequence unrelated to diagnosis of HCLv.
Homology to germline sequence for rearrangements with the indicated VH gene usage and combined data from 71 non–VH4-34–expressing rearrangements. Error bars indicate SDs from the means of the points shown. Patients with classic (○) or variant (●) hairy cells are shown.
Homology to germline sequence for rearrangements with the indicated VH gene usage and combined data from 71 non–VH4-34–expressing rearrangements. Error bars indicate SDs from the means of the points shown. Patients with classic (○) or variant (●) hairy cells are shown.
Characteristics at diagnosis
| VH4-34 status and variant phenotype . | VH4-34+ . | VH4-34− . | P . | HCLv . | HCL . | P . |
|---|---|---|---|---|---|---|
| Median age at diagnosis (n) | 55.0 (14) | 45.6 (68) | .011 | 54.8 (20) | 45.7 (62) | .016 |
| Median, homology, % (n) | 99.4 (14) | 95.6 (71) | < .0001 | 97.6 (22) | 95.9 (63) | .12 |
| Median homology, %, classic HCL only (n) | 98.7 (6) | 95.6 (57) | .006 | |||
| Median homology, %, HCLv only (n) | 99.6 (8) | 95.7 (14) | .02 | |||
| White blood cell count, > 5 × 109/L (%) | 13/14 (93) | 23/57 (40) | .0006 | 15/18 (83) | 21/53 (40) | .002 |
| White blood cell count, > 5 × 109/L, classic HCL only (%) | 6/6 (100%) | 15/47 (32%) | .002 | |||
| White blood cell count > 5 × 109/L, VH4-34− only (%) | 8/10 (80%) | 15/47 (32%) | .01 | |||
| Median white blood cell count, ×109/L (n) | 9.0 (14) | 3.8 (57) | .002 | 11.3 (18) | 3.5 (53) | < .0001 |
| Median white blood cell count, ×109/L, classic HCL only (n) | 10.5 (6) | 2.9 (47) | .003 | |||
| Median white blood cell count, ×109/L, VH4-34− only (n) | 13.8 (10) | 2.9 (47) | .0003 | |||
| Median neutrophils, ×109/L (n) | 2.23 (12) | 0.82 (49) | .004 | 3.01 (14) | 0.77 (47) | < .0001 |
| Median hemoglobin, g/dL | 13.2 (13) | 11.6 (52) | .12 | 13.2 (17) | 11.4 (48) | .006 |
| Median platelets, ×109/L (n) | 105 (13) | 68 (54) | .014 | 135 (16) | 65 (51) | .0001 |
| VH4-34 status and variant phenotype . | VH4-34+ . | VH4-34− . | P . | HCLv . | HCL . | P . |
|---|---|---|---|---|---|---|
| Median age at diagnosis (n) | 55.0 (14) | 45.6 (68) | .011 | 54.8 (20) | 45.7 (62) | .016 |
| Median, homology, % (n) | 99.4 (14) | 95.6 (71) | < .0001 | 97.6 (22) | 95.9 (63) | .12 |
| Median homology, %, classic HCL only (n) | 98.7 (6) | 95.6 (57) | .006 | |||
| Median homology, %, HCLv only (n) | 99.6 (8) | 95.7 (14) | .02 | |||
| White blood cell count, > 5 × 109/L (%) | 13/14 (93) | 23/57 (40) | .0006 | 15/18 (83) | 21/53 (40) | .002 |
| White blood cell count, > 5 × 109/L, classic HCL only (%) | 6/6 (100%) | 15/47 (32%) | .002 | |||
| White blood cell count > 5 × 109/L, VH4-34− only (%) | 8/10 (80%) | 15/47 (32%) | .01 | |||
| Median white blood cell count, ×109/L (n) | 9.0 (14) | 3.8 (57) | .002 | 11.3 (18) | 3.5 (53) | < .0001 |
| Median white blood cell count, ×109/L, classic HCL only (n) | 10.5 (6) | 2.9 (47) | .003 | |||
| Median white blood cell count, ×109/L, VH4-34− only (n) | 13.8 (10) | 2.9 (47) | .0003 | |||
| Median neutrophils, ×109/L (n) | 2.23 (12) | 0.82 (49) | .004 | 3.01 (14) | 0.77 (47) | < .0001 |
| Median hemoglobin, g/dL | 13.2 (13) | 11.6 (52) | .12 | 13.2 (17) | 11.4 (48) | .006 |
| Median platelets, ×109/L (n) | 105 (13) | 68 (54) | .014 | 135 (16) | 65 (51) | .0001 |
HCL indicates classic hairy-cell leukemia; and HCLv, variant HCL.
Leukopenia in VH4-34+ and VH4-34− HCL
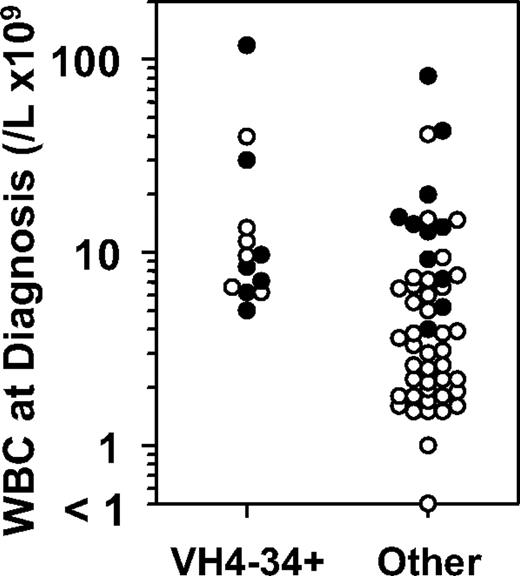
To determine whether VH4-34 positivity was associated with adverse clinical factors, those clinical factors associated with HCLv were investigated. Leukocytosis at presentation, a known characteristic of HCLv, is attributed to both absence of neutropenia and increase in circulating malignant cells.3,7 Absence of leukopenia, specifically a presenting white blood cell count of at least 5 × 109/L, was more frequently observed in VH4-34+ than VH4-34− patients (93% vs 40%, P < .001; Table 3). Although a greater proportion of variant than classic patients reached this white blood cell count at diagnosis (83% vs 40%, P = .002), VH4-34 positivity was independently related because among just classic patients a greater percentage had white blood cell counts of at least 5 × 109/L (100% vs 32%, P = .002, Table 3). As shown in Figure 3, by rank order analysis, presenting white blood cell counts were greater for VH4-34+ versus negative patients (median 9.0 vs 3.8 × 109/L, P = .002). Although the same was true of variant versus classic patients (median 11.3 vs 3.5 × 109/L, P < .001), among just classic patients, VH4-34 positivity correlated with presenting white blood cell count (P = .003). Thus, absence of leukopenia is a feature that correlates independently with both VH4-34 positivity and with HCLv. As shown in Table 3, presenting neutrophils, hemoglobin, and platelets were significantly greater with variant compared with classic disease, and although neutrophil and platelet counts were significantly greater with VH4-34+ than VH4-34− patients (P = .004-.014), VH4-34 status was not independently related to levels of these normal cells. Thus, VH4-34 positivity was correlated with lack of leukopenia, probably because of increased malignant rather than increased normal cells.
White blood cells counts at diagnosis in patients with or without VH4-34 expression, whose hairy cells are classic (○) or variant (●) HCL.
White blood cells counts at diagnosis in patients with or without VH4-34 expression, whose hairy cells are classic (○) or variant (●) HCL.
VH4-34 positivity and failure of initial cladribine therapy in patients with HCL
To determine whether VH4-34 positivity was associated with the failure of initial cladribine to achieve remission, an important characteristic of HCLv,3 we studied HCL patients who were being screened for recombinant immunotoxin trials that required inadequate response to initial or most recent purine analog therapy (Table 4). Historical data on each patient were analyzed to determine complete, hematologic, or partial remission on the basis of published criteria.9 Hematologic remission, which requires all the criteria of complete remission except for bone marrow biopsy, was important to consider because many patients did not get bone-marrow studies after cladribine. As shown in Table 5, complete, hematologic, and partial remission rates were significantly lower with VH4-34+ compared with VH4-34− patients (P = .03, .004, and < .001, respectively).
Outcome after initial cladribine for HCL (patient numbers)
| . | Total . | VH4-34+, HCLv . | VH4-34+, HCL . | VH4-34−, HCLv . | VH4-34−, HCL . |
|---|---|---|---|---|---|
| Patients evaluable for survival | 82 | 8 | 6 | 12 | 56 |
| Patients evaulable for response | 61 | 7 | 6 | 8 | 40 |
| Complete remission | 14 | 0 | 0 | 1 | 13 |
| Hematologic remission | 18 | 0 | 2 | 2 | 14 |
| Partial response | 10 | 0 | 0 | 1 | 9 |
| No response | 19 | 7 | 4 | 4 | 4 |
| . | Total . | VH4-34+, HCLv . | VH4-34+, HCL . | VH4-34−, HCLv . | VH4-34−, HCL . |
|---|---|---|---|---|---|
| Patients evaluable for survival | 82 | 8 | 6 | 12 | 56 |
| Patients evaulable for response | 61 | 7 | 6 | 8 | 40 |
| Complete remission | 14 | 0 | 0 | 1 | 13 |
| Hematologic remission | 18 | 0 | 2 | 2 | 14 |
| Partial response | 10 | 0 | 0 | 1 | 9 |
| No response | 19 | 7 | 4 | 4 | 4 |
HCL indicates classic hairy-cell leukemia; and HCLv, variant HCL.
Outcome after initial cladribine for HCL (response rates, progression-free, and overall survival)
| Response comparisons . | VH4-34+ . | VH4-34− . | P . | HCLv . | HCL . | P . |
|---|---|---|---|---|---|---|
| Complete remission rate (%) | 0/13 (0) | 14/48 (29) | .03 | 1/15 (7) | 13/46 (28) | .15 |
| Complete/hematologic remission rate (%) | 2/13 (15) | 30/48 (63) | .004 | 3/15 (20) | 29/46 (63) | .006 |
| Major response rate (%) | 2/13 (15) | 40/48 (83) | .00001 | 4/15 (27) | 38/46 (83) | .0001 |
| Median progression-free survival, mo (n) | 5.1 (13) | 22.8 (48) | .007 | 5.1 (15) | 23.1 (46) | .02 |
| Median overall survival, y (n) | 8.63 (14) | 26.22 (68) | < .0001 | >8.79 (20) | 26.22 (62) | .02 |
| Major response rate for classic HCL only (%) | 2/6 (33) | 36/40 (90) | .006 | |||
| Major response rate for those VH4-34− only (%) | 4/8 (50) | 36/40 (90) | .02 | |||
| Median overall survival, y, classic HCL only (n) | 4.70 (6) | 26.64 (56) | < .0001 | |||
| Median overall survival, y, VH4-34− only (n) | >4.74 (12) | 26.64 (56) | .22 | |||
| Median age at diagnosis, age > 50 y (n) | 55.75 (12) | 57.00 (24) | .31 | 60.8 (13) | 56.70 (23) | .1 |
| Median overall survival, age > 50 y (n) | 8.63 (12) | 21.05 (24) | .0001 | 8.79 (13) | 16.53 (23) | .11 |
| Median overall survival, age > 50 y, classic HCL only | 4.70 (5) | 17.33 (18) | < .0001 | |||
| Median overall survival, age > 50 y, VH4-34− only | 21.05 (6) | 17.33 (18) | .23 |
| Response comparisons . | VH4-34+ . | VH4-34− . | P . | HCLv . | HCL . | P . |
|---|---|---|---|---|---|---|
| Complete remission rate (%) | 0/13 (0) | 14/48 (29) | .03 | 1/15 (7) | 13/46 (28) | .15 |
| Complete/hematologic remission rate (%) | 2/13 (15) | 30/48 (63) | .004 | 3/15 (20) | 29/46 (63) | .006 |
| Major response rate (%) | 2/13 (15) | 40/48 (83) | .00001 | 4/15 (27) | 38/46 (83) | .0001 |
| Median progression-free survival, mo (n) | 5.1 (13) | 22.8 (48) | .007 | 5.1 (15) | 23.1 (46) | .02 |
| Median overall survival, y (n) | 8.63 (14) | 26.22 (68) | < .0001 | >8.79 (20) | 26.22 (62) | .02 |
| Major response rate for classic HCL only (%) | 2/6 (33) | 36/40 (90) | .006 | |||
| Major response rate for those VH4-34− only (%) | 4/8 (50) | 36/40 (90) | .02 | |||
| Median overall survival, y, classic HCL only (n) | 4.70 (6) | 26.64 (56) | < .0001 | |||
| Median overall survival, y, VH4-34− only (n) | >4.74 (12) | 26.64 (56) | .22 | |||
| Median age at diagnosis, age > 50 y (n) | 55.75 (12) | 57.00 (24) | .31 | 60.8 (13) | 56.70 (23) | .1 |
| Median overall survival, age > 50 y (n) | 8.63 (12) | 21.05 (24) | .0001 | 8.79 (13) | 16.53 (23) | .11 |
| Median overall survival, age > 50 y, classic HCL only | 4.70 (5) | 17.33 (18) | < .0001 | |||
| Median overall survival, age > 50 y, VH4-34− only | 21.05 (6) | 17.33 (18) | .23 |
HCL indicates classic hairy cell leukemia; and HCLv, variant HCL.
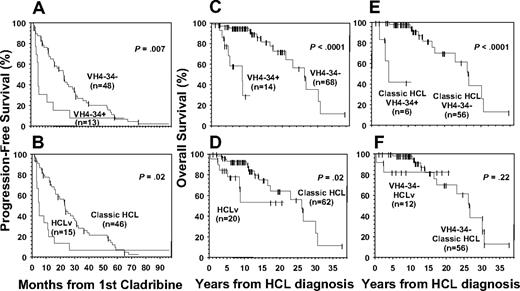
Hematologic and partial remission rates also were lower for variant compared with classic patients. The lower-than-reported observed remission rates for classic patients in the current study were expected given the skewed patient population studied. Among just classic HCL patients, VH4-34 positivity correlated significantly with lower response (P = .006, Table 5), indicating its independence from variant versus classic phenotype. The relationship of variant phenotype to response in just VH4-34− patients had less significance (P = .02). As shown in Figure 4A and B, shorter progression-free survival was associated with VH4-34 positivity (P = .007) and HCLv (P = .02), with median progression-free survivals of 5.1 versus 22.8 months depending on VH4-34 status, and 5.1 versus 23.1 months depending on HCL variant versus classic (Table 5). Thus, response rate and duration after initial cladribine was more significantly related to VH4-34 status than to variant phenotype.
Progression-free survival and overall survival in patients with respect to VH4-34 status and variant phenotype. (A-B) Progression-free survival; (C-F) overall survival. Median values are listed in Table 5. Panels C and D show data for all 82 patients with respect to VH4-34 (C) or variant (D) status, whereas panel E includes only classic HCL and panel F only VH4-34− patients.
Progression-free survival and overall survival in patients with respect to VH4-34 status and variant phenotype. (A-B) Progression-free survival; (C-F) overall survival. Median values are listed in Table 5. Panels C and D show data for all 82 patients with respect to VH4-34 (C) or variant (D) status, whereas panel E includes only classic HCL and panel F only VH4-34− patients.
VH4-34 positivity and overall survival
Because lower overall survival was reported in HCLv than in classic HCL,3 dates of death and diagnosis were examined to compare survival after diagnosis relative to VH4-34 status or variant phenotype. As shown in Table 5 and Figure 4C, VH4-34 positivity was significantly correlated with poorer survival, with predicted median survival 8.63 years in VH4-34+ versus 26.22 years in VH4-34− patients (P < .001). A difference of more than 8.79 versus 26.22 years was observed for variant versus classic patients (Figure 4D, P = .02). As shown in Table 5, VH4-34 status was independent of variant phenotype in correlating with poorer overall survival (P < .001). In contrast, overall survival was not significantly associated with variant phenotype in only VH4-34− patients (P = .22). Of the 8 VH4-34+ patients in this study who died, in 7 the immediate cause of death was HCL along with infectious complications. The eighth patient had significant HCL tumor burden and died of a massive stroke, despite being young in age (63 years) and having an absence of carotid atherosclerotic disease. Therefore, an association of thromboembolic disease to his HCL could not be excluded.
VH4-34 positivity and overall survival in older HCL patients
As was observed in comparing variant to classic patients, VH4-34+ patients were older than VH4-34− patients at diagnosis (median ages 55.0 vs 45.6, P = .011, Table 3). To determine whether lower overall survival in VH4-34+ patients was related to their older age at diagnosis, patients older than the age of 50 were considered separately, because in those 36 older patients, there was no significant difference in age with respect to VH4-34 status (P = .31) or variant phenotype (P = .1; Table 5). In these older patients, overall survival was still significantly shorter for VH4-34+ than VH4-34− patients (median 8.63 vs 21.05 years, P < .001) but not for HCLv compared with classic patients (median 8.79 vs 16.53 years, P = .11). As shown in Table 5, VH4-34 status was independent of variant phenotype in its association with poorer survival in older patients (P < .001), but variant phenotype was not associated with poor survival independent of VH4-34 status (P = .23). Thus, VH4-34 positivity was associated with lower survival from diagnosis, independent of age or variant phenotype. Moreover, lower survival of the HCLv patients was the result of VH4-34+ patients with HCLv, rather than to an independent association between HCLv and survival.
Discussion
Our original goal was to study molecular characteristics of HCLv to determine whether expressed immunoglobulin rearrangements have homology to germline and gene usage different from the “classic” or typical form of the disease. We found that VH4-34, the most common VH gene used in HCLv, had very high homology to its germline sequence and was associated with variant features such as advanced age, lack of leukopenia, poor response to initial cladribine treatment, and poor progression-free and overall survival. Moreover, these associations were observed even in patients with VH4-34+ hairy cells who are considered classic HCL rather than HCLv. Conversely, comparisons of response and overall survival between classic and variant patients lost significance when only the VH4-34− population was examined. Thus, VH4-34+ HCL is a disorder, overlapping only partially with HCLv, associated with variant features and poor prognosis. It has long been thought that patients with HCL not responding to initial purine analog treatment actually have a different disease. These results indicate that VH4-34+ HCL is a disorder accounting for many of these patients and may account for a component of the poor outcome previously reported in HCLv.
Molecular versus immunophenotype in HCL
Each of the 82 patients in this study was carefully examined to verify the diagnosis of HCL or HCLv by the use of flow cytometry, immunohistochemistry, and morphology of blood, bone marrow, spleen, and lymph nodes when available. Nevertheless, because of significant overlap in criteria, in some patients a perfect distinction between SMZL, HCLv, and HCL may not be possible. The frequent loss of antigen expression over time, as observed in this study, may add to the unreliability of available methods to classify patients with HCL and predict response to standard cladribine therapy. Our results show that unmutated VH4-34 positivity is a fixed marker in HCL which predicts for poor outcome, both response to initial therapy and survival, and this appears independent of the exact diagnosis within HCL.
VH4-34 expression in malignant lymphocytes
VH4-34 is expressed in approximately 5% to 10% of adult normal B cells,33-36 65% of diffuse large-cell lymphoma,37 25% of duodenal follicular lymphoma,38 50% of primary central nervous system lymphoma,39 4% to 22% of chronic lymphocytic leukemia (CLL),40,41 9% to 33% of acute lymphoblastic leukemia,40,42 and almost never in multiple myeloma.43-45 We and others have reported VH4-34 expression in isolated cases of HCLv and classic HCL,18,24,25 but the 2 to 5 cases per study were insufficient for understanding the impact of VH4-34 in this disease.
Relation between VH4-34 and poor prognosis
VH4-34 is reported to be overexpressed in autoimmune disease, including 55% of systemic lupus erythematosus,46 virtually all cases of cold agglutinin disease,47-52 and 89% of multiple sclerosis.53 Monoclonal antibodies produced by VH4-34–expressing B cells are reported to react to a variety of normal antigens, including red blood cells,46-49 normal and malignant B cells,50-52,54,55 and single-stranded DNA.56 Although percent homology to germline is rarely determined for VH4-34 clones binding to autoantigens, mutated and less-commonly unmutated VH4-34 rearrangements are reported, the latter from either human fetal spleen or cerebral spinal fluid in multiple sclerosis.49,57-60 It is proposed that VH4-34–expressing B cells may recognize antigen, undergo somatic mutation, and then either mediate autoimmune disease or transform to CLL or diffuse large-cell lymphoma, both known for frequent autoimmune complications.
In contrast, malignant transformation of such cells at a much later point of differentiation to multiple myeloma does not occur because of eventual clonal deletion or anergy.43 We and others18 recently reported similarities between classic HCL and SMZL regarding VH usage and rates of somatic mutation, suggesting that classic hairy cells undergo malignant transformation in the marginal zone of the germinal center. The unmutated status of VH4-34+ hairy cells in all but 1 of our 14 patients suggests transformation before the germinal center and is consistent with the complete lack of autoimmune complications in our patients. We therefore speculate that VH4-34+ lymphocytes in general may be less susceptible to checkpoints leading to clonal deletion, which leads to aggressive and resistant disease in hairy cell patients, or to the autoimmune complications seen in patients with other disorders.
Other factors potentially affecting prognosis
It has been reported that unmutated rearrangements in CLL correlate with poor prognosis.61 To determine whether poor prognosis in HCL is more closely related to unmutated than to VH4-34+ rearrangements, we compared just the VH4-34− rearrangements for progression free (n = 50) and overall (n = 71) survival but found no significant difference whether they were mutated or unmutated (P = .3-.6). A total of 36 patients had abdominal imaging data before the initial cladribine, and enlarged lymph nodes, associated with lower repose to purine analog,62-64 were observed in 2 of 2 VH4-34+/HCL classic, 0 of 5 VH4-34+/HCLv, 2 of 9 VH4-34−/HCLv, and 3 of 20 VH4-34−/HCL classic patients. These data are insufficient to suggest that VH4-34 positivity is associated with the adenopathy observed in patients with poor prognosis for response.
Clinical implications of VH4-34+ HCL
Except for 1 VH4-34+ HCL patient who had a mutated rearrangement and hematologic remission, the other patients had unmutated VH4-34 rearrangements and poorer response to initial cladribine therapy (Table 2, Figure 2). Like HCLv, VH4-34+ HCL may represent a disorder distinct from classic HCL and overlapping with HCLv. Of those VH4-34 patients who lived to try alternative treatments, 3 had complete remission to recombinant anti-CD22 immunotoxin BL22,17,65 and 2 patients had hematologic remission to combined pentostatin and rituximab. HCL patients sometimes receive pentostatin alone after failure of cladribine because of anecdotal reports of complete remission to cladribine64,66-68 or pentostatin69 alone after failure of the other purine analog. However, of 4 VH4-34+ patients in this study who received pentostatin alone after cladribine, none responded. In view of the observation that the remission rate decreases with each course of purine analog equally whether patients receive the same or the other agent,70 it is not clear whether lack of cross resistance between cladribine and pentostatin is the exception rather than the rule. We believe that newly diagnosed patients with HCL and leukocytosis, regardless of diagnosis of HCLv, should be tested for VH4-34 positivity either by PCR cloning, by flow cytometry, or by enzyme-linked immunoabsorbent assay using an anti-idiotype antibody such as 9G4.44,46,56 In our study the incidence of VH4-34 positivity was 36% in HCL patients presenting with white blood cells greater than or equal to 5 × 109/L (Figure 3). Patients with VH4-34+ HCL could be considered for alternative approaches, such as therapy with rituximab combined with purine analog, rather than with purine analog alone.71,72
An Inside Blood analysis of this article appears at the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank our clinical staff Rita Mincemoyer, Linda Ellison, Elizabeth Maestri, Barbara Debrah, and Sonya Duke for facilitating sample acquisitions and managing results. We recognize Dr Constance Yuan and Marlene Gronberg for flow cytometric analysis and Dr Seth Steinberg for statistical advice. We wish to dedicate this manuscript to the memory of Anna Orthwein, who performed research for this work.
This work was supported by the intramural program, NCI, NIH.
National Institutes of Health
Authorship
Contributions: E.A. designed and performed research, analyzed data, and wrote the paper; T.S. and M.S.-S. performed research and analyzed data; and R.J.K. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert J. Kreitman, National Cancer Institute, Laboratory of Molecular Biology, National Institutes of Health, 37/5124b, 37 Convent Dr, MSC 4255 Bethesda, MD; e-mail kreitmar@mail.nih.gov.