Abstract
We previously showed that Tie2-expressing monocytes (TEMs) have nonredundant proangiogenic activity in tumors. Here, we compared the gene expression profile of tumor-infiltrating TEMs with that of tumor-associated macrophages (TAMs), spleen-derived Gr1+Cd11b+ neutrophils/myeloid-derived suppressor cells, circulating “inflammatory” and “resident” monocytes, and tumor-derived endothelial cells (ECs) by quantitative polymerase chain reaction–based gene arrays. TEMs sharply differed from ECs and Gr1+Cd11b+ cells but were highly related to TAMs. Nevertheless, several genes were differentially expressed between TEMs and TAMs, highlighting a TEM signature consistent with enhanced proangiogenic/tissue-remodeling activity and lower proinflammatory activity. We validated these findings in models of oncogenesis and transgenic mice expressing a microRNA-regulated Tie2-GFP reporter. Remarkably, resident monocytes and TEMs on one hand, and inflammatory monocytes and TAMs on the other hand, expressed coordinated gene expression profiles, suggesting that the 2 blood monocyte subsets are committed to distinct extravascular fates in the tumor microenvironment. We further showed that a prominent proportion of embryonic/fetal macrophages, which participate in tissue morphogenesis, expressed distinguishing TEM genes. It is tempting to speculate that Tie2+ embryonic/fetal macrophages, resident blood monocytes, and tumor-infiltrating TEMs represent distinct developmental stages of a TEM lineage committed to execute physiologic proangiogenic and tissue-remodeling programs, which can be coopted by tumors.
Introduction
Experimental data suggest that chronic inflammation promotes tumor development by recruiting innate immune cells.1,2 One mechanism by which these cells may foster tumor progression is the promotion of angiogenesis.3,4 Virtually every innate immune cell type has been implicated in tumor angiogenesis, most notably mast cells, tumor-associated macrophages (TAMs), Tie2-expressing monocytes (TEMs), neutrophils, and the so-called “myeloid-derived suppressor cells” (MDSCs), a heterogeneous population comprising both immature and differentiated myeloid cells that expand in tumor-bearing hosts.4-6
Tumors recruit circulating monocytes, which extravasate and differentiate into TAMs. TAMs are thought to produce several growth factors, cytokines, and extracellular matrix remodeling molecules that together facilitate tumor angiogenesis, invasion, and metastasis.7 Although it is a common theme in tumor biology that the fate of tumor-homing monocytes is modulated directly by the tumor microenvironment,2,7 it is possible that distinct circulating monocyte subsets may contribute distinct macrophage subpopulations in tumors. Indeed, circulating monocytes, as identified by physical properties and Cd115 (Csf1 receptor) expression, can be divided into at least 2 subsets according to the expression of surface markers. In mice, expression of Gr1, Ccr2 (the receptor of Ccl2, a potent monocyte chemoattractant), Sell (L-selectin, or Cd62l), Cx3cr1 (the receptor for the chemokine fractalkine), and Cd43 distinguishes “inflammatory” (Gr1+Ccr2+Cd62l+Cx3cr1lowCd43−) from “resident” (Gr1−Ccr2−Cd62l−Cx3cr1highCd43+) monocytes.8,9 Inflammatory monocytes are considered the precursors of macrophages and dendritic cells (DCs) recruited to inflamed tissues, whereas resident monocytes patrol blood vessels and perhaps supply certain organs with a resident population of macrophage-lineage cells.8 Pahler et al10 suggested that TAMs mostly derive from Ccr2+ inflammatory monocytes; indeed, tumors grown in Ccr2 null mice, which have reduced numbers of circulating inflammatory but not resident monocytes,11 recruit few TAMs.10 In vivo imaging has recently suggested that TAMs/macrophages comprise subsets endowed with different migratory activity12 ; whether such heterogeneity of TAM behavior reflects the existence of developmentally and/or functionally distinct subsets in tumors is currently unknown. Moreover, the identification of putative monocyte/macrophage subsets in tumors is currently limited by the paucity of suitable markers that may distinguish TAM subpopulations.7
TEMs13-17 are a subset of circulating and tumor-infiltrating monocytes expressing the angiopoietin receptor Tie2.18 Our previous studies indicated that TEMs are proangiogenic when isolated from either tumors or peripheral blood, suggesting their commitment to a proangiogenic function already in the circulation. Although mouse TEMs are a minor proportion of the bulk of tumor-infiltrating macrophages, fluorescence-activated cell sorting (FACS)–isolated, tumor-derived TEMs enhanced angiogenesis to a greater extent than TAMs when coinjected with tumor cells subcutaneously in mice.14 Similarly, human blood–derived Tie2+ monocytes were more proangiogenic than Tie2-negative monocytes when coinjected with tumor cells subcutaneously in athymic mice.16 Moreover, the selective elimination of TEMs by a conditional suicide gene approach inhibited angiogenesis and tumor growth in several mouse tumor models, suggesting that TEMs have nonredundant, proangiogenic activity in tumors.14 Although TEMs express Tie2, which has long been regarded as an endothelial cell (EC)–specific molecule, they are distinct from endothelial progenitor cells3 (EPCs) and do not incorporate in the tumor endothelium.13 Yet, little is known of the molecular identity of TEMs, and their relationship with TAMs, other tumor-infiltrating myeloid cells, and circulating monocytes remains to be investigated.
To address these issues, we isolated TEMs, TAMs, and relevant myeloid cell populations, including circulating monocyte subsets and splenic Gr1+Cd11b+ neutrophils/MDSCs, and profiled their gene expression by a quantitative polymerase chain reaction (PCR)–based multigene array approach. Our data indicate that tumor-derived TEMs are a subset of TAMs expressing a distinguishing gene signature consistent with proangiogenic and tissue-remodeling activity. Remarkably, the TEM signature also distinguishes resident from inflammatory monocytes in the blood of tumor-free mice, indicating its association with a specific monocyte developmental stage or lineage. Moreover, TEM-specific markers are expressed by a significant proportion of embryonic/fetal macrophages,19,20 which appear early in development and are found at sites of angiogenesis and tissue remodeling. Overall, these data suggest the existence of a TEM lineage of monocytes that play important roles during organ and tumor development and vascularization.
Methods
Materials and methods are reported in full as supplemental data (available on the Blood website; see the Supplemental Materials link at the top of the online article). A comprehensive list of antibodies used in this study is presented as supplemental Table 1.
Transgenic mice and lentiviral vector constructs
Briefly, the Tie2-GFPmir142T lentiviral vector (LV) was generated by cloning 4 tandem copies of a 23-bp sequence with perfect complementarity to microRNA-142,21 downstream to the wPRE sequence in the Tie2-GFP LV.13 CD1/Tie2-GFPmir142T transgenic mice were generated by LV-mediated transgenesis, as described previously,14 and bred for 6 generations before use. All animal procedures were performed according to protocols approved by the Animal Care and Use Committee of the Fondazione San Raffaele del Monte Tabor and communicated to the Ministry of Health and local authorities according to the Italian law.
Real-time quantitative PCR analysis and statistical analysis
After cell sorting, cells were washed in low-protein buffer and total mRNA purified following RNeasy Micro/Mini kit guidelines (QIAGEN). RNA was quantified and retrotranscribed with SuperScript III (Invitrogen). To obtain adequate amounts of cDNA for each gene profiling experiment (interrogating 280 genes), 1 to 3 cell sorting sessions were required. Quantitative PCR analyses were performed with TaqMan assays from Applied Biosystems. We used either single-gene TaqMan assays to analyze the expression of individual genes, or multigene TaqMan low-density arrays. We used 2 custom-made and one premade (Immune Panel) TaqMan low-density arrays, each measuring the expression of 96 genes in 4 technical replicates. A total of 100 ng to 1 μg cDNA was loaded on each array. Quantitative PCR was run for 35 (low-density arrays) or 40 to 45 cycles (individual gene assays) in standard mode using an ABI7900HT apparatus (Applied Biosystems). The SDS 2.2.1 software was used to extract raw data. The difference between the threshold cycle (CT) of each gene and that of the endogenous controls β2m or Gapdh (ΔCT) was used to determine gene expression. The lower the ΔCT, the higher the gene expression level. Statistical methods and other relevant information are described in full in the supplemental data.
Results
Isolation of TEMs and TAMs from murine tumors
To gene profile TEMs and TAMs, we chose a quantitative PCR approach because it allows for identifying low-abundance transcripts from small amounts of mRNA. We used low-density quantitative PCR arrays with standardized fluidics, which ensure reproducibility, intersample comparison, and global analysis. We sorted TEMs, TAMs, and ECs from N202 mammary tumors grown subcutaneously in Tie2-GFP transgenic mice14 ; in this model, GFP expression can be conveniently used to discriminate TEMs from other tumor-infiltrating myeloid cells. However, purification of tumor-derived TEMs is technically challenging because of their scarcity and weak GFP expression level. Moreover, both TEMs and ECs express GFP in Tie2-GFP mice.14 These factors contribute to increase the risk of contamination from other cell types, ECs in particular. To address this issue, we first developed a model to assay the presence of contaminating ECs in our TEM preparations, and selected a cell sorting formula that enabled the purification of EC-free TEM fractions (supplemental data; supplemental Figure 1).
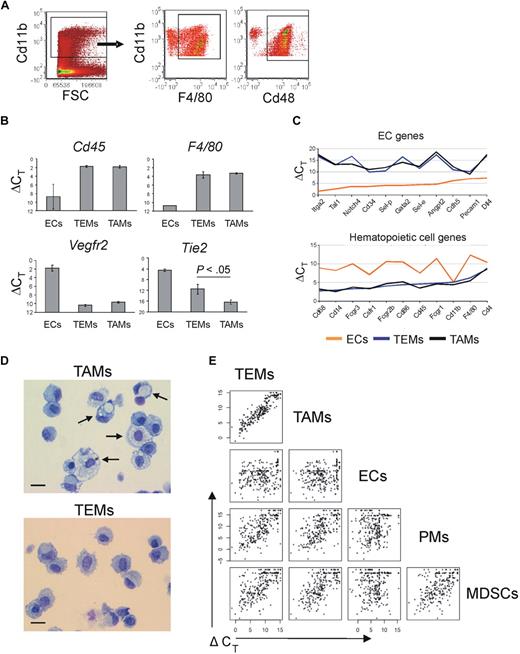
In N202 tumors, the wide majority of the tumor-infiltrating Cd11b+ cells were Emr1/F4/80+ and Cd48+ (77% ± 6% and 88% ± 5%, respectively; mean frequency of marker positive cells ± SD; n = 4), thus representing myeloid cells highly enriched in TAMs (Figure 1A). We then sorted TEMs as Tie2-GFP+Cd11b+Cd31low/−, TAMs as Tie2-GFP−Cd11b+Cd31low/−, and ECs as Tie2-GFP+Cd11b−Cd31+ cells (supplemental Figure 2). To validate the sorted cell populations, we measured expression of the hematopoietic lineage-specific marker Ptprc/Cd45, the macrophage-specific marker F4/80, the EC-specific marker Vegfr2 (Flk1), and Tie2/Tek, in TEMs, TAMs, and ECs (n = 2-4 biologic samples) by quantitative PCR (Figure 1B). Both TEMs and TAMs expressed high-level Cd45 and F4/80, which confirmed their hematopoietic origin and monocyte/macrophage identity. Of note, Tie2 expression was significantly higher in TEMs than in TAMs (t test: P < .05; n = 4).
Gene expression profile of TEMs, TAMs, MDSCs, PMs, and ECs. (A) Flow cytometry analyses of tumors grown subcutaneously in wild-type FVB mice (n = 4) and made into single-cell suspensions shows that the majority of tumor-infiltrating myeloid cells (gated as Cd11b+ cells, dot plot on the left) are highly enriched in F4/80+ or Cd48+ macrophages (dot plots on the right). One representative experiment is shown. (B) Quantitative PCR analysis of TEMs (n = 4), TAMs (n = 4), and ECs (n = 2) showing the expression level of relevant genes. Results show ΔCT values (mean ± SEM) over endogenous control Gapdh. The lower the ΔCT, the higher the expression level. TEMs and TAMs express Cd45 (pan-hematopoietic–specific marker) and F4/80 (macrophage-specific marker) to similar extent. Vegfr2 (endothelial-specific marker) is expressed robustly only by ECs. TEMs express Tie2 to a significantly higher extent than TAMs (P < .05). (C) Quantitative PCR-based, multigene array analysis of TEMs (n = 3), TAMs (n = 3), and ECs (n = 1) showing the expression level (ΔCT vs β2m) of relevant EC (top panel) and hematopoietic/myeloid (bottom panel) genes. TEMs and TAMs robustly express classic hematopoietic/myeloid genes but not EC genes. (D) Morphology (May-Grünwald-Giemsa staining) of TAMs (top panel) and TEMs (bottom panel) FACS-sorted from N202 tumors grown for 3 weeks in Tie2-GFP transgenic mice (n = 2 independent experiments). Arrows indicate large macrophages containing conspicuous cytoplasmic phagosomes. Scale bar represents 30 μm. Photos are representative of n = 25 photos/sample. (E) One-dot-one-gene representation of the expression profile (280 genes analyzed) of tumor-derived TEMs (n = 3), TAMs (n = 3), and ECs (n = 1), Gr1+Cd11b+ neutrophils/MDSCs (n = 2; isolated from the spleen of tumor bearing mice) and PMs (n = 2), analyzed by quantitative PCR as in panel C. Each one-dot-one-gene plot compares 2 cell types, as indicated. The data show that TEMs are highly related to TAMs (Pearson linear correlation: 0.926) but sharply differ from Gr1+Cd11b+ cells (0.682), PMs (0.701), and ECs (0.240).
Gene expression profile of TEMs, TAMs, MDSCs, PMs, and ECs. (A) Flow cytometry analyses of tumors grown subcutaneously in wild-type FVB mice (n = 4) and made into single-cell suspensions shows that the majority of tumor-infiltrating myeloid cells (gated as Cd11b+ cells, dot plot on the left) are highly enriched in F4/80+ or Cd48+ macrophages (dot plots on the right). One representative experiment is shown. (B) Quantitative PCR analysis of TEMs (n = 4), TAMs (n = 4), and ECs (n = 2) showing the expression level of relevant genes. Results show ΔCT values (mean ± SEM) over endogenous control Gapdh. The lower the ΔCT, the higher the expression level. TEMs and TAMs express Cd45 (pan-hematopoietic–specific marker) and F4/80 (macrophage-specific marker) to similar extent. Vegfr2 (endothelial-specific marker) is expressed robustly only by ECs. TEMs express Tie2 to a significantly higher extent than TAMs (P < .05). (C) Quantitative PCR-based, multigene array analysis of TEMs (n = 3), TAMs (n = 3), and ECs (n = 1) showing the expression level (ΔCT vs β2m) of relevant EC (top panel) and hematopoietic/myeloid (bottom panel) genes. TEMs and TAMs robustly express classic hematopoietic/myeloid genes but not EC genes. (D) Morphology (May-Grünwald-Giemsa staining) of TAMs (top panel) and TEMs (bottom panel) FACS-sorted from N202 tumors grown for 3 weeks in Tie2-GFP transgenic mice (n = 2 independent experiments). Arrows indicate large macrophages containing conspicuous cytoplasmic phagosomes. Scale bar represents 30 μm. Photos are representative of n = 25 photos/sample. (E) One-dot-one-gene representation of the expression profile (280 genes analyzed) of tumor-derived TEMs (n = 3), TAMs (n = 3), and ECs (n = 1), Gr1+Cd11b+ neutrophils/MDSCs (n = 2; isolated from the spleen of tumor bearing mice) and PMs (n = 2), analyzed by quantitative PCR as in panel C. Each one-dot-one-gene plot compares 2 cell types, as indicated. The data show that TEMs are highly related to TAMs (Pearson linear correlation: 0.926) but sharply differ from Gr1+Cd11b+ cells (0.682), PMs (0.701), and ECs (0.240).
We then analyzed the clonogenic potential of TEMs and TAMs in colony-forming cell assays. Sorted TEMs did not generate hematopoietic colonies (supplemental Table 2), indicating that they are virtually devoid of hematopoietic progenitors.
Tumor TEMs are monocytes/macrophages expressing a distinguishing gene signature
To investigate the nature of TEMs, we measured the expression of 280 genes (supplemental Table 3) previously implicated in angiogenesis, tissue remodeling, immune response, cell adhesion, chemotaxis, neural guidance, vascular morphogenesis, in TEMs (n = 3 independent experiments), TAMs (n = 3), and ECs (n = 1). We extended the gene expression analysis to peritoneal macrophages (PMs; sorted as Cd11b+F4/80+Cd31low/− cells; n = 2) and neutrophils/MDSCs isolated from the spleen of N202 tumor-bearing mice (sorted as Cd11b+Gr1+ cells; n = 2).
The expression profile of TEMs clearly indicated that they belong to the monocyte/macrophage lineage and are distinct from ECs. Indeed, TEMs robustly express typical “myeloid genes,” but low to negligible amounts of “EC genes” (Figure 1C). A list of the genes highly expressed in TEMs and grouped by function is shown in supplemental Table 4. Among the highly expressed monocyte/macrophage genes were Csf1 receptor (Csf1r/Cd115), Fc receptors (Fcgr1, Fcgr2b, Fcgr3), fractalkine receptor (Cx3cr1), macrosialin (Cd68), F4/80, Cd14, scavenger receptors (mannose receptor Mrc1, macrophage scavenger receptors Msr1 and Msr2, hemoglobin/haptoglobin scavenger receptor Cd163, membrane-bound scavenger receptor/cytokine Cxcl16), endopeptidases (metalloproteases, cathepsins), and several integrins. In addition, TEMs express monocyte/macrophage-derived cytokines (Il1a, Il1b, Il6, Tnf, Tgfb1), chemokines (Ccl4, Ccl3/Mip1a/b, Cxcl10, Cxcl9, Cxcl11), and classic proangiogenic factors (Vegfa, Vegfb, Sema4d, Pdgfb, lactadherin [Mfge8], thymidine phosphorylase [Ecgf1], placental growth factor [Pgf]).
Morphologic analysis of FACS-sorted cells (n = 2 independent experiments; Figure 1D) showed that TEMs are a homogeneous cell population with typical monocyte/macrophage morphology (> 94% of the cells with bean-shaped nucleus and large cytoplasm). On the other hand, TAMs displayed a greater extent of morphologic heterogeneity and contained both monocyte-like cells and large macrophages with features of activated phagocytes (rounded nucleus; large cytoplasm with numerous vacuoles). The cell morphology corroborated the gene expression data that both TEMs and TAMs belong to the monocyte/macrophage lineage.
A comparison between the gene expression profile of TEMs and TAMs revealed that the 2 monocyte/macrophage subsets were highly related (Figure 1E). However, TEMs differed significantly from spleen-derived Gr1+Cd11b+ neutrophils/MDSCs, PMs, and tumor-derived ECs. To rigorously identify differences between TEMs and TAMs, we implemented in R (http://www.R-project.org) a multivariate regression model, similarly to the model implemented in sas,22 to compute over the whole dataset and estimate the fold change in gene expression for each single target gene. This model jointly evaluates the role of different variables of interest providing for (1) statistical significance of the observed differences in expression level across the whole set of experimental samples, (2) identifying the experimental variables that contribute to each individual measurement, and (3) subtracting experimentally introduced biases to obtain a stringent estimate of the actual biologic differences. Statistical analysis of the data revealed several differentially expressed genes between TEMs and TAMs (29% at P < .05; Table 1). Among these, Arginase1 (Arg1) and several scavenger receptors23 (Cd163, stabilin-1 [Stab1], Mrc1, Msr2) were up-regulated, whereas Nos2, proinflammatory24 and antiangiogenic25 molecules (Il1b, Ptgs2/Cox2, Il12a, Tnf, Ccl5, Cxcl10, Cxcl11) were down-regulated in TEMs versus TAMs. With reference to the M1-M2 polarization paradigm proposed by some authors,23 the enhanced expression of scavenger receptors and the down-regulation of inflammatory mediators by TEMs would place them at the M2 extreme of the polarization window (ie, TEMs are significantly more M2-skewed than TAMs), consistent with their marked proangiogenic and protumoral activity.14,15 Among the most differentially expressed genes were also the hyaluronan receptor-1 (Lyve1), neuropilin-1 (Nrp1), stromal cell derived factor-1 (Cxcl12/Sdf-1), insulin growth factor-1 (Igf1), and Toll-like receptor-4 (Tlr4), all of which were up-regulated in TEMs. The finding of several differentially expressed genes strongly suggests that TEMs represent a distinct subset or differentiation state of TAMs.
Genes differentially expressed between tumor-derived TEMs and TAMs
| Gene . | Fold more . | Fold less . | ΔCt . | P . | Gene . | Fold more . | Fold less . | ΔCt . | P . |
|---|---|---|---|---|---|---|---|---|---|
| Up-regulated | |||||||||
| Cd163 | 15.8 | 6.3 | < .001 | Timp2 | 2.8 | 4.5 | < .01 | ||
| Lyve1 | 14.1 | 6.1 | < .001 | Plxnd1 | 2.8 | 6.2 | < .01 | ||
| Igf1 | 8.2 | 9.3 | < .001 | Efna1 | 2.7 | 12.6 | < .01 | ||
| Stab1 | 6.4 | 5.5 | < .001 | Slamf1 | 2.6 | 14.6 | < .05 | ||
| Mrc1 | 5.3 | 3.0 | < .001 | Fcgr3 | 2.6 | 3.1 | < .05 | ||
| Sema6d | 5.2 | 15.2 | < .001 | Angpt1 | 2.6 | 14.2 | < .05 | ||
| Nrp1 | 4.6 | 6.3 | < .001 | Sema3c | 2.6 | 12.3 | < .05 | ||
| Cxcl12 | 4.4 | 10.6 | < .001 | Timp3 | 2.5 | 12.2 | < .05 | ||
| Thbs3 | 4.3 | 12.1 | < .001 | Kit | 2.5 | 14.9 | < .05 | ||
| Cxcl13 | 4.3 | 5.1 | < .001 | Sdc2 | 2.5 | 9.1 | < .05 | ||
| Efnb2 | 4.2 | 12.9 | < .01 | Efnb1 | 2.5 | 12.0 | < .05 | ||
| Neo1 | 4.1 | 15.1 | < .01 | Cdh5 | 2.4 | 11.6 | < .05 | ||
| Plxna3 | 4.1 | 12.8 | < .001 | Plxnb2 | 2.4 | 6.3 | < .05 | ||
| Itga2 | 4.0 | 16.3 | < .05 | Il4ra | 2.3 | 5.1 | < .05 | ||
| Plxna1 | 3.6 | 8.1 | < .001 | Il10ra | 2.2 | 6.5 | < .05 | ||
| Msr2 | 3.5 | 4.4 | < .001 | Arg1 | 2.2 | 5.7 | < .05 | ||
| Tlr4 | 3.5 | 7.8 | < .01 | Hpse | 2.1 | 5.6 | < .05 | ||
| Plxna4 | 3.1 | 10.9 | < .01 | Serpinb2 | 2.1 | 9.8 | < .05 | ||
| Edg1 | 3.0 | 8.1 | < .01 | Itgb1 | 2.1 | 5.0 | < .05 | ||
| Fcgr2b | 2.9 | 3.9 | < .01 | ||||||
| Down-regulated | |||||||||
| Il1b | −6.6 | 2.4 | < .001 | Ptprc | −2.7 | 3.7 | < .01 | ||
| Ptgs2 | −5.0 | 5.5 | < .001 | Ctss | −2.7 | 0.8 | < .01 | ||
| Il4 | −4.0 | 12.0 | < .001 | Tgfb1 | −2.6 | 4.2 | < .05 | ||
| Nos2 | −3.9 | 8.8 | < .001 | Il18 | −2.6 | 8.2 | < .05 | ||
| Ccr7 | −3.8 | 7.6 | < .001 | Sell | −2.5 | 10.3 | < .05 | ||
| Il12a | −3.6 | 16.2 | < .01 | Ccr2 | −2.5 | 5.8 | < .05 | ||
| Smad7 | −3.6 | 8.2 | < .001 | Il12b | −2.5 | 11.8 | < .05 | ||
| Bcl2l1 | −3.2 | 6.6 | < .01 | Nfkb2 | −2.5 | 5.9 | < .05 | ||
| Stat4 | −3.1 | 10.7 | < .01 | Fas | −2.4 | 9.8 | < .05 | ||
| Tnf | −3.1 | 4.0 | < .01 | Gusb | −2.4 | 4.9 | < .05 | ||
| Il13 | −3.1 | 12.8 | < .01 | Smad3 | −2.4 | 9.0 | < .05 | ||
| Actb | −3.0 | 1.0 | < .01 | Socs1 | −2.4 | 6.6 | < .05 | ||
| Edn1 | −3.0 | 11.0 | < .01 | Bax | −2.4 | 6.2 | < .05 | ||
| Ccl5 | −3.0 | 4.3 | < .01 | Pgk1 | −2.4 | 4.1 | < .05 | ||
| H2-Ea | −3.0 | 5.1 | < .01 | Ski | −2.3 | 6.1 | < .05 | ||
| Vegfa | −2.9 | 4.5 | < .01 | Csf1 | −2.3 | 7.8 | < .05 | ||
| Il1a | −2.9 | 5.2 | < .01 | Ctla4 | −2.2 | 12.9 | < .05 | ||
| Cxcl10 | −2.8 | 2.9 | < .01 | Stat1 | −2.1 | 3.7 | < .05 | ||
| Cd80 | −2.8 | 9.1 | < .01 | Tnfrsf18 | −2.1 | 12.0 | < .05 | ||
| Ece1 | −2.7 | 13.1 | < .05 | Tbx21 | −2.1 | 12.1 | < .05 | ||
| Cxcl11 | −2.7 | 6.6 | < .01 | ||||||
| Gene . | Fold more . | Fold less . | ΔCt . | P . | Gene . | Fold more . | Fold less . | ΔCt . | P . |
|---|---|---|---|---|---|---|---|---|---|
| Up-regulated | |||||||||
| Cd163 | 15.8 | 6.3 | < .001 | Timp2 | 2.8 | 4.5 | < .01 | ||
| Lyve1 | 14.1 | 6.1 | < .001 | Plxnd1 | 2.8 | 6.2 | < .01 | ||
| Igf1 | 8.2 | 9.3 | < .001 | Efna1 | 2.7 | 12.6 | < .01 | ||
| Stab1 | 6.4 | 5.5 | < .001 | Slamf1 | 2.6 | 14.6 | < .05 | ||
| Mrc1 | 5.3 | 3.0 | < .001 | Fcgr3 | 2.6 | 3.1 | < .05 | ||
| Sema6d | 5.2 | 15.2 | < .001 | Angpt1 | 2.6 | 14.2 | < .05 | ||
| Nrp1 | 4.6 | 6.3 | < .001 | Sema3c | 2.6 | 12.3 | < .05 | ||
| Cxcl12 | 4.4 | 10.6 | < .001 | Timp3 | 2.5 | 12.2 | < .05 | ||
| Thbs3 | 4.3 | 12.1 | < .001 | Kit | 2.5 | 14.9 | < .05 | ||
| Cxcl13 | 4.3 | 5.1 | < .001 | Sdc2 | 2.5 | 9.1 | < .05 | ||
| Efnb2 | 4.2 | 12.9 | < .01 | Efnb1 | 2.5 | 12.0 | < .05 | ||
| Neo1 | 4.1 | 15.1 | < .01 | Cdh5 | 2.4 | 11.6 | < .05 | ||
| Plxna3 | 4.1 | 12.8 | < .001 | Plxnb2 | 2.4 | 6.3 | < .05 | ||
| Itga2 | 4.0 | 16.3 | < .05 | Il4ra | 2.3 | 5.1 | < .05 | ||
| Plxna1 | 3.6 | 8.1 | < .001 | Il10ra | 2.2 | 6.5 | < .05 | ||
| Msr2 | 3.5 | 4.4 | < .001 | Arg1 | 2.2 | 5.7 | < .05 | ||
| Tlr4 | 3.5 | 7.8 | < .01 | Hpse | 2.1 | 5.6 | < .05 | ||
| Plxna4 | 3.1 | 10.9 | < .01 | Serpinb2 | 2.1 | 9.8 | < .05 | ||
| Edg1 | 3.0 | 8.1 | < .01 | Itgb1 | 2.1 | 5.0 | < .05 | ||
| Fcgr2b | 2.9 | 3.9 | < .01 | ||||||
| Down-regulated | |||||||||
| Il1b | −6.6 | 2.4 | < .001 | Ptprc | −2.7 | 3.7 | < .01 | ||
| Ptgs2 | −5.0 | 5.5 | < .001 | Ctss | −2.7 | 0.8 | < .01 | ||
| Il4 | −4.0 | 12.0 | < .001 | Tgfb1 | −2.6 | 4.2 | < .05 | ||
| Nos2 | −3.9 | 8.8 | < .001 | Il18 | −2.6 | 8.2 | < .05 | ||
| Ccr7 | −3.8 | 7.6 | < .001 | Sell | −2.5 | 10.3 | < .05 | ||
| Il12a | −3.6 | 16.2 | < .01 | Ccr2 | −2.5 | 5.8 | < .05 | ||
| Smad7 | −3.6 | 8.2 | < .001 | Il12b | −2.5 | 11.8 | < .05 | ||
| Bcl2l1 | −3.2 | 6.6 | < .01 | Nfkb2 | −2.5 | 5.9 | < .05 | ||
| Stat4 | −3.1 | 10.7 | < .01 | Fas | −2.4 | 9.8 | < .05 | ||
| Tnf | −3.1 | 4.0 | < .01 | Gusb | −2.4 | 4.9 | < .05 | ||
| Il13 | −3.1 | 12.8 | < .01 | Smad3 | −2.4 | 9.0 | < .05 | ||
| Actb | −3.0 | 1.0 | < .01 | Socs1 | −2.4 | 6.6 | < .05 | ||
| Edn1 | −3.0 | 11.0 | < .01 | Bax | −2.4 | 6.2 | < .05 | ||
| Ccl5 | −3.0 | 4.3 | < .01 | Pgk1 | −2.4 | 4.1 | < .05 | ||
| H2-Ea | −3.0 | 5.1 | < .01 | Ski | −2.3 | 6.1 | < .05 | ||
| Vegfa | −2.9 | 4.5 | < .01 | Csf1 | −2.3 | 7.8 | < .05 | ||
| Il1a | −2.9 | 5.2 | < .01 | Ctla4 | −2.2 | 12.9 | < .05 | ||
| Cxcl10 | −2.8 | 2.9 | < .01 | Stat1 | −2.1 | 3.7 | < .05 | ||
| Cd80 | −2.8 | 9.1 | < .01 | Tnfrsf18 | −2.1 | 12.0 | < .05 | ||
| Ece1 | −2.7 | 13.1 | < .05 | Tbx21 | −2.1 | 12.1 | < .05 | ||
| Cxcl11 | −2.7 | 6.6 | < .01 | ||||||
The expression level of each gene in tumor-derived TEMs is indicated as fold-change versus TAMs. The ΔCt of each gene was calculated using β2m as endogenous control.
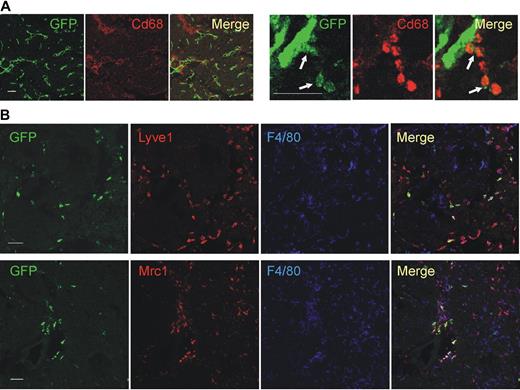
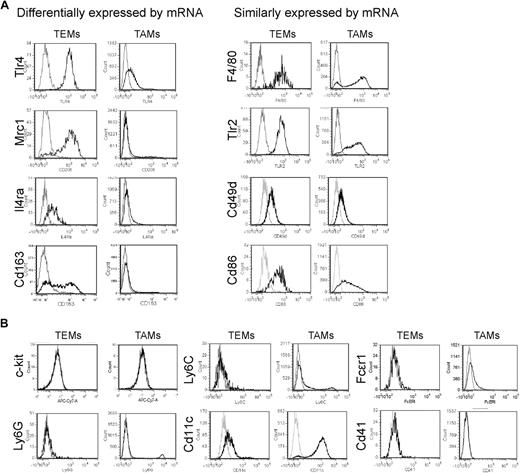
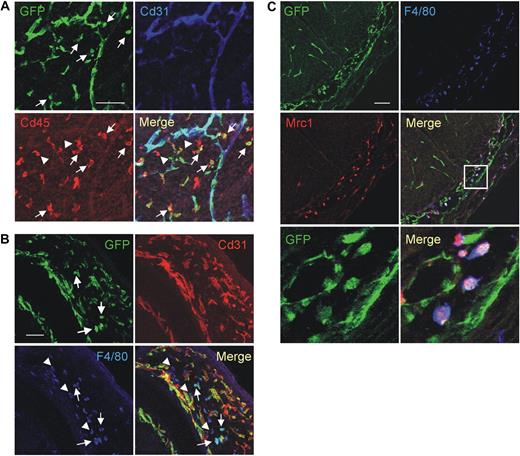
We then validated the gene expression results by protein analysis, either by immunofluorescence staining (IFS) and confocal microscopy of tumor sections (Figure 2) or by flow cytometry of tumor-derived cell suspensions (Figure 3). We used 2 different tumor models: N202 tumors grown subcutaneously in Tie2-GFP transgenic mice14 (n = 5 for IFS; n = 3-7 for flow cytometry); and mammary tumors spontaneously arising in MMTV-PyMT transgenic mice26 previously transplanted with Tie2-GFP bone marrow (BM) cells (n = 4 tumors from 4 mice for IFS), as described.27 Whereas in Tie2-GFP mice both TEMs and ECs express GFP (Figure 2A), in BM-transplanted mice only TEMs are GFP+ (Figure 2B, supplemental Figure 3). GFP+F4/80+ TEMs were frequently located in stromal tumor areas, whereas GFP−F4/80+ TAMs were evenly distributed throughout the tumor mass (Figure 2B). IFS and/or flow analyses showed differential expression of Lyve1, Mrc1, Tlr4, interleukin-4 receptor-alpha (Il4ra), Cd163, and Stab1 but not of F4/80, Tlr2, Itga4 (Cd49d), and Cd86, between TEMs and TAMs, in agreement with the RNA data (Figures 2B, 3A, supplemental Figure 3). Although the Itgax (Cd11c) mRNA was expressed similarly in TEMs and TAMs (Table 1), TEMs were Cd11clow/− and TAMs markedly Cd11c+ by flow cytometry and IFS (Figure 3B; and data not shown). TEMs were c-kit− and did not express FcϵR1, Ly6G, Ly6C, and Cd41, which are mast cell-, neutrophil-, inflammatory monocyte-, and megakaryocyte/platelet-specific markers, respectively (Figure 3B). Together with the mRNA data, these results identify a unique TEM surface marker profile, which distinguishes them from the bulk of TAMs and related myeloid-lineage cells.
Confocal immunofluorescence analysis of mouse tumors identifies a distinguishing TEM signature. (A) N202 mammary tumors (n = 5) grown subcutaneously in Tie2-GFP transgenic mice and analyzed for GFP (green) and Cd68 (red) expression. Confocal planes are shown individually and after merging (merge). The left panels show GFP+ blood vessels and abundant Cd68+ macrophages; scale bar represents 120 μm. High-magnification photos (right panels) show perivascular Tie2-GFP+Cd68+ TEMs (arrows); scale bar represents 60 μm. For each tumor, at least 3 sections were analyzed. (B) Mammary tumors spontaneously arising in MMTV-PyMT transgenic mice (n = 4) previously transplanted with Tie2-GFP BM cells and analyzed for GFP (green), Lyve1 or Mrc1 (red), and F4/80 (blue) expression. Abundant F4/80+ macrophages are evenly distributed within the tumor mass, whereas GFP+Lyve1+ or GFP+Mrc1+ TEMs cells are mainly found in stromal septa surrounding tumor cell nests. Virtually all the Tie2-GFP+ cells express Lyve1 and Mrc1. Note that some of the Tie2-GFP− Lyve1+ or Mrc1+ cells may represent host-derived, nontransgenic TEMs. Scale bar represents 60 μm. For each tumor, at least 10 sections were analyzed for each marker.
Confocal immunofluorescence analysis of mouse tumors identifies a distinguishing TEM signature. (A) N202 mammary tumors (n = 5) grown subcutaneously in Tie2-GFP transgenic mice and analyzed for GFP (green) and Cd68 (red) expression. Confocal planes are shown individually and after merging (merge). The left panels show GFP+ blood vessels and abundant Cd68+ macrophages; scale bar represents 120 μm. High-magnification photos (right panels) show perivascular Tie2-GFP+Cd68+ TEMs (arrows); scale bar represents 60 μm. For each tumor, at least 3 sections were analyzed. (B) Mammary tumors spontaneously arising in MMTV-PyMT transgenic mice (n = 4) previously transplanted with Tie2-GFP BM cells and analyzed for GFP (green), Lyve1 or Mrc1 (red), and F4/80 (blue) expression. Abundant F4/80+ macrophages are evenly distributed within the tumor mass, whereas GFP+Lyve1+ or GFP+Mrc1+ TEMs cells are mainly found in stromal septa surrounding tumor cell nests. Virtually all the Tie2-GFP+ cells express Lyve1 and Mrc1. Note that some of the Tie2-GFP− Lyve1+ or Mrc1+ cells may represent host-derived, nontransgenic TEMs. Scale bar represents 60 μm. For each tumor, at least 10 sections were analyzed for each marker.
Flow cytometric analysis of tumors identifies a distinguishing TEM signature. (A) Flow analysis of some of the genes that were either differentially expressed or not differentially expressed between TEMs and TAMs. N202 mammary tumors grown subcutaneously in Tie2-GFP transgenic were made into single-cell suspensions and TEMs and TAMs gated as Tie2-GFP+Cd11b+7AAD− and Tie2-GFP−Cd11b+7AAD− cells, respectively. In agreement with the mRNA data, TEMs but not TAMs robustly and uniformly express Tlr4, Mrc1, Il4ra, and Cd163 (left panels). Genes not differentially expressed by quantitative PCR are similarly expressed by TEMs and TAMs (right panels). For each marker, at least 3 independent analyses were performed; for each analysis, at least 3 tumors were pooled together and made into single-cell suspensions. (B) Further phenotypic characterization of TEMs and TAMs. Note that TEMs do not express c-kit or granulocyte (Ly6G), inflammatory monocyte (Ly6C), DC (Cd11c), mast cell (FcϵRI), and megakaryocyte/platelet (Cd41) markers. For each marker, at least 3 independent analyses were performed.
Flow cytometric analysis of tumors identifies a distinguishing TEM signature. (A) Flow analysis of some of the genes that were either differentially expressed or not differentially expressed between TEMs and TAMs. N202 mammary tumors grown subcutaneously in Tie2-GFP transgenic were made into single-cell suspensions and TEMs and TAMs gated as Tie2-GFP+Cd11b+7AAD− and Tie2-GFP−Cd11b+7AAD− cells, respectively. In agreement with the mRNA data, TEMs but not TAMs robustly and uniformly express Tlr4, Mrc1, Il4ra, and Cd163 (left panels). Genes not differentially expressed by quantitative PCR are similarly expressed by TEMs and TAMs (right panels). For each marker, at least 3 independent analyses were performed; for each analysis, at least 3 tumors were pooled together and made into single-cell suspensions. (B) Further phenotypic characterization of TEMs and TAMs. Note that TEMs do not express c-kit or granulocyte (Ly6G), inflammatory monocyte (Ly6C), DC (Cd11c), mast cell (FcϵRI), and megakaryocyte/platelet (Cd41) markers. For each marker, at least 3 independent analyses were performed.
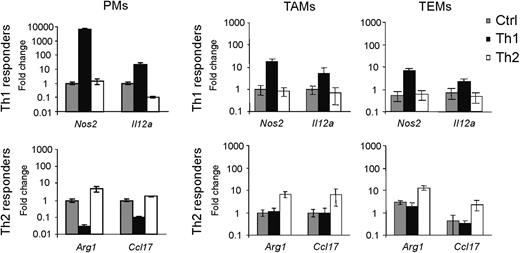
TEMs are refractory to proinflammatory stimuli
The gene expression data indicated that TEMs have a Th2/M2 phenotype. We wondered whether Th1 stimulation could modulate this phenotype. To this aim, we stimulated (or left unstimulated) tumor-derived TEMs, TAMs, and PMs with either Th1 (LPS + Ifng) or Th2 (Il4) cytokines (n = 3 independent experiments) and evaluated the expression of established responder genes6,23 (Nos2 and Il12a for Th1 responses; Arg1 and Ccl17 for Th2 responses). Th1 stimulation elicited minor responses in TEMs and TAMs, compared with PMs (Figure 4). Conversely, TEMs and TAMs were more responsive than PMs to Th2 stimulation. These results indicated that both tumor-derived TEMs and TAMs display defective Th1 responses but can respond to Th2 stimuli. Of note, both unstimulated and Th2-stimulated TEMs had higher Arg1 and lower Nos2 expression than TAMs, in agreement with the gene profile data.
TEMs, like TAMs, are refractory to proinflammatory stimuli. In vitro stimulation of PMs, TEMs, and TAMs with Th1 (LPS + Ifng) and Th2 (Il4) cytokines, or the same cells left unstimulated (Ctrl). Top and bottom panels show the expression of Th1 (Nos2, Il12a) and Th2 (Arg1, Ccl17) responder genes, respectively. The expression of each gene is indicated as fold change (mean ± 2 × SD of 3 independent experiments) over its expression in unstimulated PMs (for PMs) or unstimulated TAMs (for TEMs and TAMs); endogenous control: β2m. Note that TEMs and TAMs are much less responsive than PMs to Th1 cytokine treatment. Conversely, Th2 cytokine stimulation elicits a greater response in TEMs and TAMs than in PMs.
TEMs, like TAMs, are refractory to proinflammatory stimuli. In vitro stimulation of PMs, TEMs, and TAMs with Th1 (LPS + Ifng) and Th2 (Il4) cytokines, or the same cells left unstimulated (Ctrl). Top and bottom panels show the expression of Th1 (Nos2, Il12a) and Th2 (Arg1, Ccl17) responder genes, respectively. The expression of each gene is indicated as fold change (mean ± 2 × SD of 3 independent experiments) over its expression in unstimulated PMs (for PMs) or unstimulated TAMs (for TEMs and TAMs); endogenous control: β2m. Note that TEMs and TAMs are much less responsive than PMs to Th1 cytokine treatment. Conversely, Th2 cytokine stimulation elicits a greater response in TEMs and TAMs than in PMs.
Circulating TEMs belong to the resident monocyte subset
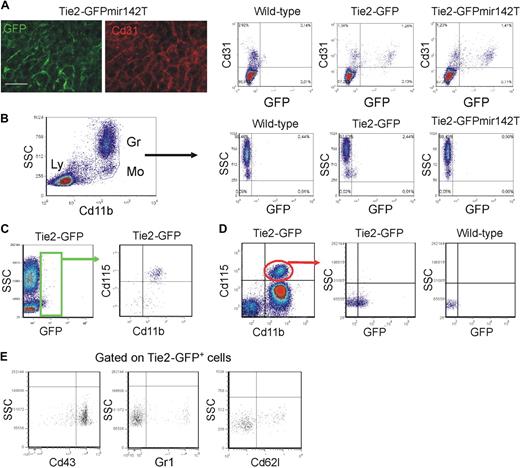
We then analyzed the circulating TEMs, which may be the precursors of tumor TEMs.14 We previously speculated that circulating TEMs belong to the myeloid lineage based on their expression of Cd11b and CD14 in mice and humans, respectively.14,16 However, several cell surface–associated markers are coexpressed by endothelial- and hematopoietic-lineage cells.5 To rigorously demonstrate that TEMs belong to the hematopoietic lineage, we used a genetic approach. MicroRNAs (miR) regulate networks of gene expression and help determining cell identity and lineage specification. miR-142 is specifically expressed in the hematopoietic lineage28 ; we previously showed that expression of a GFP reporter gene containing miR-142 target sequences was suppressed specifically in all hematopoietic-lineage cells of transgenic mice,21 indicating stringent association of miR-142 expression with hematopoietic cell specification. Here, we incorporated miR-142 target sequences (miR142T) downstream to the Tie2-GFP reporter and generated Tie2-GFPmiR142T transgenic mice by LV-mediated transgenesis. In the progeny of these mice, GFP was efficiently expressed in Cd31+ ECs (Figure 5A) but, as opposed to mice carrying the unmodified transgene, it was completely suppressed in circulating monocytes (n = 3; Figure 5B). These results indicate robust miR-142 activity in circulating TEMs, providing genetic evidence for their hematopoietic lineage and further clarifying that they are distinct from circulating ECs. In agreement with these findings, the majority of circulating GFP+ cells in Tie2-GFP mice expressed the monocyte-specific marker Csf1r/Cd115 (84% ± 5%, mean ± SD; n = 16; Figure 5C). Of note, only 30% plus or minus 6% (n = 13) of the total Cd115+ monocytes were GFP+, indicating that TEMs represent a subset of circulating monocytes (Figure 5D). The GFP+ TEMs were mostly Gr1− (84% ± 5%, n = 10), Cd62l− (74% ± 6%, n = 7), and Cd43+ (80% ± 6%, n = 10), thus expressing a resident monocyte marker profile (Figure 5F).
Circulating TEMs are bona fide hematopoietic cells distinct from circulating ECs and express a resident monocyte phenotype. (A) Analysis of mouse liver by IFS (left) and flow cytometry (right). (Left panels) Expression of GFP (green) in Cd31+ (red) liver sinusoids of Tie2-GFPmir142T mice (n = 2). Scale bar represents 60 μm. Dot plots on the right show liver tissue–derived single-cell suspensions of representative wild-type (n = 1), Tie2-GFP (n = 2), and Tie2-GFPmir142T (n = 3) mice. Note that GFP is expressed robustly in the Cd31+ ECs of both Tie2-GFPmir142T and Tie2-GFP mice. (B) Flow cytometry analysis of blood cells of representative wild-type (n = 1), Tie2-GFP (n = 2), and Tie2-GFPmir142T (n = 3) mice. Cd11b+ myeloid cells are gated (dot plot on the left) and GFP expression analyzed in this hematopoietic subset (dot plots on the right). Note that GFP is expressed in the Cd11b+ SSClow monocytes of Tie2-GFP, but not Tie2-GFPmir142T mice. Ly indicates lymphocytes; Gr, granulocytes; Mo, monocytes. (C-E) Flow cytometry analysis of the blood of Tie2-GFP transgenic mice. (C) Virtually all the Tie2-GFP+ TEMs (green gate in the dot plot on the left; n = 16) are Cd11b+Cd115+7AAD− monocytes (dot plot on the right). (D) Only a fraction (∼ 30%; n = 13) of the Cd11b+Cd115+7AAD− monocytes (dot plot on the left) are Tie2-GFP+ (dot plot in the middle). Monocytes from wild-type mice (n = 5) were used to set the gate for GFP-positive events (dot plot on the right). (E) Dot plots show the expression of surface markers that can distinguish resident from inflammatory monocytes (n = 7-10). The Cd115+GFP+7AAD− cells were gated and the expression of Cd43, Gr1, and L-selectin (Cd62l) analyzed. The majority of Tie2-GFP+Cd115+7AAD− TEMs are Cd43+Gr1−Cd62l− resident monocytes. Representative experiments are shown.
Circulating TEMs are bona fide hematopoietic cells distinct from circulating ECs and express a resident monocyte phenotype. (A) Analysis of mouse liver by IFS (left) and flow cytometry (right). (Left panels) Expression of GFP (green) in Cd31+ (red) liver sinusoids of Tie2-GFPmir142T mice (n = 2). Scale bar represents 60 μm. Dot plots on the right show liver tissue–derived single-cell suspensions of representative wild-type (n = 1), Tie2-GFP (n = 2), and Tie2-GFPmir142T (n = 3) mice. Note that GFP is expressed robustly in the Cd31+ ECs of both Tie2-GFPmir142T and Tie2-GFP mice. (B) Flow cytometry analysis of blood cells of representative wild-type (n = 1), Tie2-GFP (n = 2), and Tie2-GFPmir142T (n = 3) mice. Cd11b+ myeloid cells are gated (dot plot on the left) and GFP expression analyzed in this hematopoietic subset (dot plots on the right). Note that GFP is expressed in the Cd11b+ SSClow monocytes of Tie2-GFP, but not Tie2-GFPmir142T mice. Ly indicates lymphocytes; Gr, granulocytes; Mo, monocytes. (C-E) Flow cytometry analysis of the blood of Tie2-GFP transgenic mice. (C) Virtually all the Tie2-GFP+ TEMs (green gate in the dot plot on the left; n = 16) are Cd11b+Cd115+7AAD− monocytes (dot plot on the right). (D) Only a fraction (∼ 30%; n = 13) of the Cd11b+Cd115+7AAD− monocytes (dot plot on the left) are Tie2-GFP+ (dot plot in the middle). Monocytes from wild-type mice (n = 5) were used to set the gate for GFP-positive events (dot plot on the right). (E) Dot plots show the expression of surface markers that can distinguish resident from inflammatory monocytes (n = 7-10). The Cd115+GFP+7AAD− cells were gated and the expression of Cd43, Gr1, and L-selectin (Cd62l) analyzed. The majority of Tie2-GFP+Cd115+7AAD− TEMs are Cd43+Gr1−Cd62l− resident monocytes. Representative experiments are shown.
Common gene signature of circulating resident monocytes and tumor-infiltrating TEMs
We then analyzed gene expression (39 genes of interest, selected among those analyzed in tumor-derived cells) in Cd115+Gr1− resident and Cd115+Gr1+ inflammatory monocytes obtained from the blood of tumor-free mice (n = 4 independent experiments), and compared the results with those obtained with tumor-derived TEMs and TAMs (Table 2). Remarkably, 22 of 39 genes displayed concordant expression in resident versus inflammatory monocytes and TEMs versus TAMs (exact binomial test; P = .003). These included Arg1, Igf1, Cxcl12, Lyve1, Stab1, Cd163, Edg1, Mrc1 (up-regulated in resident monocytes and TEMs vs inflammatory monocytes and TAMs, respectively), and Sell, Ccr2, Ptgs2/Cox2 (down-regulated). Only one gene, Il12a, displayed discordant expression, whereas the remainder genes were differentially expressed in either resident versus inflammatory monocytes or TEMs versus TAMs. The association of the TEM gene expression signature, which enriches for tissue-remodeling versus proinflammatory genes, with resident monocytes suggests that resident and inflammatory blood monocytes represent 2 functionally distinct subsets that may be differentially committed to generate tumor-infiltrating TEMs and TAMs, respectively.
Genes expressed either differentially or not between resident and inflammatory monocytes
| . | Up-regulated . | . | Down-regulated . | ||||
|---|---|---|---|---|---|---|---|
| Fold more . | ΔCt . | P . | Fold less . | ΔCt . | P . | ||
| Arg1 | 58.7 | 11.5 | < .001 | Chi3l3/Ym1 | −18.0 | 6.4 | < .001 |
| Cxcl12 | 20.5 | 15.1 | < .001 | Sell | −9.5 | 8.1 | < .001 |
| Lyve1 | 14.7 | 12.8 | < .001 | Ccr2 | −5.6 | 7.6 | < .001 |
| Stab1 | 14.1 | 16.7 | < .001 | Ptgs2 | −4.5 | 18.1 | < .001 |
| Igf1 | 13.6 | 15.3 | < .001 | Not differentially expressed | |||
| Cd163 | 5.2 | 13.3 | < .001 | Angpt1 | Cx3cr1 | Itga4 | Tbx21 |
| Edg1 | 4.7 | 9.0 | < .001 | Angpt2 | Egf | Il4 | Tgfb1 |
| Il10 | 4.4 | 16.0 | < .001 | Ccr7 | Fgf2 | Vegfr2 | Tlr4 |
| Mmp9 | 3.1 | 12.3 | < .05 | Csf1R | Vegfr1 | Mmp12 | Tnf |
| Mrc1 | 2.7 | 9.9 | < .05 | Cd80 | Ifng | Nos2 | Tnfrsf18 |
| Il12a | 2.6 | 14.0 | < .05 | Cd86 | Il1b | Nrp1 | |
| Mmp2 | 2.5 | 17.6 | < .05 | ||||
| . | Up-regulated . | . | Down-regulated . | ||||
|---|---|---|---|---|---|---|---|
| Fold more . | ΔCt . | P . | Fold less . | ΔCt . | P . | ||
| Arg1 | 58.7 | 11.5 | < .001 | Chi3l3/Ym1 | −18.0 | 6.4 | < .001 |
| Cxcl12 | 20.5 | 15.1 | < .001 | Sell | −9.5 | 8.1 | < .001 |
| Lyve1 | 14.7 | 12.8 | < .001 | Ccr2 | −5.6 | 7.6 | < .001 |
| Stab1 | 14.1 | 16.7 | < .001 | Ptgs2 | −4.5 | 18.1 | < .001 |
| Igf1 | 13.6 | 15.3 | < .001 | Not differentially expressed | |||
| Cd163 | 5.2 | 13.3 | < .001 | Angpt1 | Cx3cr1 | Itga4 | Tbx21 |
| Edg1 | 4.7 | 9.0 | < .001 | Angpt2 | Egf | Il4 | Tgfb1 |
| Il10 | 4.4 | 16.0 | < .001 | Ccr7 | Fgf2 | Vegfr2 | Tlr4 |
| Mmp9 | 3.1 | 12.3 | < .05 | Csf1R | Vegfr1 | Mmp12 | Tnf |
| Mrc1 | 2.7 | 9.9 | < .05 | Cd80 | Ifng | Nos2 | Tnfrsf18 |
| Il12a | 2.6 | 14.0 | < .05 | Cd86 | Il1b | Nrp1 | |
| Mmp2 | 2.5 | 17.6 | < .05 | ||||
Bold text indicates concordant expression in resident versus inflammatory monocytes and TEMs versus TAMs. Text without bold or underlining indicates significantly up-regulated/down-regulated either in resident versus inflammatory monocytes or TEMs versus TAMs. Underlined text indicates discordant expression in resident versus inflammatory monocytes and TEMs versus TAMs.
A prominent macrophage population expresses typical TEM genes in developing embryos
The physiologic role of TEMs is unknown. We investigated the presence and distribution of TEMs during embryonic/fetal development and analyzed embryos of Tie2-GFP transgenic mice14 from E7.5 to E15.5 (n = 1-4 litters/stage) by IFS and confocal microscopy. Starting from E8.5 and throughout embryonic development, GFP was consistently expressed in Cd31+ ECs forming vascular structures and in scattered hematopoietic cells expressing Cd45, Cd11b, or F4/80 (Figure 6A-B; and data not shown). Anti-Tie2 immunostaining confirmed expression of the Tie2 receptor by the GFP+ cells (supplemental Figure 5A). The GFP+Cd45+ or GFP+F4/80+ cells were particularly abundant in the mesenchyme surrounding the developing neural organs, between somites and in interdigital zones, but were hardly found in the fetal liver, a major hematopoietic organ (Figure 6A-C; and data not shown). In several instances, GFP+F4/80+ cells were found intermixed with developing blood vessels, eg, in the meninges, mesencephalic flexure of the brain, and hyaloid cavity of the eye. A large proportion of the embryonic/fetal Cd11b+ or F4/80+ cells (such as those of the fetal liver) did not express GFP (supplemental Figure 5B-C). Of note, the GFP+ (or Tie2+) F4/80+ cells expressed typical TEM markers, including Mrc1 (Figure 6C), Cd163, and Lyve1 (supplemental Figure 5D-E, Figure 7C).
Characterization of mouse embryonic/fetal macrophages. (A) A representative E13.5 Tie2-GFP transgenic embryo analyzed for GFP (green), Cd45 (red), and Cd31 (blue) expression. The dorsal area of the embryo is shown. GFP is uniformly expressed by Cd31+ blood vessels and by scattered Tie2-GFP+Cd45+Cd31− hematopoietic cells (arrows). Note that several Cd45+Cd31− hematopoietic cells are GFP− (arrowheads). Scale bar represents 60 μm. Similar results were obtained in E8.5 to E15.5 embryos. Results are representative of n = 8 litters (each including 2 or 3 embryos) analyzed. For each embryo, at least 3 sections were analyzed. (B) A representative E14.5 Tie2-GFP transgenic embryo analyzed for GFP (green), Cd31 (red), and F4/80 (blue) expression. The brain of the embryo is shown. GFP is uniformly expressed by Cd31+ blood vessels and by scattered Tie2-GFP+F4/80+Cd31− macrophages (arrows). Note that several F4/80+Cd31− macrophages are GFP− (arrowheads). Scale bar represents 60 μm. Embryos were analyzed as described in panel A. (C) A representative E12.5 Tie2-GFP transgenic embryo analyzed for GFP (green), Mrc1 (red), and F4/80 (blue) expression. The spinal cord of the embryo is shown. GFP is expressed by Mrc1+F4/80+ macrophages that are intermixed with GFP+Mrc1−F4/80− blood vessels in the meninges (inset). Scale bar represents 60 μm. The 2 bottom panels show a high-power magnification of the inset shown above. Note the perivascular GFP+Mrc1+F4/80+ macrophages. Embryos were analyzed as described in panel A.
Characterization of mouse embryonic/fetal macrophages. (A) A representative E13.5 Tie2-GFP transgenic embryo analyzed for GFP (green), Cd45 (red), and Cd31 (blue) expression. The dorsal area of the embryo is shown. GFP is uniformly expressed by Cd31+ blood vessels and by scattered Tie2-GFP+Cd45+Cd31− hematopoietic cells (arrows). Note that several Cd45+Cd31− hematopoietic cells are GFP− (arrowheads). Scale bar represents 60 μm. Similar results were obtained in E8.5 to E15.5 embryos. Results are representative of n = 8 litters (each including 2 or 3 embryos) analyzed. For each embryo, at least 3 sections were analyzed. (B) A representative E14.5 Tie2-GFP transgenic embryo analyzed for GFP (green), Cd31 (red), and F4/80 (blue) expression. The brain of the embryo is shown. GFP is uniformly expressed by Cd31+ blood vessels and by scattered Tie2-GFP+F4/80+Cd31− macrophages (arrows). Note that several F4/80+Cd31− macrophages are GFP− (arrowheads). Scale bar represents 60 μm. Embryos were analyzed as described in panel A. (C) A representative E12.5 Tie2-GFP transgenic embryo analyzed for GFP (green), Mrc1 (red), and F4/80 (blue) expression. The spinal cord of the embryo is shown. GFP is expressed by Mrc1+F4/80+ macrophages that are intermixed with GFP+Mrc1−F4/80− blood vessels in the meninges (inset). Scale bar represents 60 μm. The 2 bottom panels show a high-power magnification of the inset shown above. Note the perivascular GFP+Mrc1+F4/80+ macrophages. Embryos were analyzed as described in panel A.
A distinguishing TEM signature expressed by mouse embryonic/fetal macrophages. (A) Flow cytometry analysis of representative E14.5 Tie2-GFP transgenic embryos (> 8 embryos pooled together). Virtually all GFP+ cells express the Tie2 receptor. Whereas a small proportion of the Cd11b+ cells are GFP+, a substantial proportion of the F4/80+ cells are GFP+. The Cd11b− or F4/80−, GFP+ cells mostly represent ECs. Similar results were obtained in E8.5 to E14.5 embryos. Results are representative of n = 11 litters analyzed. (B) Flow cytometric analysis of representative E12.5 Tie2-GFP transgenic embryos (> 8 embryos pooled together), analyzed as whole embryos (total cells), or embryos without fetal liver (total cells without fetal liver), or fetal liver alone (fetal liver). In whole embryos, the majority of Lyve1+ cells are F4/80+ macrophages. In embryos without fetal liver, virtually all the Lyve1+ cells are F4/80+ macrophages. In the fetal liver, the majority of the Lyve1+ cells are F4/80− cells, probably representing lymphatic ECs. The Lyve1+F4/80+ macrophages (green gate in the dot plot at bottom left) display enhanced GFP expression (histogram at bottom right, open green line) compared with the Lyve1− cells (red gate and filled red line). (C) A representative E12.5 wild-type embryo analyzed for Lyve1 (green), Tie2 receptor (red), and F4/80 (blue) expression. The dorsal area of the embryo is shown. Note the Lyve1+Tie2+F4/80+ macrophages scattered among Tie2+Lyve1−F4/80− blood vessels. Scale bar represents 60 μm. Results are representative of n = 2 litters (each including 3 embryos) analyzed. For each embryo, at least 3 sections were analyzed. (D) Morphology (May-Grünwald-Giemsa staining) of Tie2-GFP−Cd11b+ (left) and Tie2-GFP+Cd11b+ (right) cells sorted from E13.5 Tie2-GFP transgenic embryos depleted of the fetal liver. Scale bar represents 30 μm. Photos are representative of n = 25 photos/sample. (E) A representative E12.5 Tie2-GFPmir142T transgenic embryo analyzed for GFP (green) and Mrc1 (red) expression. The spinal cord of the embryo is shown. Note that the Mrc1+ macrophages do not express GFP in this mouse model, whereas ECs do. The bottom right panel shows a high-power magnification of the inset indicated in the bottom left panel. Similar results were obtained in all analyzed embryos (E12.5-E15.5). Results are representative of n = 4 litters (each including 3 embryos) analyzed. For each embryo, at least 3 sections were analyzed. (F) Flow cytometric analysis of representative E12.5 Tie2-GFP and Tie2-GFPmir142T transgenic embryos (> 8 embryos pooled together). Note that in Tie2-GFPmit142T embryos (n = 8 litters analyzed at different stages) there are no Cd11b+ or F4/80+ cells expressing GFP.
A distinguishing TEM signature expressed by mouse embryonic/fetal macrophages. (A) Flow cytometry analysis of representative E14.5 Tie2-GFP transgenic embryos (> 8 embryos pooled together). Virtually all GFP+ cells express the Tie2 receptor. Whereas a small proportion of the Cd11b+ cells are GFP+, a substantial proportion of the F4/80+ cells are GFP+. The Cd11b− or F4/80−, GFP+ cells mostly represent ECs. Similar results were obtained in E8.5 to E14.5 embryos. Results are representative of n = 11 litters analyzed. (B) Flow cytometric analysis of representative E12.5 Tie2-GFP transgenic embryos (> 8 embryos pooled together), analyzed as whole embryos (total cells), or embryos without fetal liver (total cells without fetal liver), or fetal liver alone (fetal liver). In whole embryos, the majority of Lyve1+ cells are F4/80+ macrophages. In embryos without fetal liver, virtually all the Lyve1+ cells are F4/80+ macrophages. In the fetal liver, the majority of the Lyve1+ cells are F4/80− cells, probably representing lymphatic ECs. The Lyve1+F4/80+ macrophages (green gate in the dot plot at bottom left) display enhanced GFP expression (histogram at bottom right, open green line) compared with the Lyve1− cells (red gate and filled red line). (C) A representative E12.5 wild-type embryo analyzed for Lyve1 (green), Tie2 receptor (red), and F4/80 (blue) expression. The dorsal area of the embryo is shown. Note the Lyve1+Tie2+F4/80+ macrophages scattered among Tie2+Lyve1−F4/80− blood vessels. Scale bar represents 60 μm. Results are representative of n = 2 litters (each including 3 embryos) analyzed. For each embryo, at least 3 sections were analyzed. (D) Morphology (May-Grünwald-Giemsa staining) of Tie2-GFP−Cd11b+ (left) and Tie2-GFP+Cd11b+ (right) cells sorted from E13.5 Tie2-GFP transgenic embryos depleted of the fetal liver. Scale bar represents 30 μm. Photos are representative of n = 25 photos/sample. (E) A representative E12.5 Tie2-GFPmir142T transgenic embryo analyzed for GFP (green) and Mrc1 (red) expression. The spinal cord of the embryo is shown. Note that the Mrc1+ macrophages do not express GFP in this mouse model, whereas ECs do. The bottom right panel shows a high-power magnification of the inset indicated in the bottom left panel. Similar results were obtained in all analyzed embryos (E12.5-E15.5). Results are representative of n = 4 litters (each including 3 embryos) analyzed. For each embryo, at least 3 sections were analyzed. (F) Flow cytometric analysis of representative E12.5 Tie2-GFP and Tie2-GFPmir142T transgenic embryos (> 8 embryos pooled together). Note that in Tie2-GFPmit142T embryos (n = 8 litters analyzed at different stages) there are no Cd11b+ or F4/80+ cells expressing GFP.
We then performed flow cytometry on embryos from E8.5 to E15.5 (n = 1 or 2 samples/stage, by pooling ≥ 8 embryos/stage). As expected, the GFP+ cells, which comprise both ECs and F4/80+ macrophages, uniformly expressed the Tie2 receptor (Figure 7A). Whereas only 10% plus or minus 5% of the total myeloid-lineage cells (as stained by Cd11b) expressed GFP (n = 5 at different stages), a greater proportion of the F4/80+ cells were GFP+ (27% ± 4%; n = 7). Several F4/80+ macrophages coexpressed Lyve1 (Figure 7B); the F4/80+Lyve1+ cells, which were mostly excluded from the fetal liver, were virtually all GFP+ (Figure 7B). We confirmed these findings by IFS of wild-type embryos (n = 3 at E8.5, E10.5, and E13.5), which showed colocalization of Lyve1 and Tie2 receptor by a fraction of the F4/80+ macrophages (Figure 7C).
The early time of appearance, morphology, and distribution of the GFP+F4/80+ cells within the embryo were consistent with those of embryonic/fetal macrophages.19,29,30 FACS sorting (after pooling 8 E13.5 embryos depleted of the fetal liver) and morphologic analysis (Figure 7D) showed that the embryonic/fetal GFP+Cd11b+ cells were a homogeneous population of large phagocytes (> 95% macrophages with rounded nucleus, large cytoplasm filled with phagosomes and vacuoles). Flow cytometry indicated that the GFP+F4/80+ macrophages did not express the hematopoietic progenitor markers c-kit and Sca-1 (n = 2 samples analyzed at E13.5 and E14.5; supplemental Figure 6), corroborating the notion that they represent mature macrophages. FACS-sorted GFP−Cd11b+ cells were also highly enriched in macrophages (> 80%) but contained a measurable fraction of immature myeloid elements.
We did not detect GFP+ macrophages in Tie2-GFPmiR142T embryos by IFS (Figure 7E); of note, these embryos showed uniform and robust vascular GFP marking. In agreement with the IFS data, flow cytometry analyses showed that the Cd11b+ or F4/80+ myeloid cells of Tie2-GFPmiR142T embryos were uniformly GFP− (Figure 7F), whereas the Cd11b−GFP+ (or F4/80−GFP+) ECs from either Tie2-GFPmiR142T or Tie2-GFP transgenic mice displayed similar GFP mean fluorescence activity. These data indicate robust activity of miR-142 in embryonic/fetal Tie2+F4/80+ cells, further confirming that they represent bona fide hematopoietic-lineage cells.
Finally, we analyzed gene expression (25 selected genes, selected among those analyzed in tumor-derived cells) in GFP+Cd11b+ and GFP−Cd11b+ macrophages obtained from E13.5 Tie2-GFP transgenic embryos (n = 2 independent experiments, each by pooling 15 E13.5 embryos depleted of the fetal liver) and compared the results with those obtained with tumor-derived TEMs and TAMs (Table 3). Fourteen of 25 genes displayed concordant expression in GFP+ versus GFP− embryonic/fetal macrophages and tumor-derived TEMs versus TAMs (exact binomial test; P = .02). These included Igf1, Lyve1, Arg1, Mrc1, Cd163, and Stab1 (up-regulated in GFP+ embryonic/fetal macrophages and TEMs versus GFP− embryonic/fetal macrophages and TAMs, respectively) and Sell (down-regulated). Remarkably, when we extended the analysis to include gene expression data obtained from circulating monocytes (Table 2), 12 of 24 analyzed genes were found to be coordinately expressed in GFP+ embryonic/fetal macrophages, resident monocytes, and tumor-derived TEMs versus GFP− embryonic/fetal macrophages, inflammatory monocytes, and TAMs (exact binomial test; P < .001).
Genes expressed either differentially or not between Tie2-GFP+ and Tie2-GFP−, Cd11b+ embryonic/fetal macrophages
| . | Up-regulated . | . | Down-regulated . | ||||
|---|---|---|---|---|---|---|---|
| Fold more . | ΔCt . | P . | Fold less . | ΔCt . | P . | ||
| Mmp12 | 39.3 | 2.3 | < .001 | Sell | −35.1 | 8.8 | < .001 |
| Igf1 | 25.0 | 7.4 | < .001 | Chi3l3/Ym1 | −15.2 | 6.1 | < .001 |
| Lyve1 | 21.0 | 3.7 | < .001 | Mmp9 | −8.3 | 6.6 | < .001 |
| Arg1 | 11.6 | 4.9 | < .001 | Egf | −7.3 | 12.3 | < .01 |
| Mrc1 | 7.2 | 2.6 | < .01 | Angpt1 | −4.4 | 11.5 | < .05 |
| Cd163 | 6.4 | 7.0 | < .01 | Not differentially expressed | |||
| Stab1 | 3.7 | 1.0 | < .05 | Angpt2 | Edg1 | Il12a | Nrp1 |
| Cx3cr1 | 3.5 | 1.0 | < .05 | Csf1r | Fgf2 | Vegfr2 | Tfrc |
| Cxcl12 | Vegfr1 | Mmp2 | Tlr4 | ||||
| . | Up-regulated . | . | Down-regulated . | ||||
|---|---|---|---|---|---|---|---|
| Fold more . | ΔCt . | P . | Fold less . | ΔCt . | P . | ||
| Mmp12 | 39.3 | 2.3 | < .001 | Sell | −35.1 | 8.8 | < .001 |
| Igf1 | 25.0 | 7.4 | < .001 | Chi3l3/Ym1 | −15.2 | 6.1 | < .001 |
| Lyve1 | 21.0 | 3.7 | < .001 | Mmp9 | −8.3 | 6.6 | < .001 |
| Arg1 | 11.6 | 4.9 | < .001 | Egf | −7.3 | 12.3 | < .01 |
| Mrc1 | 7.2 | 2.6 | < .01 | Angpt1 | −4.4 | 11.5 | < .05 |
| Cd163 | 6.4 | 7.0 | < .01 | Not differentially expressed | |||
| Stab1 | 3.7 | 1.0 | < .05 | Angpt2 | Edg1 | Il12a | Nrp1 |
| Cx3cr1 | 3.5 | 1.0 | < .05 | Csf1r | Fgf2 | Vegfr2 | Tfrc |
| Cxcl12 | Vegfr1 | Mmp2 | Tlr4 | ||||
Bold text indicates concordant expression in embryo TEMs versus embryonic macrophages and tumor TEMs versus TAMs. Text without bold or underlining indicates significantly up-regulated/down-regulated either in embryo TEMs versus embryonic macrophages or tumor TEMs versus TAMs. Underlined text indicates discordant expression in embryo TEMs versus embryonic macrophages and tumor TEMs versus TAMs.
Discussion
Our gene expression data demonstrate that TEMs are distinct from endothelial-lineage cells and represent a subset of tumor-infiltrating macrophages expressing a distinguishing gene signature, which is consistent with enhanced proangiogenic and tissue-remodeling activity and lower proinflammatory activity. The “TEM gene signature” also distinguishes resident from inflammatory monocytes in the peripheral blood and identifies a population of embryonic/fetal macrophages that appear early during mouse development (starting from E8.5) and are preferentially associated with areas of active tissue remodeling and angiogenesis.
Several studies have reported that mononuclear cells expressing a reporter gene from Tie2 transcription regulatory elements are recruited to tumors and ischemic tissues.13,15,31-34 Based on the assumption that Tie2 expression is specific of ECs, these Tie2+ cells were often interpreted as EPCs, even if they expressed hematopoietic/myeloid markers33 and were often found outside of the vessel wall.31,33 By implementing stringent combinations of gene marking and imaging approaches, we previously showed that these Tie2+ mononuclear cells indeed represent myeloid cells.14,15 Here, we conclusively demonstrate that Tie2-expressing cells recruited to tumors do not belong to the EC lineage and are bona fide monocytes/macrophages. By comparing tumor-derived TEMs, TAMs, and ECs, we show that the gene expression profile of TEMs is similar to that of TAMs but clearly distinct from that of ECs. Further data support the non-EC nature of TEMs. Indeed, TEMs (1) robustly express hematopoietic-specific markers (eg, Cd45, F4/80, and miR-142, the latter shown using a microRNA-regulated reporter transgenic mouse line), (2) express low to negligible levels of EC-specific genes (eg, Vegfr2 and VE-cadherin/Cdh5), and (3) do not incorporate in the tumor endothelium and often reside in perivascular spaces. These findings strongly support the notion that Tie2-expressing cells repeatedly observed in tumors13-15,27,34,35 are not EPCs. Whether true EPCs contribute to tumor angiogenesis is still a matter of debate, with recent reports showing a contribution limited to selected experimental conditions, such as tumors treated by vascular-disrupting agents or chemotherapy.5,13,36,37
Although TEMs do not physically incorporate in the tumor endothelium, they seem required for tumor angiogenesis.15 Within the panel of interrogated genes, many genes previously implicated in tumor angiogenesis were among the most differentially expressed between TEMs and TAMs. One such gene is Lyve1 (14-fold up), which encodes for the hyaluronan receptor-1 expressed on lymphatic ECs and subsets of macrophages. Because TEMs also express Stab1 (stabilin-1, a hyaluronan receptor; 6.4-fold up) and Cd163 (hemoglobin/haptoglobin scavenger receptor; 15.8-fold up), it is probable that the previously described Lyve1+Stab1+Cd163+ macrophages observed in tumors, wounds, and remodeling adipose tissue38,39 indeed represent TEMs. Tlr4, a Toll-like receptor implicated in tumorigenesis,40 is also preferentially expressed by TEMs (3.5-fold up). Whereas Lyve1 binds hyaluronic acid (HA), Tlr4 binds to its low-molecular-weight fragmentation products. The preferential expression of Lyve1, Stab1, and Tlr4 by TEMs among tumor macrophages suggests a role of TEM-HA interactions in tumor angiogenesis. Indeed, HA fragments stimulate MMP production, cell migration, and angiogenesis in tumors,41 and up-regulate Irak-M expression by monocytes, deactivating them and down-regulating their expression of the antiangiogenic molecules Tnfa and Il12a.42 Interestingly, Tlr4 expression by macrophages has been found to mediate remodeling of the lung microenvironment and favor subsequent colonization by tumor cells.43 The expression of Nrp1 (Neuropilin-1, a Vegf coreceptor; 4.6-fold up) by TEMs might enhance angiogenesis by clustering Vegf in contact with Vegfr2+ ECs, as previously described.44 Efnb2 (Ephrin-B2; 4.2-fold up) encodes for a transmembrane ligand of Ephrin receptors expressed on ECs; the bidirectional signaling between ephrin-B2 and Ephrin receptors modulates angiogenesis and the development of arteries and veins.45 Semaphorin 6D (Sema6d; 5.2-fold up), a transmembrane protein that binds to membrane-bound plexinB1, may also have proangiogenic activity in tumors, as it activates Vegfr2-mediated signal transduction.46 The enhanced expression of Nrp1, Efnb2, and Sema6d by TEMs may suggest activation of ECs by cell-to-cell contacts. Another gene up-regulated in TEMs is Cxcl12 (4.4-fold up), encoding for the stromal cell–derived factor-1 (Sdf1); Cxcl12 released by TEMs may promote angiogenesis by attracting Cxcr4+ ECs and other proangiogenic cells in the tumor microenvironment.47 Angpt1 (angiopoietin-1, a Tie2 ligand; 2.6-fold up) expressed by TEMs may also promote tumor angiogenesis, as Angpt1-expressing hematopoietic cells stimulate EC sprouting in embryonic tissues.44 TEMs display up-regulated expression of Edg1 (sphingosine-1-phosphate receptor; 3.0-fold up). Edg1 null embryos are hemorrhagic and die in utero, a phenotype associated with impaired vascular maturation and defective recruitment of perivascular cells to angiogenic blood vessels.48 Moreover, sphingosine-1-phosphate induces macrophages to acquire an anti-inflammatory phenotype49 (high Arg1, low Nos2 activity). Vegfa, which is among the most expressed genes in TEMs, was however down-regulated in TEMs versus TAMs (2.9-fold down), suggesting that TEMs are exposed to a less hypoxic microenvironment in tumors, consistent with their preferential localization in peritumoral areas and their exclusion from necrotic regions. Interestingly, the ELR-negative CXC chemokines Cxcl10 and Cxcl11 were down-regulated in TEMs versus TAMs (2.8- and 2.7-fold down, respectively). These chemokines are transcribed on Irf3/Stat1 activation and are potent inhibitors of angiogenesis.25,50 Overall, the enhanced expression of several proangiogenic molecules by TEMs, together with the down-regulation of potent antiangiogeneic mediators (also including Il12a, 3.6-fold down; Il12b, 2.5-fold down; Tnfa, 3.1-fold down), may account for their nonredundant proangiogenic activity in tumors.15
Both mouse and human monocytes can be grouped into functional subsets.8,9,51-53 We found that circulating TEMs belong to the Gr1−Cd62l−Cd43+ resident monocyte subset. In addition to our previous data showing that TEMs are proangiogenic,14,15 a recent report indicated that resident monocytes promote tissue angiogenesis in the postischemic myocardium.54 Together, these data may suggest a developmental and functional relationship between circulating resident monocytes and tumor-infiltrating TEMs. Our new findings further show that resident monocytes isolated from the blood of tumor-free mice and tumor-derived TEMs display a coordinated expression profile, which points to a commitment of TEM phenotype/function in the peripheral blood. Arg1, Cxcl12, Lyve1, Igf1, Stab1, Cd163, Mrc1, and Edg1, all significantly up-regulated in tumor-derived TEMs versus TAMs, were markedly up-regulated in resident versus inflammatory monocytes. Expression of Il12a (an antiangiogenic molecule) was down-regulated in TEMs versus TAMs but unexpectedly up-regulated in resident versus inflammatory monocytes. However, 2 recent reports have shown that both mouse resident and human blood monocytes rapidly down-regulate Il12 expression on their extravasation in vivo53 or when exposed in vitro to experimental conditions mimicking the hypoxic tumor microenvironment.17 The coordinated expression profiles of resident monocytes/TEMs and inflammatory monocytes/TAMs suggest that monocyte heterogeneity in the peripheral blood may reflect the existence of precursor populations that are committed to distinct, nonredundant functions in tumors and growing/regenerating tissues. Yet, the developmental relationship between the 2 main monocyte subsets remains to be clarified. It has been proposed that resident monocytes derive from Gr1+ inflammatory monocytes,11,55 implying that the 2 phenotypes may represent distinct developmental stages along the monocyte lineage. Further studies will clarify whether TEMs represent a distinct resident monocyte subpopulation.
Although it has been predicted that TAMs comprise distinct functional subsets,7 very little is known of the actual functional heterogeneity of TAMs in tumors. Live imaging analysis12 of monocyte/macrophage subpopulations expressing the newly identified cell-surface markers (eg, Mrc1, Lyve1, Tlr4) may better clarify macrophage heterogeneity and dynamics in tumors. It has been established that tumors induce macrophages to acquire an immunosuppressive function.23 Moreover, tumor-derived factors promote the expansion of MDSCs, a heterogeneous myeloid population that can suppress T-cell function,6 in BM, blood, and lymphoid organs. Inhibition of effective antitumor T cell–dependent immunity by TAMs and MDSCs may occur in several ways, including production of indoleamine dioxygenase and nitric oxide, inhibition of DC maturation, and attraction of T regulatory cells to the tumor site.6 Interestingly, among the down-regulated genes in TEMs versus TAMs were Ccr7 (3.8-fold down), Cd80 (2.8-fold down), Tbx21 (T-box 21 transcription factor; 2.1-fold down), and H2-Ea (class II major histocompatibility complex, antigen E alpha; 3.0-fold down), which are all expressed by antigen-presenting cells. Conversely, Il4ra, which encodes for the type I interleukin-4 receptor previously shown to be expressed by immunosuppressive MDSCs,56 was up-regulated in TEMs (2.3-fold up). Because tumor TEMs do not express Cd11c and express low levels of mature antigen-presenting cell markers (compared with TAMs), it can be envisaged that they represent deactivated or immature cells. Together, our findings suggest that TEMs have little ability, if any, to commence immune responses in tumors, and may well represent an innate immunosuppressive cell type.
Both mRNA and protein expression data indicated that a substantial fraction of the F4/80+ embryonic/fetal macrophages express a TEM phenotype (Tie2+Lyve1+Mrc1+Cd163+). This finding points to a functional similarity between embryonic/fetal macrophages, which have unique developmental and biologic features among phagocytes,19,20,29 and adult proangiogenic TEMs. Whereas TEMs represent a minor proportion of the tumor-infiltrating F4/80+ cells and are mostly restricted to viable, angiogenic tumor areas,15 TEM-like cells appear to be a major macrophage subset in embryos. Of note, Ojalvo et al recently reported that the gene expression profile of TAMs significantly overlaps with enriched transcripts in fetal macrophages.30 By further narrowing down the gene expression analysis to tumor-, blood-, and embryo-derived monocyte/macrophage subpopulations (ie, by comparing the Tie2(GFP)+ with Tie2(GFP)− fraction of each subpopulation), we identified stringent concordance in gene expression/regulation among the Tie2+ myeloid subpopulations analyzed. The identification of “embryonic/fetal TEMs” extends their role to a physiologic process characterized by dramatic rates of cell turnover, tissue morphogenesis, and remodeling, which occur in the absence of exogenous inflammatory stimuli. mRNA data indicated that Tie2-GFP+ embryonic/fetal macrophages display greatly enhanced expression of Mmp12 (metalloelastase; 39-fold up vs GFP− counterpart), suggesting the importance of this protease in physiologic tissue remodeling57 by embryonic/fetal TEMs.
It is tempting to speculate that TEMs play primary functions in developing tissues, where they might execute proangiogenic and proinvasive programs that are coopted by tumors.15 Taken together, our findings suggest that TEMs represent a circulating reservoir of monocytes actively recruited to extravascular tissues by noninflammatory signals produced not only by tumors, but also by developing or regenerating tissues. Future studies will be devoted to the identification of the signals that govern the lineage determination of TEMs and their recruitment to sites of active tissue morphogenesis.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Roberta Mazzieri, Chiara Porta, and Silvia Nucera for help with some experiments; Alessio Palini for cell sorting; Elisa Allievi (CFCM facility, San Raffaele) for mouse transgenesis; Maurilio Ponzoni (Department of Pathology, San Raffaele) for morphologic analysis of cytospins; Sergij Goerdt (Mannheim, Germany) for providing the anti–stabilin-1 antibody; Elisabetta Dejana (FIRC Institute of Molecular Oncology, Milan, Italy); Claire Lewis (University of Sheffield, United Kingdom) for helpful discussions; and Luca Libelli for excellent secretarial assistance.
This work was supported by the Associazione Italiana per la Ricerca sul Cancro, European Union (Tumor-Host Genomics), and the Italian Ministry of Health (Challenge in Oncology) (L.N.).
Authorship
Contribution: F.P. designed research, performed research, analyzed data, and drafted the manuscript; M.A.V. designed research, performed research, and analyzed data; D.B. performed research; A.N. and C.D.S. developed statistical models and analyzed gene expression data; D.M. managed mouse colonies; A.S. provided vital reagents and analyzed data; and L.N. and M.D.P. conceived and coordinated research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Luigi Naldini, Angiogenesis and Tumor Targeting Research Unit, San Raffaele Institute, Olgettina, 58, 20132 Milan, Italy; e-mail: naldini.luigi@hsr.it; or Michele De Palma, Angiogenesis and Tumor Targeting Research Unit, San Raffaele Institute, Olgettina, 58, 20132 Milan, Italy; e-mail: depalma.michele@hsr.it.
References
Author notes
*F.P. and M.A.V. contributed equally to this study.
†L.N. and M.D.P. share senior authorship.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal