Abstract
Venomous snakes produce an array of toxic compounds, including procoagulants to defend themselves and incapacitate prey. The Australian snake Pseudonaja textilis has a venom-derived prothrombin activator homologous to coagulation factors V (FV) and Xa (FXa). Here we show that the FV component (pt-FV) has unique biologic properties that subvert the normal regulatory restraints intended to restrict an unregulated procoagulant response. Unlike human FV, recombinant pt-FV is constitutively active and does not require proteolytic processing to function. Sequence comparisons show that it has shed a large portion of the central B-domain, including residues that stabilize the inactive procofactor state. Remarkably, pt-FV functions in the absence of anionic membranes as it binds snake-FXa with high affinity in solution. Furthermore, despite cleavage in the heavy chain, pt-FV is functionally resistant to activated protein C, an anticoagulant. We speculate this stability is the result of noncovalent interactions and/or a unique disulfide bond in pt-FV linking the heavy and light chains. Taken together, these findings provide a biochemical rationale for the strong procoagulant nature of venom prothrombinase. Furthermore, they illustrate how regulatory mechanisms designed to limit the hemostatic response can be uncoupled to provide a sustained, disseminated procoagulant stimulus for use as a biologic toxin.
Introduction
The circulation of blood coagulation factors as precursors and their localization to the site of vascular injury after activation are important factors that maintain normal hemostasis and restrict indiscriminate clotting.1 Bypassing these regulatory paradigms can be life-threatening, yet organisms have evolved strategies that exploit these systems for a selective advantage. For example, the venom of numerous snake species comprises a diverse array of proteases that activate and overwhelm the host hemostatic system in an uncontrolled fashion.2 Some of the most potent venoms come from the Australian elapid family, including Pseudonaja textilis, Oxyuranus microlepidotus, and Oxyuranus scutellatus.3 Their venom is considered the most toxic in the world and is unusual as it contains 2 coagulation proteins: factor V (FV) and a factor Xa (FXa)–like enzyme.4-10 These proteins make up approximately 20% to 40% of the total venom and form a powerful procoagulant complex.5,7 This complex converts prothrombin to thrombin, and its activity is enhanced, to various degrees, by calcium and phospholipids, but not by the cofactor FVa.4-7,11
Venom-derived FV from these snakes share approximately 44% homology with mammalian FV and have a similar domain structure (A1-A2-B-A3-C1-C2).8,10,12 However, we were surprised to learn that their B-domains are remarkably short, approximately 46 versus approximately 800 residues in mammals. (Amino acid numbering for pt-FV is as follows: the mature sequence starts at +1 and the remaining sequence is numbered consecutively to 1430. The pt-FV B-domain is defined as the region removed after thrombin cleavage [residues 743-788]). New genomic data show that, although the length of the B-domain is variable and poorly conserved, especially in lower vertebrates (eg, ∼ 500 residues in Fugu rubripes13,14 ), a shortened version of this magnitude is unprecedented. In addition, the functional implications of a naturally occurring truncated B-domain are not understood. The generally accepted role of the B-domain is to somehow prevent or inhibit the constitutive activity of the precursor, FV. Conversion to the mature cofactor (FVa) involves proteolytic removal of the B-domain mediated by thrombin.15 Once activated, FVa associates with FXa on an activated cellular surface in the presence of calcium ions to form prothrombinase, the physiologic activator of prothrombin.1,15 More recently, using a series of human FV B-domain truncated variants, we found that discrete sequences within the B-domain contribute to the mechanism by which FV persists as an inactive procofactor.16,17 This suggests that the B-domain may play a direct role in keeping FV inactive in a sequence-specific manner. These findings, combined with the unusual size of venom-derived FV, raise the possibility that these snake species have shed regulatory components essential to keep FV inactive.
In the present study, we used recombinant venom-derived FV from the common brown snake, P textilis (pt-rFV; native protein from venom, pt-FV) to investigate the biologic properties of this unique form of FV and to examine the mechanistic basis for the strong procoagulant nature of the venom-derived prothrombinase complex. Our work demonstrates that pt-rFV has been modified into a robust procoagulant acquiring multiple gain-of-function elements not present in mammalian FV. Unraveling the mechanistic and structural basis of these unique procoagulant properties will likely further our understanding of FV function. These findings also highlight how naturally occurring changes or adaptations can enhance the existing properties of a protein to presumably confer a selective advantage.
Methods
Reagents
The inhibitors benzamidine and 4-amidinophenylmethanesulfonyl fluoride hydrochloride were from Sigma-Aldrich, and dansylarginine-N-(3-ethyl-1,5-pentanediyl)amide (DAPA) was from Haematologic Technologies. Glutamyl-glycyl-arginyl chloromethyl ketone (EGR-CH2Cl) was obtained from EMD Biosciences. The peptidyl substrate H-D-phenylalanyl-L-pipecolyl-L-arginyl-p-nitroanilide (S2238) was from Diapharma. All tissue culture reagents were from Invitrogen except insulin-transferrin-sodium selenite (Roche Applied Science). Small unilamellar phospholipid vesicles (PCPS) composed of 75% (wt/wt) hen egg L-α-phosphatidylcholine and 25% (wt/wt) porcine brain L-α-phosphatidylserine (Avanti Polar Lipids) were prepared and characterized as described.18
Proteins
Human prothrombin was isolated from plasma as described previously.19 Prethrombin-1 and prethrombin-2 were prepared and purified by established procedures,20 and thrombin was obtained from Haematologic Technologies. A constitutively active partial B-domainless form of human FV (hFV-810), rFVa, rFX, and rFXa were prepared, purified, and characterized as described.16,21 The purified enzymatic subunit of pseutarin C (pt-FXa) was a generous gift from QRxPharma (Sydney, Australia). Human activated protein C (APC) was obtained from Dr Sriram Krishnaswamy (Children's Hospital of Philadelphia). Molecular weights and extinction coefficients (E0.1%, 280 nm) of the various proteins used are summarized as described.16,22 The molecular weight of pt-rFV was determined to be 170 000, and the extinction coefficient was assumed to be similar to rFV-810. For pt-FXa, values for the human protein were used. Unless otherwise noted, all functional assays were performed at 25°C in 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 0.15 M NaCl, 2 mM CaCl2, 0.1% polyethylene glycol 8000, pH 7.5 (assay buffer).
Analytical ultracentrifugation
Molecular weights were determined in a Beckman Optima XL-I analytical ultracentrifuge (Beckman Coulter) using interference optics. Sedimentation velocity was measured at 20°C in 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 150 mM NaCl, 5 mM CaCl2, pH 7.4, at 50 400 × g with an AN60Ti rotor. Protein concentrations were: pt-rFV, 0.60 mg/mL (3.6 μM) and pt-FXa, 0.18 mg/mL (3.9 μM). For these experiments, pt-FXa was active site–blocked with EGR-CH2Cl essentially as described.23 Sedimentation coefficients and molecular weights were determined by g(s*) analysis.24
Construction of rFV variants
The P textilis FV cDNA derived from the venom gland10 was modified with flanking XmaI restriction sites and then subcloned into the pED expression plasmid.16 The cDNA was further modified by exchanging sequences encoding the signal sequence of P textilis FV with that of human FV using the technique of splicing by overlap extension. The final construct, pED-FV-ptex, encoded for the human FV signal peptide (residues −28 to −1) followed by the mature venom-derived pt-FV protein (residues 1-1430). Sequence analysis indicated that the P textilis FV cDNA was the same as reported by Rao et al,8 except for codons 50 (Lys) and 1305 (Phe), which are identical to the homologous residues of O scutellatus and O microlepidotus.10 A pt-FV variant with Arg742 and Arg788 replaced by Gln (pt-FV-QQ) was generated with the QuickChange site-directed mutagenesis kit (Stratagene) using appropriate mutagenic complementary oligonucleotides. These amino acids are homologous to Arg709 and Arg1545 in human FV and represent thrombin cleavage sites. After mutagenesis, the entire cDNA was sequenced to confirm the presence of the desired mutations and to ensure that there were no polymerase-induced errors.
Expression and purification of pt-rFV
Plasmids encoding pt-FV or pt-FV-QQ were introduced into baby hamster kidney cells, and high producing stable clones were established as described.16 Cells were expanded into triple flasks and cultured in Dulbecco modified Eagle medium/F12 media (no phenol red) supplemented with insulin-transferrin-sodium selenite and 2.5 mM CaCl2. Conditioned media was collected for 4 to 6 days, centrifuged, and stored at −20°C in the presence of 10 μM 4-amidinophenylmethanesulfonyl fluoride hydrochloride and 1 mM benzamidine. For purification, media was thawed at 37°C and loaded onto a 30 mL Q-Sepharose FF column (GE Healthcare) equilibrated in 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 0.15 M NaCl, 5 mM CaCl2, pH 7.4. The column was washed with the same buffer and eluted with a 0.15- to 1-M NaCl gradient. Fractions containing pt-rFV were pooled and dialyzed versus 20 mM 2-(n-morpholino)ethanesulfonic acid (MES), 2 mM CaCl2, pH 6.0, and then loaded on a Poros HS/20 column (10 × 100 mm; Applied Biosystems) equilibrated with the same buffer. The column was washed with 20 mM MES, 2 mM CaCl2, pH 6.0, and then eluted with a 0 to 0.6 M NaCl gradient. FV containing fractions were pooled and dialyzed versus 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 0.15 M NaCl, 2 mM CaCl2, pH 7.4, and the protein was stored at −80°C. The final yield was approximately 4 mg of pt-rFV per liter of conditioned media.
Protein characterization
Protein purity was assessed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using precast 4% to 12% gradient gels (Invitrogen) under nonreducing and reducing conditions (50 mM dithiothreitol) using the 3-(n-morpholino)propanesulfonic acid buffer system followed by staining with Coomassie Brilliant Blue R-250. N-terminal sequence analysis was performed in the laboratory of Dr Alex Kurosky and Steve Smith at the University of Texas Medical Branch at Galveston or Dr Jan Pohl, Centers for Disease Control (Atlanta, GA). Chemical γ-carboxyglutamic acid (Gla) analysis on pt-FXa was carried out in our laboratory as described.21 This analysis yielded 10.9 (± 0.34) mol Gla/mol protein of 11 mol Gla/mol protein predicted from sequence alignments.9,10
Kinetics of protein substrate cleavage
Steady-state initial velocities of macromolecular substrate cleavage were determined discontinuously at 25°C as described.21 The kinetic parameters of prothrombinase or FVa-FXa-catalyzed prothrombin or prethrombin-1 activation (Km and Vmax) were determined in assay buffer by measuring the initial rate of thrombin formation at increasing concentrations of macromolecular substrate. Assay mixtures contained PCPS (50 μM), DAPA (3 μM), the various pt-rFV(a) cofactor species (20 nM), and various concentrations of prothrombin (0-5.0 μM) or prethrombin-1 (0-15 μM). The reaction was initiated with pt-FXa (0.1 nM). When prethrombin-2 was used as protein substrate, the following reaction conditions were used: PCPS (50 μM), DAPA (3 μM), and prethrombin-2 (1.4 μM) were incubated with the various FV(a) cofactors (0-40 nM), and the reaction was initiated with pt-FXa (5 nM). For certain experiments, PCPS was omitted from the reaction mixture (see the legends for Figures 2–3). For experiments in which pt-rFVa was used, pt-rFV (600 nM) was incubated with 2 nM thrombin at 37°C for 15 minutes followed by the addition of 2.4 nM hirudin.
APC inactivation of pt-rFV
The inactivation of the various cofactor proteins (500 nM; pt-rFV, pt-rFVa, rhFVa, or h-FV-810) in the presence of PCPS (50 μM) was initiated by APC addition (10 or 750 nM). Aliquots of the reaction mixture were withdrawn at the indicated time intervals and assessed by SDS-PAGE (4%-12% gel) and through assessment of functional activity. In this assay, residual cofactor activity was assessed by measuring the effect of FVa on prothrombin activation essentially as described.21 Assay mixtures contained 1.4 μM prothrombin, 50 μM PCPS, 3 μM DAPA, 1 nM FV(a), and 1 nM pt-FXa. With rhFVa or hFV-810, human rFXa was used instead. Under these conditions, the initial rate of the reaction was proportional to the concentration of the cofactor.
Data analysis
Data were analyzed according to the referenced equations by nonlinear least squares regression as previously detailed.17 Reported estimates of error represent plus or minus 2 SDs. The dissociation constants (Kd) for the interaction between pt-FXa and the venom cofactors were obtained from the dependence of the initial rate of prethrombin-2 activation on the concentrations of the cofactor species as described16 with the stoichiometry fixed at 1. Initial velocity measurements of prothrombin or prethrombin-1 cleavage by prothrombinase were analyzed by fitting the data to the Henri-Michaelis-Menten equation to yield fitted values for Km and Vmax.
Results
Expression and physical characterization of pt-rFV
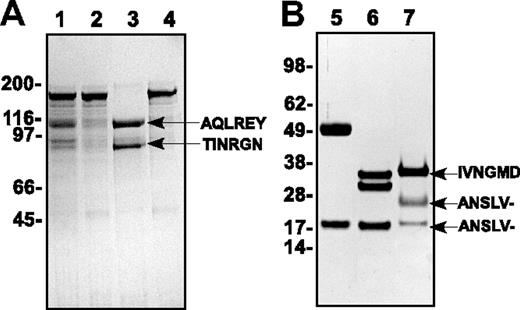
The abundance of proteases in snake venom has precluded the isolation of a homogeneous uncleaved preparation of pt-FV.4-7 To circumvent this, we expressed and purified pt-rFV from baby hamster kidney cells. Wild-type protein and a variant lacking both thrombin cleavage sites (pt-rFV-QQ; residues 742 and 788) migrated primarily as single bands (∼ 160-180 kDa) on a reducing SDS-PAGE (Figure 1A). Sedimentation velocity studies indicated that pt-rFV was homogeneous with a molecular weight of 170 (± 1) × 103 and a sedimentation coefficient (s20, w) of 8.30 (± 0.01) × 10−13 s (data not shown). Whereas pt-rFV-QQ was not cleaved by thrombin, the wild-type protein was processed to pt-rFVa at the predicted sites (Figure 1).8,10
SDS-PAGE analysis of purified proteins. Proteins (3 μg/lane) were subjected to SDS-PAGE under reducing conditions and visualized by staining with Coomassie Brilliant Blue R-250. Lane 1 represents pt-rFV; lane 2, pt-rFV-QQ; lane 3, pt-rFV plus thrombin; lane 4, pt-rFV-QQ plus thrombin; lane 5, rFX; lane 6, rFXa; and lane 7, pt-FXa. The apparent molecular weights of the standards are indicated on the left. N-terminal sequence results of the indicated protein bands are shown. The dash (–) following the Val (panel B) indicates that the yield was too low to accurately assign an amino acid and likely represents the presence of γ-carboxyglutamic acid.
SDS-PAGE analysis of purified proteins. Proteins (3 μg/lane) were subjected to SDS-PAGE under reducing conditions and visualized by staining with Coomassie Brilliant Blue R-250. Lane 1 represents pt-rFV; lane 2, pt-rFV-QQ; lane 3, pt-rFV plus thrombin; lane 4, pt-rFV-QQ plus thrombin; lane 5, rFX; lane 6, rFXa; and lane 7, pt-FXa. The apparent molecular weights of the standards are indicated on the left. N-terminal sequence results of the indicated protein bands are shown. The dash (–) following the Val (panel B) indicates that the yield was too low to accurately assign an amino acid and likely represents the presence of γ-carboxyglutamic acid.
FV purified from the crude venom of P textilis and O scutellatus has low activity in the presence of bovine FXa.6,7 Our results using human FXa and pt-rFV were consistent with these observations. The low activity appeared to be principally the result of the finding that pt-rFV or thrombin-cleaved pt-rFV (pt-rFVa) bound with a reduced affinity to membrane-bound human FXa (Kd > 400 nM) using established fluorescent binding measurements (data not shown).16 To characterize pt-rFV, FXa purified from the venom of P textilis (pt-FXa) was used in all subsequent experiments. SDS-PAGE analysis of pt-FXa is presented in Figure 1 along with N-terminal sequence analysis, which gave the expected results. The appearance of the light chain as a doublet is likely due to the result of heterogeneous O-glycosylation.9,10
Pt-rFV is a constitutively active cofactor
Because of the strikingly short B-domain in pt-FV, we used a series of activity measurements to determine whether this domain functions to maintain the protein as a procofactor. As expected, pt-rFVa assembled with pt-FXa-membranes and rapidly activated prothrombin (Figure 2A). Steady-state kinetic constants with venom-derived prothrombinase were similar to those observed for human prothrombinase (Table 1).16,22 Human FVa (data not shown) or a B-domain truncated derivative of FV, hFV-810,16 also assembled with pt-FXa-membranes and activated prothrombin with comparable kinetic parameters (Table 1). In contrast to inactive human FV,15 single-chain pt-rFV or pt-rFV-QQ rapidly stimulated prothrombin activation and gave kinetic constants comparable with pt-rFVa (Figure 2A; Table 1). Progress curves for either pt-rFV or pt-rFV-QQ increased linearly over time with no obvious lag in thrombin generation (data not shown). These data indicate that, unlike human FV, pt-FV is synthesized as a constitutively active cofactor and does not require proteolytic removal of the B-domain to express procoagulant function.
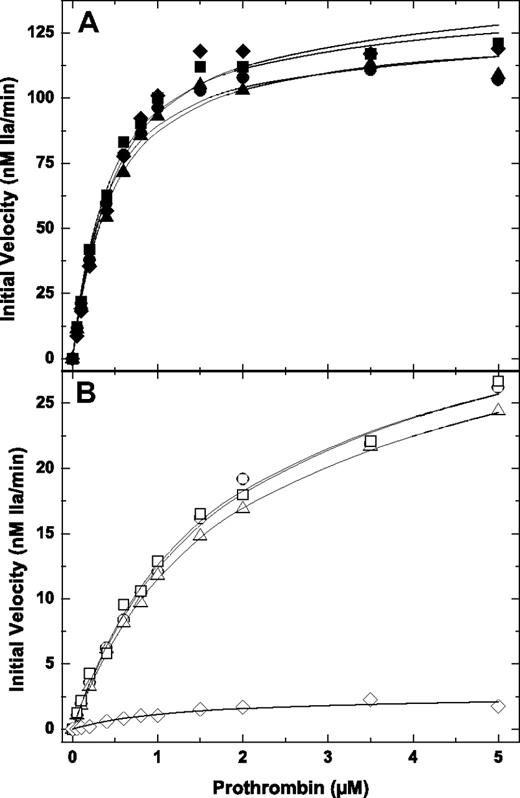
Prothrombin activation in the presence or absence of PCPS. The initial velocity of thrombin generation was determined at increasing concentrations of prothrombin in the presence (A) or absence (B) of 50 μM PCPS and 3 μM DAPA with 0.1 nM pt-FXa and 20 nM hFV-810 (⧫ or ◊), pt-rFV (● or ○), pt-rFV-QQ (▲ or △), or pt-rFVa (■ or □). The lines were drawn after analysis of all datasets to a rectangular hyperbola, and the fitted kinetic constants can be found in Table 1. The data are representative of 2 similar experiments.
Prothrombin activation in the presence or absence of PCPS. The initial velocity of thrombin generation was determined at increasing concentrations of prothrombin in the presence (A) or absence (B) of 50 μM PCPS and 3 μM DAPA with 0.1 nM pt-FXa and 20 nM hFV-810 (⧫ or ◊), pt-rFV (● or ○), pt-rFV-QQ (▲ or △), or pt-rFVa (■ or □). The lines were drawn after analysis of all datasets to a rectangular hyperbola, and the fitted kinetic constants can be found in Table 1. The data are representative of 2 similar experiments.
Kinetic constants for macromolecular substrate cleavage
| . | Enzyme/substrate . | |||||||
|---|---|---|---|---|---|---|---|---|
| Prothrombinase*/prothrombin . | FXa-FVa/prothrombin . | Prothrombinase/prethrombin-1 . | FXa FVa/prethrombin-1 . | |||||
| Km, μM . | kcat, min−1 . | Km, μM . | kcat, min−1 . | Km, μM . | kcat, min−1 . | Km, μM . | kcat, min−1 . | |
| pt-rFV | 0.39 ± 0.04 | 1250 ± 38 | 1.83 ± 0.12 | 352 ± 11 | 1.12 ± 0.08 | 1610 ± 28 | 1.09 ± 0.11 | 1570 ± 38 |
| pt-rFV-QQ | 0.44 ± 0.05 | 1270 ± 39 | 1.87 ± 0.05 | 334 ± 4 | 1.17 ± 0.06 | 1670 ± 24 | 1.20 ± 0.11 | 1560 ± 34 |
| pt-rFVa | 0.41 ± 0.03 | 1350 ± 31 | 1.74 ± 0.15 | 347 ± 1 | 1.38 ± 0.06 | 2210 ± 26 | 1.38 ± 0.06 | 2170 ± 43 |
| hFV-810 | 0.48 ± 0.07 | 1400 ± 62 | NA | NA | ND | ND | ND | ND |
| . | Enzyme/substrate . | |||||||
|---|---|---|---|---|---|---|---|---|
| Prothrombinase*/prothrombin . | FXa-FVa/prothrombin . | Prothrombinase/prethrombin-1 . | FXa FVa/prethrombin-1 . | |||||
| Km, μM . | kcat, min−1 . | Km, μM . | kcat, min−1 . | Km, μM . | kcat, min−1 . | Km, μM . | kcat, min−1 . | |
| pt-rFV | 0.39 ± 0.04 | 1250 ± 38 | 1.83 ± 0.12 | 352 ± 11 | 1.12 ± 0.08 | 1610 ± 28 | 1.09 ± 0.11 | 1570 ± 38 |
| pt-rFV-QQ | 0.44 ± 0.05 | 1270 ± 39 | 1.87 ± 0.05 | 334 ± 4 | 1.17 ± 0.06 | 1670 ± 24 | 1.20 ± 0.11 | 1560 ± 34 |
| pt-rFVa | 0.41 ± 0.03 | 1350 ± 31 | 1.74 ± 0.15 | 347 ± 1 | 1.38 ± 0.06 | 2210 ± 26 | 1.38 ± 0.06 | 2170 ± 43 |
| hFV-810 | 0.48 ± 0.07 | 1400 ± 62 | NA | NA | ND | ND | ND | ND |
The data are representative of 2 independent measurements. The errors in the fitted constants represent ± 2 SD.
ND indicates not determined. NA, for this experiment, rates were very low precluding an accurate assessment of kinetic parameters; however, we estimate the Km to be approximately 3 μM. The kcat value is difficult to estimate because the Kd between snake FXa and hFV-810 in solution is very high (Table 2); thus, the total enzyme concentration in this experiment is not known with certainty.
Prothrombinase is defined as FXa and FVa bound to a negatively charged membrane in the presence of calcium.
Pt-rFV functions in the absence of anionic membranes
A central view of blood coagulation is that the interaction of coagulation proteins with negatively charged cellular surfaces is critical for normal hemostasis and has a substantial influence on reaction rates.1 As expected, removal of anionic membranes from reaction mixtures containing hFV-810 or hFVa (data not shown) reduced the rate of prothrombin activation more than 150-fold (Figure 2B). No thrombin generation was observed under these conditions in the absence of the cofactor (data not shown). Surprisingly, initial rates of prothrombin activation using pt-rFV, pt-rFV-QQ, or pt-rFVa in the absence of membranes were reduced only approximately 10-fold compared with the enzyme in the presence of membranes (Figure 2B; Table 1).
A difficulty in interpreting these data is that prothrombin binds membranes and thereby contributes to enhancing rates of thrombin generation. To address this, we measured the rate of prethrombin-1 activation in the presence and absence of membranes. Prethrombin-1 is a derivative of prothrombin lacking the membrane-binding Gla and kringle-1 domains.20 Consistent with results using prothrombin, the kinetics of prethrombin-1 activation using pt-rFV, pt-rFV-QQ, or pt-rFVa were similar to those using human prothrombinase (Figure 3A; Table 1).22 Surprisingly, however, removal of membranes from the reaction mixture had no appreciable effect on the rate of prethrombin-1 activation as the kinetic parameters for solution phase or pt-FXa-pt-rFV in the presence of membranes were equivalent (Figure 3B; Table 1). These data indicate that the decreased rate of prothrombin activation with pt-FXa-pt-rFV was not the result of decreased enzyme function but rather the elimination of membrane binding by the substrate. Furthermore, these data show that the venom-derived enzyme complex, either in solution or in the presence of membranes, functions in an equivalent manner, a finding not mimicked with mammalian FXa-FVa.
Prethrombin-1 activation in the presence or absence of PCPS. The initial velocity of thrombin generation was determined at increasing concentrations of prethrombin-1 in the presence (A) or absence (B) of 50 μM PCPS and 3 μM DAPA with 0.1 nM pt-FXa and 20 nM pt-rFV (● or ○), pt-rFV-QQ (▲ or △), or pt-rFVa (■ or □). The lines were drawn after analysis of all datasets to a rectangular hyperbola, and the fitted kinetic constants can be found in Table 1. The data are representative of 2 similar experiments.
Prethrombin-1 activation in the presence or absence of PCPS. The initial velocity of thrombin generation was determined at increasing concentrations of prethrombin-1 in the presence (A) or absence (B) of 50 μM PCPS and 3 μM DAPA with 0.1 nM pt-FXa and 20 nM pt-rFV (● or ○), pt-rFV-QQ (▲ or △), or pt-rFVa (■ or □). The lines were drawn after analysis of all datasets to a rectangular hyperbola, and the fitted kinetic constants can be found in Table 1. The data are representative of 2 similar experiments.
Pt-FXa and pt-rFV form a high-affinity complex
The data shown in Figures 2 and 3 above suggest that pt-FXa was fully saturated with pt-rFV. To assess this directly, we used a functional binding measurement with prethrombin-2 as a substrate to infer the equilibrium dissociation constant for pt-FXa-pt-rFV in the presence or absence of membranes (Table 2). As expected, in the presence of PCPS, we found that the dissociation constant for cofactor binding to pt-FXa was approximately 3 nM regardless of the cofactor species, a value similar to the human proteins.16 In marked contrast to the bovine system (Kd for FXa-FVa in solution ∼ 1-3 μM), the dissociation constant of pt-FXa for pt-rFV, pt-FV-QQ, or pt-rFVa increased only 2- to 3-fold in the absence of membranes (Kd ∼ 8 nM).25,26 Sedimentation velocity experiments showed that pt-rFV-pt-FXa formed a 1:1 stoichiometric complex in solution with a molecular weight of 213 (± 2) × 103 and a sedimentation coefficient (s20, w) of 9.33 (± 0.01) × 10−13 s (data not shown). In contrast to the venom-derived cofactors, we were not able to reliably establish a binding constant for solution phase hFV-810-pt-FXa (Table 2), suggesting that binding to soluble pt-FXa involves unique features on pt-rFV that are not present on hFV-810. Together, these data indicate that the pt-FXa-pt-FV complex has bypassed the normal requirement for negatively charged membranes to form a high-affinity 1:1 complex.
Binding constants for prothrombinase and FXa-FVa assembly
| Cofactor species . | Kd, nM (+PCPS) . | Kd, nM (−PCPS) . |
|---|---|---|
| pt-rFV | 3.06 ± 0.45 | 8.08 ± 1.23 |
| pt-rFV-QQ | 2.88 ± 0.37 | 9.39 ± 1.54 |
| pt-rFVa | 3.84 ± 0.78 | 7.54 ± 0.62 |
| hFV-810 | 3.76 ± 0.87 | >100* |
| Cofactor species . | Kd, nM (+PCPS) . | Kd, nM (−PCPS) . |
|---|---|---|
| pt-rFV | 3.06 ± 0.45 | 8.08 ± 1.23 |
| pt-rFV-QQ | 2.88 ± 0.37 | 9.39 ± 1.54 |
| pt-rFVa | 3.84 ± 0.78 | 7.54 ± 0.62 |
| hFV-810 | 3.76 ± 0.87 | >100* |
The data are representative of 2 independent measurements. The errors in the fitted constants represent plus or minus 2 SDs. Equilibrium dissociation constants are inferred from kinetic experiments detailed in “Kinetics of protein substrate cleavage.”
We were not able to accurately determine a value; a lower limit estimate is provided.
Pt-rFV is functionally resistant to APC
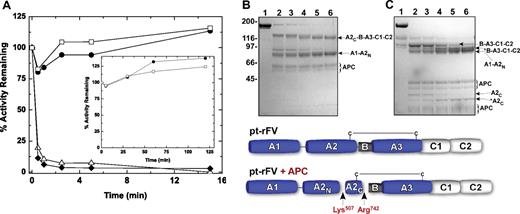
APC is an important anticoagulant in the hemostatic system and inactivates both FVa and FVIIIa.27 For human FVa, cleavage at Arg306, Arg506, and Arg679 results in A2 domain dissociation, loss of FXa binding, and cofactor function.15 Sequence analysis of pt-FV indicates that these sites are not strictly conserved, suggesting it may be resistant to APC.8 Using an APC concentration (10 nM) and reaction times (≤ 15 minutes) that resulted in the complete loss of hFVa or hFV-810 function, both pt-rFV and pt-rFVa retained full activity (Figure 4A), and SDS-PAGE analysis showed only limited cleavage of these proteins (data not shown). However, using higher concentrations of APC (750 nM), pt-FV was significantly proteolyzed over an extended time period (Figure 4B-C). N-terminal sequence analysis revealed that APC cleaves pt-rFV in the A2 domain at Lys507 and Arg742, which are equivalent to Arg506 and Arg709 in the human sequence. It is probable that APC cleaves pt-FV at additional site(s), especially in the A2C and B domains; however, we were not able to make that determination. Surprisingly, despite proteolysis of pt-rFV, full procoagulant activity was retained (Figure 4A inset). Analysis of both reducing and nonreducing gels revealed that a large fraction of pt-rFV (Figure 4B-C) is held together by one or more disulfide bonds between A2C (either Cys540 and/or Cys642) and the light chain. This was also confirmed after treatment of pt-rFV with thrombin (data not shown). We speculate that noncovalent interactions along with this unique disulfide bond(s) may stabilize fragments generated by APC. Together, these data show that despite APC cleavage at sites known to reduce cofactor activity, pt-rFV is functionally resistant to this anticoagulant.
Inactivation of FV(a) by APC. Reaction mixtures containing 50 μM PCPS and 500 nM pt-rFV (●), pt-rFVa (□), hFV-810 (⧫), or rFVa (△) were incubated with either 10 nM or 750 nM (inset) APC. At selected time intervals, samples were removed for cofactor activity (A) or SDS-PAGE under nonreducing (B) or reducing conditions (C). Protein bands subjected to N-terminal sequence analysis are marked and annotated according to the scheme below the gels: A2C-B-A3-C1-C2, 508-1430; A1-A2N, 1-507; B-A3-C1-C2, 743-1430; A2C, 508-742. We speculate that *B-A3-C1-C2 and *A2C are cleaved at their C-terminal ends because their N-terminal sequence was determined to be the same as B-A3-C1-C2 and A2C, respectively. APC can be visualized and is denoted by a brace. The functional measurements and gels are representative of 2 similar experiments.
Inactivation of FV(a) by APC. Reaction mixtures containing 50 μM PCPS and 500 nM pt-rFV (●), pt-rFVa (□), hFV-810 (⧫), or rFVa (△) were incubated with either 10 nM or 750 nM (inset) APC. At selected time intervals, samples were removed for cofactor activity (A) or SDS-PAGE under nonreducing (B) or reducing conditions (C). Protein bands subjected to N-terminal sequence analysis are marked and annotated according to the scheme below the gels: A2C-B-A3-C1-C2, 508-1430; A1-A2N, 1-507; B-A3-C1-C2, 743-1430; A2C, 508-742. We speculate that *B-A3-C1-C2 and *A2C are cleaved at their C-terminal ends because their N-terminal sequence was determined to be the same as B-A3-C1-C2 and A2C, respectively. APC can be visualized and is denoted by a brace. The functional measurements and gels are representative of 2 similar experiments.
Discussion
Most blood coagulation factors are synthesized as inactive precursors that only express activity after discrete proteolysis. The conformational changes that ensue allow these proteins to assemble on cellular surfaces localized at the site of vessel injury where they optimally function.1 Furthermore, in most instances, negative regulatory pathways are in place to limit their activity. Here we show that venom FV from P textilis circumvents these paradigms of normal hemostasis through a variety of changes transforming it into a potent procoagulant. These procoagulant enhancements likely provide this snake with a selective advantage by facilitating massive disseminated coagulation after envenomation of prey.3,28
Our data support the conclusion that pt-FV is synthesized in an active state and, unlike human FV, does not require proteolytic removal of the B-domain to express procoagulant activity. This represents the first reported example of a constitutively procoagulant form of FV in nature. Based on our prior studies with human FV, we speculate that the molecular basis for this finding is principally related to the short B-domain of pt-FV. The FV B-domain is an unusual domain with no known homology to any other protein, including the B-domain of the homologous clotting factor, FVIII. In most vertebrates, it corresponds to more than 40% of the mass of the protein, has a relatively low sequence homology, and displays a peculiar heavily glycosylated structure with numerous short tandem repeats of unknown function.29 Despite this lack of conservation, one of its roles is to inhibit constitutive cofactor activity as it is well established that proteolytic removal of the B-domain activates FV.15,29 Studies using an FV derivative with a shortened B-domain (FVdes811-1491; FV-810) support a model in which this region serves an inhibitory function by rendering binding sites on the heavy and/or light chain inaccessible to FXa and/or prothrombin.16,30,31 Using a panel of progressively finer FV B-domain truncated variants, we also found that a key cluster of B-domain residues plays an important role in maintaining the inactive procofactor state.17 Strikingly, part of this sequence cluster (963-1008) is unusually basic (18 of 46 residues are Arg or Lys) and remarkably well conserved among mammals and even lower vertebrates, pointing to its functional importance. In contrast, pt-FV and FV derived from the venom of O scutellatus and O microlepidotus lack this conserved sequence.8,10 We speculate that the absence of this basic region, and possibly other structural features, switches the functional state of pt-FV from an inactive procofactor to a constitutively active cofactor. Thus, pt-FV represents a clear biologic correlate to structure/function studies with human FV and is a naturally occurring example of a protein that has acquired a new functional state through loss of inhibitory sequences. Despite this loss, pt-FV has retained 2 of 3 thrombin cleavage sites flanking the heavy and light chains (Arg742 and Arg788). Whereas our work shows that cleavage at these sites is without functional consequence, their retention raises the possibility that they may have some important in vivo biologic function yet to be fully appreciated. Another possibility is that there simply was no selective pressure to change them. Other than perhaps stabilizing the molecule, there would appear to be no real advantage in eliminating these sites, thereby making the molecule resistant to thrombin.
A key underlying feature in any hemostatic response is that the assembly of coagulation proteins is localized to damaged vascular and peripheral blood cells at the site of injury.1 This promotes enzyme complex assembly and contributes to a profound enhancement in reaction rate.1 Work detailed here shows that pt-FV has bypassed these requisite restraints as it binds pt-FXa with high affinity in solution and functions equivalently to the membrane-bound complex. From the standpoint of evolving a strong procoagulant as a biologic weapon, bypassing the membrane binding step promotes a disseminated rather than a localized response after envenomation. However, the molecular basis for these findings is not clear. In the mammalian system, FXa and FVa have a micromolar affinity in solution, whereas the introduction of anionic membranes reduces the affinity to the nanomolar range.1,25,26 It is generally thought that the membrane surface promotes the assembly of FXa and FVa in at least 2 possible ways.1 The increased affinity upon membrane binding could be the result of the reduction in the degrees of freedom for protein reorientation once bound to a membrane surface. This reduction of dimensionality would then increase the frequency of productive collisions. Alternatively, the binding of FVa or FXa to a membrane surface could cause a conformational change in either of these proteins that fundamentally changes the nature of their interaction. Work from the Lentz laboratory provides evidence that phosphatidylserine head group ligation induces conformational changes that allosterically modulate both FVa and FXa, thereby dramatically enhancing the affinity of FVa for FXa in solution.32-34 Our findings with pt-FV-pt-FXa would also seem to support the conformational change model. In the case of the venom-derived proteins, these conformational transitions are probably induced through a variety of changes to their primary structure, which mimics the structural configuration of membrane-bound FVa-FXa.
As a further procoagulant enhancement, pt-FV is functionally resistant to APC despite cleavage in the heavy chain at Lys507 and Arg742. We speculate that noncovalent interactions contribute to this stabilization as it has been previously shown in the bovine system that a portion of the A2 domain remains associated after cleavage at Arg506.35 Our data are consistent with this as they indicate that cleavage at the Arg506 equivalent site in pt-FV has no obvious deleterious effect on cofactor function. Furthermore, it is also possible that the disulfide bond between the A2c and the A3 domains (Figure 4) may play some role in stabilizing pt-FV and preventing dissociation of the C-terminal heavy chain region from the rest of the molecule. Overall, our observations with pt-rFV following APC proteolysis support a model in which A2 domain dissociation,35 as opposed to the cleavage event at Arg506 itself,36 primarily contributes to the loss of FVa cofactor activity.
In conclusion, pt-FV is a potent procoagulant with unique biologic functions not yet found in any other naturally occurring form of FV. The accrual of these enhanced functions provides a rational basis for the potent hemostatic toxicity of the venom. Uncoupling the preservation of the procofactor state and bypassing the need for membrane binding to function are central underlying modifications allowing for the enzyme complex to rapidly activate prothrombin in solution and contribute to disseminated clotting. Taken together, pt-FV represents an exceptional example of a protein that has adapted into a potent biologic weapon for host defense and envenomation of prey.
Presented in part at the 50th annual meeting of the American Society of Hematology, San Francisco, CA, December 6-9, 2008.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Sriram Krishnaswamy for assistance with sedimentation velocity studies, useful suggestions, and critical review of the manuscript; Dr Alex Kurosky and Steve Smith, University of Texas Medical Branch at Galveston; and Dr Jan Pohl, Centers for Disease Control and Prevention (Atlanta, GA) for N-terminal sequence analysis.
This work was supported by the National Institutes of Health (grants R01 HL-88010 and P01 HL-74124; R.M.C.), and the Australian Research Council and QRxPharma (Sydney, Australia; P.P.M., J.d.J., M.F.L.).
National Institutes of Health
Authorship
Contribution: M.H.A.B. and R.M.C. conducted research, analyzed data, and wrote the paper; M.B. conducted research; and L.S.P., J.d.J., P.P.M., and M.F.L. contributed vital new reagents and edited the paper.
Conflict-of-interest disclosure: M.F.L., J.d.J., and P.P.M. received research funding from QRxPharma (Sydney, Australia). The remaining authors declare no competing financial interests.
Correspondence: Rodney M. Camire, Children's Hospital of Philadelphia, Division of Hematology, 302 Abramson Research Center, 3615 Civic Center Blvd, Philadelphia, PA 19104; e-mail: rcamire@mail.med.upenn.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal