In this issue of Blood, Jones and colleagues use a genotype-phenotype approach to identify novel quantitative trait loci associated with platelet signaling pathways.
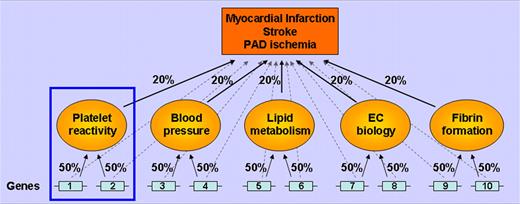
Interindividual variation in platelet reactivity likely contributes to occlusion of coronary or cerebral arteries upon atherosclerotic plaque rupture in some individuals, whereas other individuals repair such lesions without occluding the vessel. It is expected that few (or fewer) genes are involved in any one of the intermediate traits (eg, platelet reactivity) compared with a larger spectrum of genes that contribute to the final manifestion of the clinical disease (eg, pathologic arterial thrombosis; see figure). This is the rationale supporting genetic association studies of intermediate phenotypes because the power to detect gene associations is enhanced when the potential number of genes responsible for the phenotype is reduced or refined. Thus, this improves the fraction of variance explained by any single factor or gene.1 Whereas it is relatively easy to measure substances (such as blood lipids) in a high-throughput manner, it is vastly more complex to assess ex vivo functions of living tissues. Thus, for platelet genomic research, there is a “catch-22”: there is value in using ex vivo platelet function as an intermediate end point, and yet it is challenging to study the thousands of samples typically required to achieve the analytic power needed to satisfy accepted statistical thresholds.
The utility of intermediate traits as end points. This greatly simplified schematic assumes there are 10 genes contributing equally to the clinical end point (ie, 10% each, faint dashed lines). Another level of complexity to the pathophysiology is superimposed by including 5 intermediate traits, each contributing 20% to the clinical outcome. In this example, genes 1 and 2 have a much larger effect on platelet reactivity (50%) than on the clinical end point (10%). Of course, environmental factors also play a major role in these end points (not shown).
The utility of intermediate traits as end points. This greatly simplified schematic assumes there are 10 genes contributing equally to the clinical end point (ie, 10% each, faint dashed lines). Another level of complexity to the pathophysiology is superimposed by including 5 intermediate traits, each contributing 20% to the clinical outcome. In this example, genes 1 and 2 have a much larger effect on platelet reactivity (50%) than on the clinical end point (10%). Of course, environmental factors also play a major role in these end points (not shown).
In this issue of Blood, Jones et al demonstrate how moderate sample sizes—in the few hundreds—have value as a screening tool when complemented by “biologic filters” that help prioritize gene lists and, importantly, “wet bench” studies confirming the computational associations.2 The authors developed a list of 97 genes believed to be important in the function of platelets and other hematopoietic cells. These 97 genes were resequenced in 48 European samples, novel SNPs were identified with low minor allele frequencies, and 1327 SNPs were selected that tagged most of the genetic variation in these genes. DNA from 500 healthy subjects with known platelet responses to cross-linked collagen-related peptide (CRP) and ADP3 was genotyped for these 1327 SNPs. Statistical analyses revealed 17 novel associations with platelet function, including genes encoding cell surface receptors (CD36, GP6, ITGA2, PEAR1, and P2Y12), kinases (JAK2, MAP2K2, MAP2K4, MAPK14) and other signaling molecules (GNAZ, VAV3, ITPR1, FCERG1). The associations presented in Table 1 of the article and other supplemental materials will be of great interest to the platelet biology community.
The authors focused on SNPs in 3 genes of interest, PEAR1, VAV3, and ITPR1, for validation at the protein and physiologic levels. Genetic variation in PEAR1 has previously been associated with collagen-induced platelet aggregation,4 and Jones et al found that PEAR1 SNPs showed the strongest of all associations with the platelet response to CRP, as well as strong associations with ADP-induced fibrinogen binding. PEAR1 genotypes were associated with PEAR1 levels (the minor alleles showing increased expression). A minor allele of a VAV3 SNP was associated with increased protein levels, and ITPR1 genotypes were associated with platelet calcium flux in response to ADP.
The devil is in the phenotypic details of genotype-phenotype association studies. Over the past few years, a number of groups have developed different cohorts to systematically address the complex relationship between genetic variation and platelet phenotypes. Of note, the Wellcome Trust platelet assays were supplemented with aspirin, hirudin, and, for CRP-stimulated samples, apyrase,2 such that these platelet phenotypes are expected to display maximum sensitivity to GPVI-mediated platelet activation and be insensitive to the amplifying effects of thromboxane A2 and ADP. This likely contributes to the large effect size seen for GP6 SNPs on CRP-induced platelet reactivity. This aspect of the assay design is neither better nor worse than designs that lack such inhibitors—but is expected to uncover some gene associations different from those found by genomic studies using alternative assay designs. This study comprised healthy subjects of Northern European ancestry, and cannot be extrapolated to patients or different ethnic groups. It should be noted that the follow-up experiments with PEAR1, VAV3, and ITPR1 SNPs also defined associations and not causality. Genome-wide association studies (GWASs) are currently a “topical” genomics technology and have successfully identified common genetic variants associated with the risk of numerous common diseases. But the great majority of variants have had odds ratios less than 1.5,5 and many thousands of subjects have been required to achieve statistical significance. Although unbiased, GWASs identify genomic regions, not genes themselves, and the findings usually lack biologic context. Therefore, well-conducted, candidate gene studies such as the one by Jones et al still have a place in the armamentarium of platelet genomic studies.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal