Abstract
Granzyme A (GzmA) in killer cells induces caspase-independent programmed cell death. In this study, we show that GzmA cleaves the DNA damage sensor poly(adenosine 5′-diphosphate-ribose) polymerase-1 (PARP-1) after Lys498 in its automodification domain, separating the DNA binding domain from the catalytic domain, which interferes with repair of GzmA-induced DNA damage and enhances susceptibility to GzmA-mediated death. Overexpressing K498A PARP-1 reduces GzmA-mediated death and drives dying cells to necrosis rather than apoptosis. Conversely, inhibiting or genetically disrupting PARP-1 enhances cell vulnerability. The N-terminal GzmA cleavage fragment of PARP-1 acts as a PARP-1 dominant negative, binding to DNA and blocking DNA repair. Disrupting PARP-1, which is also a caspase target, is therefore required for efficient apoptosis by both caspase-independent and caspase-dependent pathways.
Introduction
Cytotoxic T lymphocytes (CTLs) and natural killer cells defend against intracellular pathogens and tumors by releasing proapoptotic mediators from cytotoxic granules to trigger apoptosis. Granzymes A (GzmA) and B (GzmB), the most abundant granule serine proteases, independently induce death when delivered into the target cell by perforin (PFN).1 GzmB activates apoptosis by cleaving caspases and some key caspase pathway substrates. GzmA, a tryptase, activates caspase-independent death morphologically identical to apoptosis, characterized by single-stranded DNA damage, mitochondrial dysfunction, and loss of cell membrane integrity. GzmA does not activate caspases; caspase-resistant cells overexpressing bcl-2 are sensitive to GzmA.2,3 GzmA enters the mitochondrion to cleave NDUFS3 in mitochondrial complex I to trigger increased reactive oxygen species and dissipation of transmembrane potential, but does not permeabilize the mitochondrial outer membrane to release cytochrome c.4-6 Reactive oxygen species triggers nuclear translocation of the SET complex, a macromolecular complex, which is key to GzmA-mediated DNA damage.7-9 GzmA cleaves SET, which inhibits the endonuclease NM23-H1 in the SET complex. After SET is disrupted, NM23-H1 makes a single-stranded DNA nick, which is extended by Trex1, a SET complex exonuclease
GzmA and GzmB cleave distinct sets of substrates.1 Lamin B is the only common known substrate of GzmA, GzmB, and the caspases.10 GzmA binds its substrates as a dimer through an extended exosite, which results in exquisite specificity.11,12 When GzmA is incubated with cellular lysates, very few proteins are digested.6,13 The list of validated physiologically relevant intracellular substrates of GzmA is short (although most likely incomplete): SET, Ape1, high mobility group protein B2 (HMGB2), lamins, histone H1, some core histones, Ku70, and NDUFS3.6,8,10,14-17 The known substrates, except for lamins and NDUFS3, are DNA-binding proteins that function in nucleosome assembly (histones, the histone chaperone SET) or DNA repair (Ape1, Ku70, and HMGB2, which recognizes distorted DNA structures). Three GzmA substrates (SET, HMGB2, and Ape1) are in the SET complex.8,14,16,18
Cell death stimuli induce repair as they activate cell death. The relative balance between repair and death determines the target cell fate. Ape1 and Ku70 cleavage interfere with DNA repair and enhance GzmA-mediated cell death.17,18 Target cells that express GzmA-resistant Ape1 are partly resistant to GzmA. Similarly silencing Ape1 increases vulnerability to GzmA. Ku70 inactivation also facilitates GzmA-induced DNA damage and cell death.17
Because GzmA disrupts 2 DNA repair pathways (base excision repair via Ape1 and double-strand break repair via Ku70), we investigated whether GzmA might target poly(adenosine 5′-diphosphate [ADP]-ribose) polymerase-1 (PARP-1), a sensor of single-stranded and doubled-stranded DNA damage. PARP-1, a caspase substrate,19-21 belongs to a family of 18 mammalian enzymes that catalyze the synthesis from nicotinamide adenine dinucleotide (NAD+) of poly(ADP) ribose (PAR), a branched polymer of repeated ADP-ribose subunits linked by glycosidic bonds, which is covalently added to target proteins.22,23 PARP-1 is responsible for most poly(ADP) ribosylase activity in mammalian cells. PARP-1 adds PAR to a variety of nuclear proteins, notably core and linker histones, high mobility group (HMG) proteins, p53, and XRCC-1, and (most importantly) automodifies itself.22,23 In fact, more than 90% of cellular PAR is attached to PARP-1. Adding a negatively charged polymer to these basic proteins inhibits their interactions with nucleic acids.24 Binding to damaged DNA activates PARP-1, which automodifies itself and enhances recruitment of DNA repair factors to DNA damage sites. PARP-1 and Ku complex may compete for binding to double-strand breaks (DSB).25
PARP-1 has an N-terminal DNA binding domain (DBD) containing 2 Zn finger motifs, a bipartite nuclear localization signal, an internal automodification domain containing a BRCT motif for protein-protein interactions, and a C-terminal catalytic domain22-24 (Figure 1A). PARP-1 binds to a variety of DNA structures, including single-stranded breaks (SSB), DSB, crossovers, cruciforms, and supercoils. Because it recognizes many altered DNA structures, PARP-1 participates in multiple DNA repair pathways, including SSB repair, double-strand break repair, base excision repair, and repair at stalled replication forks, most likely by increasing accessibility to damage sites and modifying recruited repair factors.22,23,26 PARP-1 enzymatic activity is low in unstressed cells, but dramatically increases with DNA damage.23
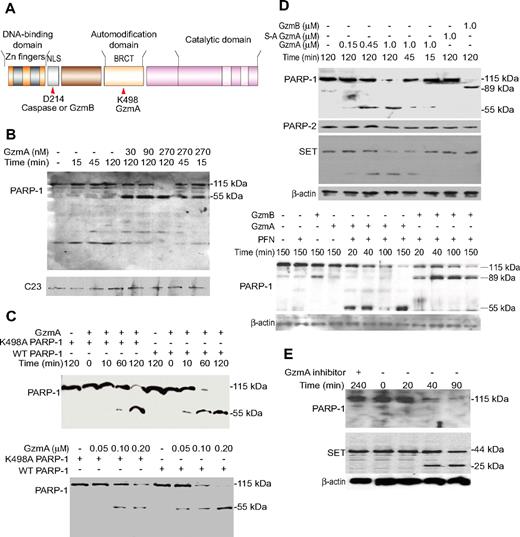
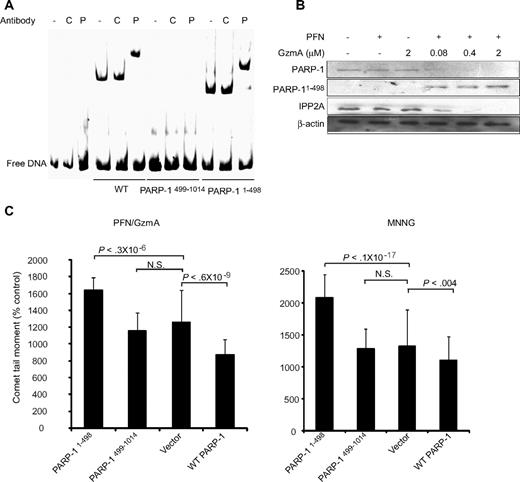
PARP-1 is a GzmA substrate. (A) Schematic of functional domains of PARP-1, indicating the caspase and GzmB cleavage site within the nuclear localization signal and GzmA cleavage site within the automodification domain (site of PARylation). (B) GzmA treatment of isolated nuclei produces a 55-kDa C-terminal fragment of PARP-1. HeLa nuclei were incubated with GzmA at the indicated concentrations for the indicated times and analyzed by immunoblot probed with PARP-1 C-terminal antiserum or for C23, a loading control. (C) When recombinant PARP-1 was incubated with GzmA, a 55-kDa C-terminal cleavage product is again seen by immunoblot. N-terminal sequencing of the 55-kDa fragment identified the cleavage site after Lys498. K498A PARP-1 is less susceptible to cleavage, but at longer times (top) or higher GzmA concentrations (bottom) it is also cleaved to a similarly sized fragment. (D) Treatment of K562 cells with GzmA and PFN leads to PARP-1, but not PARP-2, degradation. β-Actin was probed as a loading control, and the GzmA substrate SET was probed as a positive control. GzmB cleaves PARP-1 to generate an 89-kDa C-terminal fragment; S-AGzmA does not cleave either PARP (top). GzmA and GzmB (0.5 μM) cleave PARP-1 with similar kinetics (bottom). (E) PARP-1 is degraded within 40 minutes of CTL attack, with similar kinetics as SET. The serine protease inhibitor diisocoumarin (DCI) blocks PARP-1 and SET cleavage. (B-E) These are immunoblots; the PARP-1 blots are probed with an antibody that recognizes the C-terminal region of PARP-1.
PARP-1 is a GzmA substrate. (A) Schematic of functional domains of PARP-1, indicating the caspase and GzmB cleavage site within the nuclear localization signal and GzmA cleavage site within the automodification domain (site of PARylation). (B) GzmA treatment of isolated nuclei produces a 55-kDa C-terminal fragment of PARP-1. HeLa nuclei were incubated with GzmA at the indicated concentrations for the indicated times and analyzed by immunoblot probed with PARP-1 C-terminal antiserum or for C23, a loading control. (C) When recombinant PARP-1 was incubated with GzmA, a 55-kDa C-terminal cleavage product is again seen by immunoblot. N-terminal sequencing of the 55-kDa fragment identified the cleavage site after Lys498. K498A PARP-1 is less susceptible to cleavage, but at longer times (top) or higher GzmA concentrations (bottom) it is also cleaved to a similarly sized fragment. (D) Treatment of K562 cells with GzmA and PFN leads to PARP-1, but not PARP-2, degradation. β-Actin was probed as a loading control, and the GzmA substrate SET was probed as a positive control. GzmB cleaves PARP-1 to generate an 89-kDa C-terminal fragment; S-AGzmA does not cleave either PARP (top). GzmA and GzmB (0.5 μM) cleave PARP-1 with similar kinetics (bottom). (E) PARP-1 is degraded within 40 minutes of CTL attack, with similar kinetics as SET. The serine protease inhibitor diisocoumarin (DCI) blocks PARP-1 and SET cleavage. (B-E) These are immunoblots; the PARP-1 blots are probed with an antibody that recognizes the C-terminal region of PARP-1.
PARP-1 is cleaved by GzmB and caspase-3 after DEVD214 within its DBD19-21 (Figure 1A). PARP-1 is also implicated in caspase-independent death. During necrosis, lysosomal cathepsins cleave PARP-1 to a 55-kDa C-terminal fragment.27 When DNA damage is not overwhelming, PARP-1 participates in detecting and repairing DNA; when DNA damage is extensive, PARP-1 activation can deplete cellular NAD+ and interfere with cellular adenosine triphosphate (ATP) pools, leading to necrotic death.28-31 Extensive PARP-1 activation also triggers mitochondrial release of apoptosis-inducing factor to activate apoptosis.32 Therefore, eliminating PARP-1 may be essential to maintain cellular ATP to carry out ATP-dependent programmed cell death. What happens to PARP-1 helps decide whether a cell survives or dies by necrosis or apoptosis. The role of PARP-1 in regulating life or death depends on cellular context and is influenced by the proliferation rate, dependence on glycolysis, and availability of energy substrates.
In this study, we show that PARP-1 is cleaved and inactivated by GzmA after Lys498 within the PARP-1 automodification domain. The N-terminal GzmA cleavage product of PARP-1 is not only enzymatically inactive, but also interferes with DNA repair. Cells expressing GzmA-resistant mutant PARP-1 are less susceptible to GzmA-mediated cell death, whereas cells expressing the N-terminal GzmA cleavage fragment are more sensitive.
Methods
Recombinant and purified proteins
Recombinant human GzmA, S-AGzmA (inactive Ser184 to Ala mutant human GzmA), and mouse GzmB were prepared, as described.14,33 All experiments using cells from knockout mice were performed with the approval of the Animal Care and Use Committee of the Immune Disease Institute and Harvard Medical School. Human PARP-1 was expressed in Escherichia coli as a His6 tag fusion protein from pET5a-PARP-1, a gift from P. Kannan (Case University), and purified, as described.34,35 The PARP-1 cleavage site residue Lys498 to Ala was mutated by polymerase chain reaction (PCR) mutagenesis. pET vectors expressing PARP-11-498 and PARP-1499-1014 were constructed by PCR. The SET complex was purified from K562 cell lysates, as described.7
Gzm and PFN treatment
Cells were treated with Gzms and/or PFN, as previously described.2
PARP function assay
The PARP assay was from Ariumi et al.36
DNA damage assays
The Klenow fragment of DNA polymerase (New England Biolabs) was used to label DNA breaks as described.2 The comet assay was performed as described.37
Additional methods are available as supplemental materials on the Blood website (see the Supplemental Materials link at the top of the online article).
Results
GzmA treatment of isolated nuclei induces PARP-1 cleavage
To determine whether PARP-1 is a GzmA substrate, we treated isolated HeLa nuclei with GzmA and analyzed PARP-1 expression by immunoblot. GzmA treatment led to PARP-1 degradation in a dose- and time-dependent fashion, yielding a 55-kDa C-terminal product (Figure 1B). PARP-1 fragmentation was observed at GzmA concentrations as low as 30 nM.
GzmA cuts recombinant PARP-1 after Lys498
To test whether PARP-1 is a direct GzmA substrate and identify the cleavage site, we treated recombinant PARP-1 with GzmA. Recombinant PARP-1 was also cleaved by GzmA, but not by S-AGzmA, to produce a 55-kDa C-terminal fragment (Figure 1C; supplemental Figure 1). Another band migrating just below the C-terminal band (supplemental Figure 1) is presumed to be the N-terminal fragment. The C-terminal fragment was analyzed by N-terminal sequencing, and the cleavage site was identified as Lys498 in APRGK498SGAALSKK within the automodification domain, which is consistent with the GzmA tryptase activity. Cleavage after Lys498 should result in 2 similarly sized fragments (498 and 516 amino acids), in agreement with the nearly equal migration pattern on sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis gels of the fragments. Cleavage at this site separates the N-terminal DNA binding domains from the C-terminal catalytic domain, which need to function together to PARylate DNA-bound substrates.38 Therefore, GzmA cleavage may disrupt PARP-1 function.
To validate the cleavage site, we expressed a mutant form of PARP-1 in which the GzmA cleavage site Lys498 was mutated to Ala. GzmA cleaved K498A PARP-1, but less efficiently (Figure 1C). A longer incubation time or a higher GzmA concentration was required. The apparent molecular weight of the mutant PARP-1 cleavage fragment was indistinguishable from that of the wild-type (WT) protein, suggesting that GzmA cuts K498A PARP-1 at another nearby basic site, of which there are several (R496, K505, K506). Because dimeric GzmA recognizes an extended exosite rather than a well-defined catalytic pocket, this is not the first example of GzmA cleaving a substrate with reduced efficiency at an alternate nearby site when the preferred site has been mutated. Cleavage site mutants of SET, Ku70, and NDUFS3 are also inefficient GzmA substrates.6,17
PARP-1 is degraded in cells treated with PFN and GzmA
To verify that PARP-1 is cleaved in intact cells, we treated K562 cells with Gzm and PFN (Figure 1D). As expected, PFN and GzmB produced an 89-kDa C-terminal fragment, generated by cleavage after Asp214. Treating cells with GzmA alone (data not shown) or PFN alone did not affect PARP-1 (Figure 1D top). Within 15 minutes of adding PFN plus 1 μM GzmA, a PARP-1 cleavage fragment was detected. By 45 minutes there was significant loss of PARP-1. At a lower dose of GzmA (150 nM), PARP-1 was reduced after 2 hours. The same size C-terminal cleavage fragment of PARP-1 was detected in treated cells, as was seen in nuclei or in vitro. The GzmA concentration and time required to cleave PARP-1 were comparable with that required to cleave SET and to induce cell death and DNA damage (data not shown).2 Furthermore, PARP-1 cleavage kinetics by GzmA and GzmB were similar (Figure 1D bottom). Cleavage was specific. Homologous PARP-2 was not a substrate. Moreover, PFN delivery of enzymatically inactive S-AGzmA did not alter PARP-1.
PARP-1 is degraded during CTL attack
To verify that PARP-1 is degraded during CTL lysis of target cells, we analyzed PARP-1 protein in cell lysates after coincubating killer cells, generated from GzmB−/− P14 T-cell receptor transgenic mouse splenocytes, with concanavalin A (ConA)–bearing EL4 target cells (Figure 1E). Within 40 minutes of initiating CTL attack, the PARP-1 signal was reduced, and after 90 minutes it was barely detectable. However, no PARP-1 degradation product was seen, although the SET cleavage product was detected. This suggests that the C-terminal cleavage product is labile within cells, a feature of other GzmA substrates, including HMGB2, histone H1, and NDUFS3.6,15,16 When CTLs were preincubated with the serine protease inhibitor diisocoumarin at a concentration that did not alter viability, PARP-1 was protected, providing further evidence that PARP-1 is a direct physiologic GzmA substrate.
PARP-1 binds to S-AGzmA in a DNA-dependent interaction
To determine whether PARP-1 directly associates with GzmA within cells, HeLa cell lysates were incubated with S-AGzmA. The goal was to stabilize the transient interaction expected of a protease with its substrate by looking for a more stable interaction with the inactive enzyme. PARP-1 antibodies specifically and efficiently coprecipitated S-AGzmA (Figure 2A). However, recombinant PARP-1 did not coprecipitate with S-AGzmA, unless DNA was added (Figure 2B). Although DNA was required for binding, binding persisted when the bead-bound samples, precipitated from either cell lysates (Figure 2A) or recombinant proteins (Figure 2B), were treated with DNase I before solubilization in SDS–polyacrylamide gel electrophoresis sample buffer. We interpret these findings to mean that S-AGzmA recognizes PARP-1, but efficiently only when complexed with DNA. DNA bound to PARP-1 and S-AGzmA was most likely protected from digestion when DNase was added after the proteins had associated. Because GzmA cleaved recombinant PARP-1 in the absence of added DNA (Figure 1C), but coprecipitation experiments required added DNA (Figure 2B), it is likely that GzmA binds to free PARP-1, but that the interaction is enhanced on DNA-complexed PARP-1.
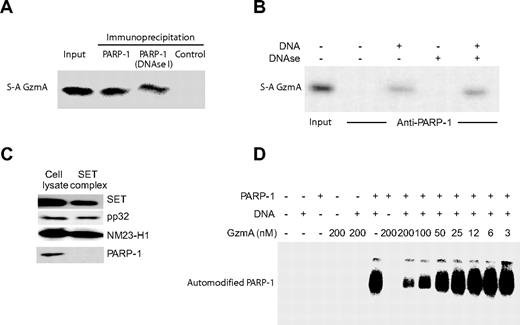
GzmA binds to PARP-1. (A) PARP-1 in cell lysates binds to S-AGzmA. Precleared HeLa cell lysates were incubated with S-AGzmA and antisera to PARP-1 or control antisera, and immune complexes were captured on protein G beads. GzmA binding was assayed by immunoblot probed with His-tag antibody. DNase treatment after PARP-1/GzmA incubation does not disrupt the association of S-AGzmA and PARP-1. (B) Recombinant PARP-1 binds S-AGzmA in vitro only in the presence of DNA. Recombinant PARP-1 attached to PARP-1 antibody–conjugated beads was incubated with S-AGzmA for 3 hours on ice in the presence or absence of 100-bp PCR product DNA. S-AGzmA was pulled down with PARP-1 only in the presence of DNA. However, if DNase I was added before extraction in SDS sample buffer, binding was not disrupted. (C) PARP-1 is not a component of the SET complex. Purified SET complex and K562 cell lysates were probed for PARP-1 and for the known SET complex components SET, pp32, and NM23-H.8 (D) GzmA disrupts the PARylation activity of PARP-1. Recombinant PARP-1 was preincubated with the indicated amount of GzmA at 37°C for 1 hour and then incubated with radiolabeled NAD+ before electrophoresis and autoradiography. The radiolabeled band corresponds to automodified PARP-1.
GzmA binds to PARP-1. (A) PARP-1 in cell lysates binds to S-AGzmA. Precleared HeLa cell lysates were incubated with S-AGzmA and antisera to PARP-1 or control antisera, and immune complexes were captured on protein G beads. GzmA binding was assayed by immunoblot probed with His-tag antibody. DNase treatment after PARP-1/GzmA incubation does not disrupt the association of S-AGzmA and PARP-1. (B) Recombinant PARP-1 binds S-AGzmA in vitro only in the presence of DNA. Recombinant PARP-1 attached to PARP-1 antibody–conjugated beads was incubated with S-AGzmA for 3 hours on ice in the presence or absence of 100-bp PCR product DNA. S-AGzmA was pulled down with PARP-1 only in the presence of DNA. However, if DNase I was added before extraction in SDS sample buffer, binding was not disrupted. (C) PARP-1 is not a component of the SET complex. Purified SET complex and K562 cell lysates were probed for PARP-1 and for the known SET complex components SET, pp32, and NM23-H.8 (D) GzmA disrupts the PARylation activity of PARP-1. Recombinant PARP-1 was preincubated with the indicated amount of GzmA at 37°C for 1 hour and then incubated with radiolabeled NAD+ before electrophoresis and autoradiography. The radiolabeled band corresponds to automodified PARP-1.
Three GzmA substrates (Ape1, HMG2, and SET) are in the SET complex, an oxidative DNA damage repair complex.7,8,16,18 Because PARP-1 senses DNA damage, we examined whether PARP-1 might also be in the complex. Cell lysates and the purified SET complex were probed for PARP-1. Whereas PARP-1 was readily detected in lysates, it was not in the purified SET complex (Figure 2C).
GzmA disrupts PARP-1 automodification
To study the effect of GzmA on PARP-1 activity, GzmA was incubated with purified native PARP-1 together with [32P]NAD+ in the presence or absence of linear double-stranded DNA (Figure 2D). In the absence of GzmA, PARP-1 became activated and incorporated [32P]NAD+. In the absence of DNA, no automodification occurred. When GzmA was added, [32P]NAD+ incorporation was reduced in a dose-dependent manner. At a GzmA concentration of 200 nM, no automodification was detected. Therefore, GzmA disrupts PARP-1 automodification (and also presumably PARylation of other proteins).
GzmA reduces PARylation in cells
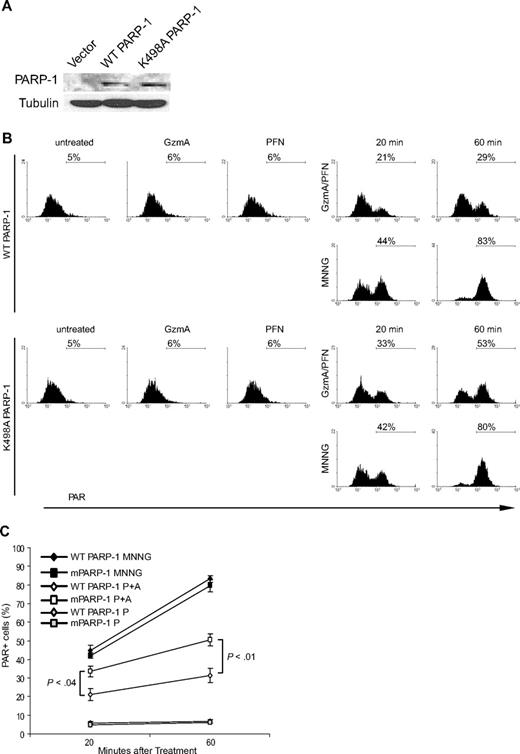
We next investigated GzmA's effect on PARP-1 function within cells. An antibody that recognizes PAR-modified proteins was used to monitor PARP-1 activity by flow cytometry in PARP-1–deficient mouse embryonic fibroblasts (MEFs) stably transfected with WT or K498A PARP-1 (Figure 3). K498A and WT PARP-1 comparably autoincorporated [32P]NAD+ in vitro, suggesting that the cleavage site mutation did not alter catalytic activity (data not shown). WT and K498A PARP-1 were similarly expressed in the transduced MEFs (Figure 3A). In the absence of DNA damage, only approximately 5% of cells stained for PAR. When MEFs were treated with the alkylating agent N-methyl-N′-nitro-N-nitrosoguanidine (MNNG), PAR levels increased so that by 1 hour approximately 80% of cells were PAR+. PAR levels were comparable in MEFs expressing either WT or K498A PARP-1. When these MEFs were treated with GzmA or PFN, PAR staining was unaltered. However, after treatment with both PFN and GzmA, PAR levels increased within 20 minutes and peaked within 60 minutes (Figure 3B-C and data not shown). In MEFs expressing WT PARP-1, 31% of cells were PAR+ at the peak, whereas in MEFs expressing K498A PARP-1, PARylation was more efficient because 50% of cells stained for PAR. Therefore, the PARP-1 response to GzmA-mediated DNA damage was blunted by GzmA cleavage of PARP-1, because it is enhanced by expressing a relatively GzmA-resistant form of PARP-1.
GzmA inhibition of PARylation within cells is partially rescued by expressing K498A PARP-1. PARP-1−/− MEF cells, transiently transfected to express either WT or K498A PARP-1, were treated with MNNG or PFN and/or GzmA for 20 or 60 minutes. Flow cytometry in cells stained with PAR antibody measured PARP activation in response to DNA damage. (A) Immunoblot shows comparable expression of WT and K498A PARP-1 in PARP-1−/− MEFs using antibody to C-terminal PARP-1. (B) Shows representative flow cytometry plots, and (C) the mean and SD of the percentage of PAR+ cells in 3 independent experiments. Neither PFN nor GzmA alone triggers PAR synthesis. PARP-1 activation to alkylating DNA damage is equivalent in cells expressing WT and K498A PARP-1. However, PARP-1 activity in response to DNA damage by GzmA (A) and PFN (P) is significantly enhanced by mutating the GzmA-favored cleavage site. The gray symbols refer to control cells just treated with PFN (P).
GzmA inhibition of PARylation within cells is partially rescued by expressing K498A PARP-1. PARP-1−/− MEF cells, transiently transfected to express either WT or K498A PARP-1, were treated with MNNG or PFN and/or GzmA for 20 or 60 minutes. Flow cytometry in cells stained with PAR antibody measured PARP activation in response to DNA damage. (A) Immunoblot shows comparable expression of WT and K498A PARP-1 in PARP-1−/− MEFs using antibody to C-terminal PARP-1. (B) Shows representative flow cytometry plots, and (C) the mean and SD of the percentage of PAR+ cells in 3 independent experiments. Neither PFN nor GzmA alone triggers PAR synthesis. PARP-1 activation to alkylating DNA damage is equivalent in cells expressing WT and K498A PARP-1. However, PARP-1 activity in response to DNA damage by GzmA (A) and PFN (P) is significantly enhanced by mutating the GzmA-favored cleavage site. The gray symbols refer to control cells just treated with PFN (P).
GzmA-mediated DNA damage is enhanced by inhibiting PARP-1 and reduced by overexpressing PARP-1
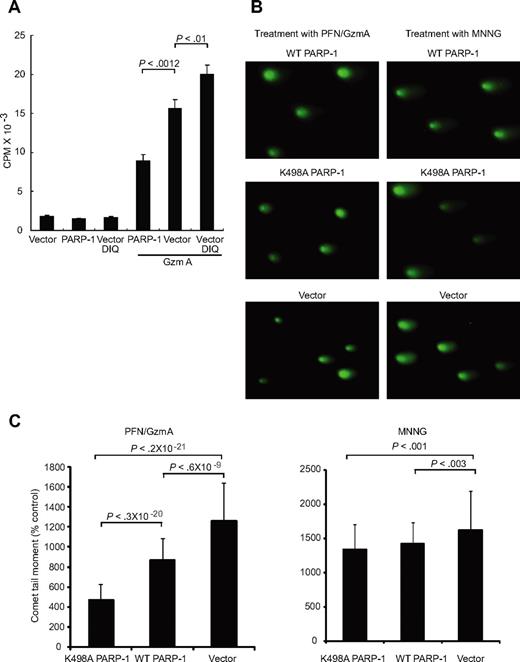
Because PARP-1 is required for efficient DNA repair, PARP-1 inactivation might enhance GzmA-induced DNA damage and cell death. To test this, we altered PARP-1 function in HeLa cells either by inhibiting PARP using the PARP-1 inhibitor 1,5-dihydroxyisoquinoline or by transfecting cells to overexpress PARP-1. GzmA- and PFN-induced DNA damage was assayed by measuring 32P-ATP incorporation at DNA breaks by the Klenow fragment of DNA polymerase2 (Figure 4A). Altering PARP-1 activity did not affect baseline DNA labeling in untreated cells or cells treated with either GzmA or PFN alone (Figure 4A; data not shown). However, DNA damage by GzmA and PFN was reduced almost by half by overexpressing PARP-1 (P = .001). In contrast, PARP-1 inhibition enhanced GzmA- and PFN-induced DNA damage by 25% (P < .01). Therefore, PARP-1 helps repair GzmA-induced DNA damage. By cleaving PARP-1, GzmA interferes with DNA repair and enhances DNA damage.
DNA repair by PARP-1 is disrupted by GzmA. (A) GzmA-induced DNA nicks, radiolabeled with Klenow, in HeLa cells are enhanced by inhibiting PARP-1 using the chemical inhibitor 1,5 dihydroxyisoquinoline (DIQ) and reduced by overexpressing WT PARP-1. (B) DNA damage in response to MNNG or GzmA and PFN, assessed by comet assay, is reduced by overexpressing WT PARP. Overexpression of K498A PARP-1 significantly reduces GzmA-mediated DNA damage compared with overexpression of WT PARP-1, but has a similar effect as WT PARP-1 in response to the alkylating agent. Representative comet assay images (B) and the mean and SD of the comet tail moment (C) from 3 independent experiments are shown.
DNA repair by PARP-1 is disrupted by GzmA. (A) GzmA-induced DNA nicks, radiolabeled with Klenow, in HeLa cells are enhanced by inhibiting PARP-1 using the chemical inhibitor 1,5 dihydroxyisoquinoline (DIQ) and reduced by overexpressing WT PARP-1. (B) DNA damage in response to MNNG or GzmA and PFN, assessed by comet assay, is reduced by overexpressing WT PARP. Overexpression of K498A PARP-1 significantly reduces GzmA-mediated DNA damage compared with overexpression of WT PARP-1, but has a similar effect as WT PARP-1 in response to the alkylating agent. Representative comet assay images (B) and the mean and SD of the comet tail moment (C) from 3 independent experiments are shown.
Mutating the PARP-1 cleavage site reduces DNA damage in GzmA-treated cells
To confirm that GzmA cleavage of PARP-1 enhances GzmA-mediated DNA damage, we quantified DNA damage by comet assay in cells overexpressing WT or K498A PARP-1. In a comet assay, treated cells are embedded in agarose on microscope slides, lysed under denaturing conditions that separate the 2 DNA strands, and subjected to an electrical field. Cut DNA migrates faster than chromosomal DNA. Nuclear DNA containing fragmented DNA takes on the appearance of a comet due to the rapid migration of the shorter DNA fragments. The size and length of the comet correlate with the extent of DNA damage. A representative experiment is shown in Figure 4B, and the mean results of 3 experiments in Figure 4C. Overexpressing WT or K498A PARP-1 slightly reduced the mean comet tail moment after MNNG exposure (P < .003 and P < .001, respectively), but mutation of the GzmA-favored cleavage site did not significantly alter the extent of DNA damage. DNA damage was not significantly increased after either GzmA or PFN treatment (data not shown). However, overexpression of K498A PARP-1 had a greater impact on DNA damage by GzmA and PFN than overexpression of WT PARP-1. The comet tail moment after GzmA and PFN treatment was approximately half as much in cells overexpressing K498A PARP-1 compared with cells overexpressing WT PARP-1. Therefore, PARP-1 cleavage after Lys498 interferes with DNA damage repair specifically in response to GzmA.
The N-terminal fragment of GzmA-cleaved PARP-1 binds DNA and interferes with DNA repair
The N-terminal 24-kDa fragment of PARP-1 generated during caspase-dependent apoptosis contains the PARP-1 DNA binding Zn finger domains, inhibits the catalytic activity of uncleaved PARP-1, and blocks DNA repair.38-40 To determine whether the N-terminal PARP-1 fragment generated by GzmA, which also contains the DBD, has similar dominant-negative behavior, we first determined whether the GzmA-generated N- and C-terminal PARP-1 fragments (PARP-11-498 and PARP-1499-1014) bind DNA by electrophoretic mobility shift assay (Figure 5A). Both full-length PARP-1 and its N-terminal fragment bound to DNA, whereas the C-terminal fragment did not. Gel shifts were confirmed by observing a supershift with PARP-1 antibody. The PARP-11-498 cleavage fragment was not labile in cells. It could be detected after GzmA and PFN treatment using an antibody specific for N-terminal PARP-1 (Figure 5B).
The N-terminal fragment of PARP-1, produced by GzmA cleavage, interferes with DNA repair. (A) The N-terminal, but not C-terminal, fragment of GzmA-cleaved PARP-1 binds to DNA by gel shift. The protein-DNA complex is supershifted by PARP-1 (P) antiserum, but not control (C) antiserum. (B) The N-terminal fragment persists in cells treated with GzmA and PFN. Cell lysates were analyzed by immunoblot probed with an antibody that recognizes the N terminus of PARP-1. (C) Overexpressing PARP1-498 in HeLa cells enhances DNA damage induced by either GzmA and PFN (left) or MNNG (right) by comet assay, whereas overexpressing WT PARP enhances DNA repair. PARP499-1014, which lacks the DNA binding domains, has no effect. N.S. = not significant. Representative data from at least 3 independent experiments are shown.
The N-terminal fragment of PARP-1, produced by GzmA cleavage, interferes with DNA repair. (A) The N-terminal, but not C-terminal, fragment of GzmA-cleaved PARP-1 binds to DNA by gel shift. The protein-DNA complex is supershifted by PARP-1 (P) antiserum, but not control (C) antiserum. (B) The N-terminal fragment persists in cells treated with GzmA and PFN. Cell lysates were analyzed by immunoblot probed with an antibody that recognizes the N terminus of PARP-1. (C) Overexpressing PARP1-498 in HeLa cells enhances DNA damage induced by either GzmA and PFN (left) or MNNG (right) by comet assay, whereas overexpressing WT PARP enhances DNA repair. PARP499-1014, which lacks the DNA binding domains, has no effect. N.S. = not significant. Representative data from at least 3 independent experiments are shown.
We next evaluated the effect of overexpressing the N- and C-terminal PARP-1 fragments of PARP-1 on repair of DNA damage induced by MNNG or GzmA and PFN (Figure 5C). Not surprisingly, overexpression of WT PARP-1 reduced the comet moment of HeLa cells treated with either MNNG or GzmA and PFN. DNA damage by either treatment increased significantly in cells expressing the N-terminal PARP-1 fragment compared with cells transfected with vector or WT PARP-1. Expressing the C-terminal PARP-1 cleavage fragment did not significantly affect DNA damage. Therefore, the GzmA-generated N-terminal PARP-1 fragment, like the N-terminal 24-kDa caspase fragment, enhances DNA damage, most likely by binding to damaged DNA and interfering with binding of full-length PARP-1.
Cells overexpressing K498A PARP-1 are relatively resistant to GzmA and more likely to die by necrosis than apoptosis
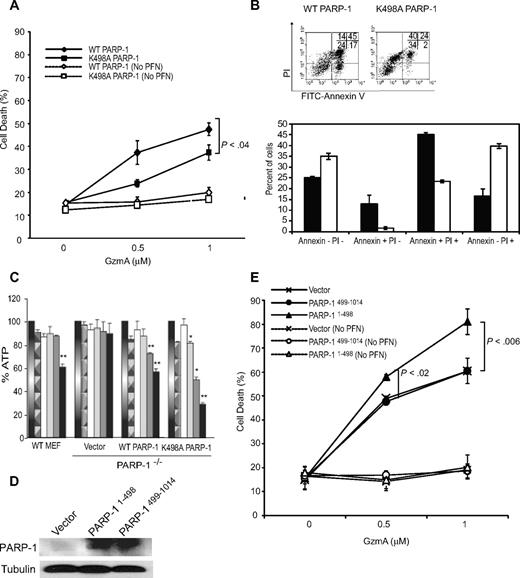
Because PARP-1 cleavage enhances DNA damage, it might also enhance GzmA-induced cell death. To test this, we compared GzmA-mediated cell death in PARP-1−/− MEFs overexpressing WT or K498A PARP-1 (Figures 3A, 6A-B). Background cell death after treatment with either GzmA or PFN alone was not significantly different from untreated cells (data not shown). Cells transduced with K498A PARP-1 were significantly more GzmA resistant than cells transduced with WT PARP-1. During apoptosis, cells initially are annexin V−propidium iodide (PI)− and then become sequentially annexin V+ PI−, annexin V+ PI+, and finally annexin V− PI+. Necrotic cells are mostly annexin V− PI+. Cells that overexpressed K498A PARP-1 were not only more likely to survive (annexin V−PI−) than cells overexpressing WT PARP-1 (35% ± 2% vs 25% ± 1% viable cells, P < .001), but cells that died were much more likely to die by necrosis than by apoptosis, containing significantly more PI+ cells that did not stain with annexin V (Figure 6B). After GzmA and PFN treatment, the proportion of apoptotic annexin V+ WT PARP-1–expressing cells was 58% ± 4%, whereas there were fewer than half as many annexin V–staining cells (25% ± 1%, P < .0001) when mutant PARP-1 was expressed. When we compared the effect of GzmA and PFN on WT MEFs and PARP-1−/− MEFs, WT MEFs were slightly more sensitive than PARP-1−/− cells because of an increase in necrotic annexin V− PI+ cells (supplemental Figure 2A-B). Similar results were obtained when WT MEFs and PARP-1−/− cells were used as targets for GzmB−/− CTLs in Cr release assays (supplemental Figure 2C). WT MEFs were significantly more sensitive than PARP-1−/− MEFs to GzmB−/− CTLs. The resistance of PARP-1−/− cells to GzmA was partially rescued by transfecting the knockout cells with a PARP-1 expression plasmid. Taken together, these results suggest that PARP-1 plays an important role in determining whether cell death occurs by necrosis or apoptosis. GzmA cleavage of PARP-1 shifts cell death from necrosis to apoptosis.
GzmA cleavage of PARP-1 enhances apoptosis and reduces necrosis of target cells. PARP-1−/− MEFs, transiently transfected with expression plasmids encoding WT or K498A PARP-1 (A-C) or PARP-1 fragments (D-E), were treated with GzmA and PFN and assayed for cell death by fluorescein isothiocyanate-annexin V and PI staining or ATP content. In (A) and (E), mean and SD of the proportion of annexin V and/or PI+ cells from 3 independent experiments are shown. (B) Representative annexin V and PI dot plots (top) and the mean and SD of 3 experiments (bottom) 4 hours after treating cells overexpressing WT (■) or K498A (□) PARP-1 with PFN and 2 μM GzmA. Expression of GzmA-resistant K498A PARP-1 increased the percentage of viable annexin V−PI− cells, but the dead cells were more likely to die of necrosis (annexin V−PI+) than apoptosis (annexin V+). The differences for each subgroup of annexin V and PI staining between WT and K498A PARP-1–expressing cells were all significant (P < .01). (C) GzmA treatment depletes cellular ATP in a PARP-1–dependent manner. ATP was measured 1 hour after treatment with nothing (gradient bar), PFN alone (patterned bar), GzmA alone (white), or both (increasing concentrations of Gzm indicated by color change from gray to black). ATP depletion requires PARP-1 because ATP levels were unchanged in PARP-1−/− MEFs. Cleavage of PARP-1 reduces ATP depletion because cells expressing GzmA-resistant K498A PARP-1 are more depleted of ATP than cells overexpressing WT PARP-1. Data shown are the mean and SD of 3 independent experiments (*P < .01; **P < .005). (D) Shows the expression of PARP-1 fragments by immunoblot probed with a mixture of antisera that recognize N-terminal and C-terminal PARP-1. (E) Cells expressing N-terminal PARP-1 had increased susceptibility to GzmA. Cells were transfected to overexpress WT (♦) or K498A (■) PARP-1 (A) or PARP-11-498 (▴) or PARP-1499-1014 (•) or with vector (×). Cell death of control cells exposed to GzmA, but not PFN, is indicated by the corresponding open symbols and dashed lines.
GzmA cleavage of PARP-1 enhances apoptosis and reduces necrosis of target cells. PARP-1−/− MEFs, transiently transfected with expression plasmids encoding WT or K498A PARP-1 (A-C) or PARP-1 fragments (D-E), were treated with GzmA and PFN and assayed for cell death by fluorescein isothiocyanate-annexin V and PI staining or ATP content. In (A) and (E), mean and SD of the proportion of annexin V and/or PI+ cells from 3 independent experiments are shown. (B) Representative annexin V and PI dot plots (top) and the mean and SD of 3 experiments (bottom) 4 hours after treating cells overexpressing WT (■) or K498A (□) PARP-1 with PFN and 2 μM GzmA. Expression of GzmA-resistant K498A PARP-1 increased the percentage of viable annexin V−PI− cells, but the dead cells were more likely to die of necrosis (annexin V−PI+) than apoptosis (annexin V+). The differences for each subgroup of annexin V and PI staining between WT and K498A PARP-1–expressing cells were all significant (P < .01). (C) GzmA treatment depletes cellular ATP in a PARP-1–dependent manner. ATP was measured 1 hour after treatment with nothing (gradient bar), PFN alone (patterned bar), GzmA alone (white), or both (increasing concentrations of Gzm indicated by color change from gray to black). ATP depletion requires PARP-1 because ATP levels were unchanged in PARP-1−/− MEFs. Cleavage of PARP-1 reduces ATP depletion because cells expressing GzmA-resistant K498A PARP-1 are more depleted of ATP than cells overexpressing WT PARP-1. Data shown are the mean and SD of 3 independent experiments (*P < .01; **P < .005). (D) Shows the expression of PARP-1 fragments by immunoblot probed with a mixture of antisera that recognize N-terminal and C-terminal PARP-1. (E) Cells expressing N-terminal PARP-1 had increased susceptibility to GzmA. Cells were transfected to overexpress WT (♦) or K498A (■) PARP-1 (A) or PARP-11-498 (▴) or PARP-1499-1014 (•) or with vector (×). Cell death of control cells exposed to GzmA, but not PFN, is indicated by the corresponding open symbols and dashed lines.
GzmA induces PARP-1–dependent ATP depletion, which is enhanced by overexpressing K498A PARP-1
The shift from apoptosis to necrosis when K498A PARP-1 was substituted for WT PARP-1 might be due to ATP depletion by uncleaved, activated PARP-1 in the setting of extensive DNA damage. To test this hypothesis, we compared the effect of GzmA and PFN on cellular ATP content in WT MEFs with PARP-1−/− MEFs stably expressing WT or K498A PARP-1 (Figure 6C). In WT MEFs, cellular ATP significantly decreased 1 hour after GzmA and PFN treatment at the highest GzmA concentration tested. This effect required both GzmA and PFN, as well as PARP-1, because there was no significant change in cellular ATP in PARP-1−/− cells. Overexpression of WT PARP-1 in PARP-1−/− cells restored ATP depletion to the level of WT MEFs, whereas overexpression of K498A PARP-1 in PARP-1−/− cells significantly increased ATP depletion. These results support the idea that GzmA cleavage of PARP-1 helps maintain ATP, which is required for programmed cell death.
Cells overexpressing the N-terminal PARP-1 fragment are more susceptible to GzmA-mediated death
To determine whether the GzmA cleavage fragments enhance GzmA-mediated death, cell death was measured by 4-hour 51Cr release assay after GzmA and PFN treatment of PARP-1−/− MEFs stably overexpressing either PARP-11-498 or PARP499-1014 (Figure 6D). Overexpression of PARP-11-498, but not PARP499-1014, increased GzmA-mediated cell death (Figure 6E). Therefore, the N-terminal cleavage fragment of PARP-1 augments cell death, possibly by its recruitment to damaged DNA and interference with binding of DNA repair factors.
PARP-1 expression protects target cells from CTL-mediated death, and N-terminal PARP-1 enhances cell death
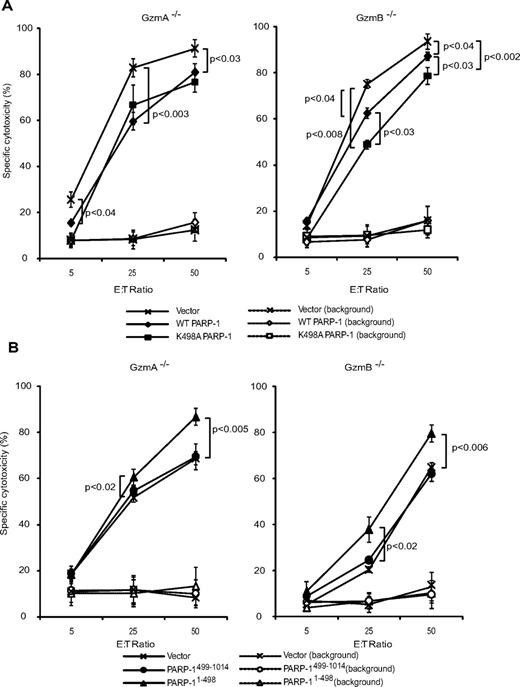
Although treating cells with GzmA and PFN is a good surrogate for CTL killing, we wanted to verify that PARP-1 targeting is important during CTL attack. This was tested by 51Cr release assay using mouse CTL effector cells, generated by activating GzmA−/− or GzmB−/− splenocytes, to kill ConA-coated PARP-1−/− MEFs transfected to overexpress WT PARP-1, K498A PARP-1, PARP-11-498, or PARP-1499-1014. PARP-1−/− cells were more sensitive to both GzmA−/− and GzmB−/− CTLs than PARP-1−/− MEFs overexpressing WT PARP-1 (Figure 7A). Cells overexpressing K498A PARP-1 were slightly, but significantly, more resistant than cells overexpressing WT PARP-1 to GzmB−/− CTLs. Not surprisingly, the K498A mutation did not affect susceptibility to GzmA−/− CTLs. Although the differences in cell death were small for each experiment, they were reproducible and also statistically significant for each experiment (each experiment was done in triplicate and repeated at least 3 times). The small effect of manipulating PARP-1 can be explained by the fact that multiple factors contribute to whether a cell can recover from an apoptotic stimulus. As expected, PARP-1−/− targets expressing dominant-negative PARP-11-498 were more vulnerable to GzmA−/− or GzmB−/− CTLs, whereas target cells overexpressing PARP-1499-1014, which is unable to bind DNA, were killed to the same extent as PARP-1−/− MEFs (Figure 7B). These results further support the assignment of Lys498 as the preferred GzmA PARP-1 cleavage site and demonstrate the importance of PARP-1 cleavage in executing GzmA-mediated death.
Expression of WT PARP-1 provides some protection from cell death triggered by GzmB−/− and GzmA−/− CTLs, whereas mutation of the preferred PARP-1 GzmA cleavage site protects only against GzmB−/− CTLs. Cell death of PARP-1–transduced ConA-coated PARP-1−/− MEFs, attacked by GzmB-expressing CTLs (GzmA−/−, left) or GzmA-expressing CTLs (GzmB−/−, right), was measured by 51Cr release assay. (A) PARP-expressing cells are more resistant to CTL attack. K498A PARP-1–expressing cells are more resistant than WT PARP-1–expressing cells to attack by GzmA-expressing CTLs, but not by GzmB-expressing cells. (B) Expression of PARP1-498 increases susceptibility to both GzmB−/− and GzmA−/− CTLs. Cells were transfected to overexpress WT (♦) or K498 (■) PARP-1 (A) or PARP-11-498 (▴) or PARP-1499-1014 (•) (B) or with vector (×). Cell death of control cells, without CTLs, is indicated by the corresponding open symbols and dashed lines.
Expression of WT PARP-1 provides some protection from cell death triggered by GzmB−/− and GzmA−/− CTLs, whereas mutation of the preferred PARP-1 GzmA cleavage site protects only against GzmB−/− CTLs. Cell death of PARP-1–transduced ConA-coated PARP-1−/− MEFs, attacked by GzmB-expressing CTLs (GzmA−/−, left) or GzmA-expressing CTLs (GzmB−/−, right), was measured by 51Cr release assay. (A) PARP-expressing cells are more resistant to CTL attack. K498A PARP-1–expressing cells are more resistant than WT PARP-1–expressing cells to attack by GzmA-expressing CTLs, but not by GzmB-expressing cells. (B) Expression of PARP1-498 increases susceptibility to both GzmB−/− and GzmA−/− CTLs. Cells were transfected to overexpress WT (♦) or K498 (■) PARP-1 (A) or PARP-11-498 (▴) or PARP-1499-1014 (•) (B) or with vector (×). Cell death of control cells, without CTLs, is indicated by the corresponding open symbols and dashed lines.
Discussion
PARP-1 is cleaved into 24-kDa N-terminal and 89-kDa C-terminal fragments during caspase-dependent apoptosis.19 In this study, we report that during caspase-independent cell death, GzmA cuts PARP-1 after Lys498 into 55-kDa fragments, disconnecting the C-terminal catalytic domain from the N-terminal DBD and destroying its PARylation activity. The N-terminal PARP-1 cleavage fragment acts as a dominant negative, binding to DNA and interfering with DNA repair. The importance of PARP-1 cleavage for GzmA-dependent death was demonstrated in cells treated with both GzmA and PFN and subjected to GzmB−/− CTLs by showing reduced DNA damage and cell death in cells overexpressing K498A PARP-1, and increased DNA damage and cell death when PARP-1 is inhibited or the N-terminal cleavage fragment is expressed. Moreover, cells expressing K498A PARP-1 were more likely to die by necrosis than apoptosis, because PARP-1 activation led to ATP depletion. Therefore, PARP-1 inactivation is critical to GzmA-mediated cell death.
The role of PARP-1 in the GzmA pathway is complex. Although PARP-1 mobilization facilitates cellular recovery by promoting DNA repair, activation of PARP-1 depletes cellular ATP and interferes with cellular recovery. In fact, in this study, manipulation of PARP-1 expression had opposing effects depending on the potency of the cell death stimulus. Under conditions in which most cells survive (supplemental Figure 2), PARP-1 expression promoted cell death: WT MEFs were more susceptible to GzmA and to GzmB−/− CTLs than PARP-1−/− MEFs. However, under conditions in which most cells die (Figure 7A), PARP-1 overexpression protected MEFs from cell death from GzmB−/− CTLs.
Our finding that PARP-1 is a GzmA substrate, taken together with previous reports showing that PARP-1 is a target of caspase-3 and GzmB, suggests that PARP-1 inactivation may be critical for all programmed cell death pathways. The only other known shared substrate of GzmA, GzmB, and caspases is lamin B.1,10 Substrates common to caspase-independent and caspase-dependent death most likely identify critical elements for apoptosis. Our results suggest that inactivating PARP-1 is one, and disrupting the nuclear envelope via lamin B cleavage is another. Of note, lysosomal cathepsins activated during necrotic death cleave PARP-1 near the GzmA cleavage site at or near Leu525 to also produce a 55-kDa C-terminal fragment.27
Because PARP-1 senses SSB (actually more efficiently than DSB) and GzmA induces SSB, it should not be surprising that GzmA would inactivate the key sensor for the DNA damage it causes. Both PARP-1 and the SET complex are mobilized by oxidative stress.4,23 Recent studies have shown that massive PARP-1 activation promotes cellular necrosis by depleting cellular energy stores. In fact, cells expressing mutant PARP-1 that is relatively resistant to GzmA were more depleted of ATP and more likely to die by necrosis than apoptosis. Necrotic cells stimulate inflammation, whereas apoptotic cells are phagocytosed by scavenger cells and cleared without inflammation.41 By activating apoptosis and not necrosis, immune killer cells are able to eliminate infected and transformed cells without causing bystander cell damage.1 By disabling PARP-1, adequate ATP levels are maintained to preserve cell membrane integrity, prevent rapid necrosis, and execute the slower energy-dependent steps of programmed cell death.28-30 In fact, differences in PARP-1 cleavage, which is activated by one death receptor pathway, but not another (Fas vs tumor necrosis factor), may help explain why Fas engagement triggers apoptosis, whereas tumor necrosis factor signaling typically leads to necrosis.42 PARP-1 activation also enhances inflammation by triggering the release of the DNA-binding protein and proinflammatory mediator HMGB1 from cells.43 Therefore, inactivating PARP-1 is also expected to reduce this potential source of inflammation. This could be investigated by determining whether HMGB1 is released from GzmA-treated cells that overexpress K498A PARP-1.
Although our study suggests that GzmA inactivates PARP-1 to facilitate apoptotic versus necrotic cell death and thereby reduce inflammation, a recent study44 and some earlier studies1 suggest that GzmA might have important proinflammatory effects. In the recent study, the GzmA concentration required to induce inflammation was substantially less than the concentration needed to induce cell death. However, the proinflammatory GzmA effect used recombinant GzmA produced in bacteria, but did not conclusively demonstrate removal of the potent bacterial inflammatory mediator lipopolysaccharide. Therefore, it will be important to confirm this study. Because the GzmA concentration at the immunologic synapse is most likely orders of magnitude greater than in the extracellular milieu, it is difficult to predict the relative importance of these 2 activities in vivo.
All the death-inducing proteases separate the PARP-1 DBD from its catalytic domain. In this study, we found that the N-terminal GzmA cleavage fragment binds DNA and interferes with DNA repair by intact PARP-1 in response to another DNA-damaging agent (MNNG). This is also a characteristic of the shorter caspase/GzmB N-terminal PARP-1 fragment, and is most likely also a property of the cathepsin products. The dominant-negative property of the cleavage products most likely stems from their competition not only with full-length PARP-1, but also with other factors that recognize and repair DNA damage,25 although we have not formally tested that in this study. However, this is likely because expressing the N-terminal fragment not only augmented DNA damage in PARP-1–expressing HeLa cells (Figure 5C), but also enhanced cell death induced by both GzmA and PFN (Figure 6E) and CTLs (Figure 7B) in PARP-1−/− MEFs. Binding of PARP-1 catalytically inactive fragments to DNA damage sites should serve to amplify cell death by interfering with DNA repair initiated by any residual intact PARP-1 or by PARP-2, the only other PARP family member known to be activated by DNA damage,23 which is not cleaved by GzmA (Figure 1D).
Although GzmA cleaves recombinant PARP-1 in the absence of DNA, binding of inactive GzmA to PARP-1 is augmented by DNA, because the 2 recombinant molecules can only be coprecipitated in the presence of added DNA. However, once S-AGzmA has bound to PARP-1 in the presence of DNA, adding a DNase to the bound complex does not disrupt the complex. This suggests that GzmA preferentially recognizes and cleaves PARP-1 that has localized to damaged or distorted DNA. GzmA recognition of PARP-1 may be enhanced by a conformational change in PARP-1 that occurs when it binds DNA.
PARP-1 has important roles beyond DNA repair.23,45-47 PARylation of histones, particularly histone H1, releases the linker histone from nucleosomes, reducing chromatin compaction. PARP-1 also binds to nucleosomes in much the same way as H1 and competes for the same binding site.46 PARP-1 automodification leads to its release from nucleosomes and their decondensation. H1 has been suggested to bind to highly condensed heterochromatin, whereas PARP-1 may preferentially associate with inactive chromatin poised for activation. PARP-1 is also recruited to promoters that are activated by topoisomerase II, which forms DSB within certain promoters.47 PARP-1 recruitment and automodification may then act to open up chromatin for transcription. Interestingly, 2 other GzmA substrates, HMGB2 and Ku70,16,17 have also been implicated in topoisomerase II–PARP-1–dependent transcriptional activation.47 We speculate that PARP-1 may be involved in activating genes involved in the response to oxidative stress or DNA damage, and that GzmA may specifically inhibit this pathway. Although the role of PARP-1 in altering chromosome accessibility and transcriptional regulation is still poorly understood, PARP-1 inactivation by GzmA and other death-inducing proteases most likely interferes with PARP-1's other cell-protective functions.
PARP inhibitors are currently being evaluated clinically for their ability to enhance cancer cell death in conjunction with alkylating agents.48 Inhibiting PARP-1 increases sensitivity to DNA-alkylating agents, topoisomerase I inhibitors, and ionizing radiation. Based on our results, PARP inhibitor drugs would also be expected to enhance cancer cell susceptibility to immune surveillance via Gzm-mediated cell death by natural killer cells and tumor-specific CTLs.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grant AI-45587 (to J.L.) and a Leukemia & Lymphoma Society fellowship (to D.C.).
National Institutes of Health
Authorship
Contribution: P.Z., D.M., D.C., and D.Z. designed and performed the research, and analyzed the data. P.Z. and D.M. also wrote the paper. A.S. analyzed data and wrote the paper. J.L. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Judy Lieberman, Immune Disease Institute, 200 Longwood Ave, Boston, MA 02115; e-mail: lieberman@idi.harvard.edu.
References
Author notes
*P.Z. and D.M. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal