Abstract
Here we report stable gene transfer in cord blood-derived CD34+ hematopoietic stem cells using a hyperactive nonviral Sleeping Beauty (SB) transposase (SB100X). In colony-forming assays, SB100X mediated the highest efficiency (24%) of stable Discosoma sp red fluorescent protein (DsRed) reporter gene transfer in committed hematopoietic progenitors compared with both the early-generation hyperactive SB11 transposase and the piggyBac transposon system (1.23% and 3.8%, respectively). In vitro differentiation assays further demonstrated that SB100X-transfected CD34+ cells can develop into DsRed+ CD4+CD8+ T (3.17%-21.84%; median, 7.97%), CD19+ B (3.83%-18.66%; median, 7.84%), CD56+CD3− NK (3.53%-79.98%; median, 7.88%), and CD33+ myeloid (7.59%-15.63%; median, 9.48%) cells. SB100X-transfected CD34+ cells achieved approximately 46% engraftment in NOD-scid IL2γcnull (NOG) mice. Twelve weeks after transplantation, 0.57% to 28.96% (median, 2.79%) and 0.49% to 34.50% (median, 5.59%) of total human CD45+ cells in the bone marrow and spleen expressed DsRed, including CD19+ B, CD14+ monocytoid, and CD33+ myeloid cell lineages. Integration site analysis revealed SB transposon sequences in the human chromosomes of in vitro differentiated T, B, NK, and myeloid cells, as well as in human CD45+ cells isolated from bone marrow and spleen of transplanted NOG mice. Our results support the continuing development of SB-based gene transfer into human hematopoietic stem cells as a modality for gene therapy.
Introduction
Genetic correction of hematopoietic stem cells (HSCs) has been shown to be curative in the treatment of inherited immunodeficiencies, such as X-linked severe combined immune deficiency (X-SCID) and adenosine deaminase deficiency.1,2 Other genetic and acquired diseases are now being considered as candidates for HSC-based gene therapy, including lysosomal storage diseases, hemophilias, β-thalassemia and sickle cell disease, Wiskott-Aldrich syndrome, and chemotherapy-induced myelosuppression.3,4 Gene therapy targeting HSCs has shown promise in the treatment of HIV infections and cancer.5-8
The potential of HSC-based gene therapy is enormous as HSCs are able to self-renew and undergo differentiation into progenitor populations, ultimately leading to the generation of mature cells of multiple lineages with diverse functions. Therefore, ex vivo stable gene transfer into HSCs followed by transplantation could result in the long-term persistence of genetically modified HSCs in the recipient, providing a potential cure to a number of disorders affecting components of the hematopoietic system.1-4 However, the application of this technology is restricted because of the limitations of the currently available methods of gene transfer. Most methods require the use of viruses, and transduction with recombinant retroviruses, such as γ-retroviruses (eg, Moloney murine leukemia virus [MLV]), lentiviruses (eg, HIV-1, HIV-2, SIV), and spumaviruses (eg, human foamy virus) are the preferred choice. Use of other viral vectors, such as adeno-associated viruses, adenoviruses, and herpesviruses, have had limited success in HSC gene transfer.3,4
At present, γ-retroviruses have been used in all approved clinical HSC gene therapy trials.2-4 Because preferential integration near promoter regions or in actively transcribed genes occurs with the use of γ-retroviruses9 and lentiviruses,10 respectively, insertional mutagenesis through transcriptional upregulation of cellular proto-oncogenes and/or inactivation of tumor suppressor genes is a major risk to this method of gene transfer. The serious risk of insertional mutagenesis was illustrated when 4 of 20 patients with X-SCID developed T-cell acute lymphoblastic leukemia after infusion of CD34+ HSCs that had been transduced with an MLV-based γ-retroviral vector carrying the therapeutic gene.11-13 Because of this risk, nonvirally mediated gene transfer to HSCs should be developed.
Nonviral gene transfer systems for HSCs provide considerable advantages over viral vectors in clinical use. Their major advantages include the simplicity of gene transfer, low cost, ease of handling, potential for large-scale production, and importantly, biosafety. Three main types of nonintegrating DNA plasmids are currently available for HSC gene transfer: conventional expression vectors, Epstein-Barr virus-based, and scaffold/matrix attachment region-based episomal vectors.14 However, the use of nonintegrating, nonviral vectors does not result in stable transgene expression in HSCs, making this a less attractive method for HSC gene transfer.
DNA transposons have recently emerged as an alternative tool for HSC gene transfer because they possess the advantages of both retroviruses and nonviral vectors, namely, biosafety because of random chromosomal integration permitting long-term transgene expression, simplicity of gene transfer, a lack of immunogenicity, and no requirement for cell cycling for gene transfer to occur.15 The Sleeping Beauty (SB)15,16 and piggyBac transposon/transposase systems17 have been extensively studied in the past years and have been shown to mediate transposition in a wide range of vertebrate cells and tissues, including cultured mammalian cells,16-22 mouse liver and lung tissues,23,24 mouse embryonic stem cells,25 human primary T cells,26-28 and reprogramming-induced pluripotent stem cells.29-31 They have also been successfully used in mammalian germ-line transgenesis32 and insertional mutagenesis for cancer gene discovery.33,34
The utility of the original SB10 transposase system for stable gene transfer to HSCs has been limited, and it has not been determined whether SB10-engineered CD34+ HSCs remain competent for multilineage differentiation with stable transgene expression and retain their in vivo repopulating capacity.35 Recently, a hyperactive SB transposase mutant (SB100X) was engineered that results in superior gene transfer in vertebrate cells, including CD34+ cells.36 In this report, we demonstrate that the SB100X transposase is superior to the previously engineered hyperactive SB11 and to the piggyBac system for mediating stable gene transfer in CD34+ HSCs. We show that SB100X transposase-transfected CD34+ cells can stably express a reporter transgene and differentiate into T, B, natural killer (NK), and myeloid cells in vitro. Importantly, we show that these cells maintain stable transgene expression as well as their capacity to repopulate and differentiate into both lymphoid and myeloid lineages in vivo. This work also provides molecular evidence that stable transgene expression in the differentiated progeny of CD34+ cells is the result of transposition events by the hyperactive SB100X transposase.
Methods
Construction of transposons and transposase-encoding plasmids
SB and piggyBac transposon and transposase-expressing vectors were constructed using standard molecular cloning techniques. Briefly, the SB and piggyBac transposon terminal repeat sequences from pT2/DsRed26 and pXL-Bac II37,38 (kindly provided by Prof Malcolm Fraser Jr, University of Notre Dame, Notre Dame, IN), respectively, were cloned onto both ends of a DsRed red fluorescent reporter gene terminating with a BGH polyadenylation signal (Invitrogen) under the control of the CAGGS promoter. SB11 and piggyBac transposase genes were individually cloned into a minimal expression vector (pKCMV) containing only an origin of replication, kanamycin resistance gene, and cytomegalovirus (CMV) promoter. SB100X transposase was expressed from a CMV promoter on the pCMV(CAT)T7 expression plasmid.
Nucelofection of cord blood-derived CD34+ cells
Umbilical cord blood (UCB) was obtained from the Duke University Cord Blood Center, St Louis Blood Center, New York Blood Center, and the Red Cross in the Twin Cities after University of Minnesota Institutional Review Board approval and informed consent obtained in accordance with the Declaration of Helsinki. UCB was also purchased from the National Disease Research Interchange. After Ficoll-Hypaque (Mediatech Cellgro) gradient separation, UCB mononuclear cells were collected and enriched for CD34+ cells using Miltenyi MACS separation techniques (Miltenyi Biotec). CD34+ cells were enriched to more than 93% purity, and no CD3+, CD19+, and CD56+ cells in purified CD34+ cell populations were detected by flow cytometric analysis. CD34+ cells were washed with 1 × phosphate-buffered saline/0.5% bovine serum albumin (Sigma-Aldrich) and resuspended at 0.5 to 1 × 106 cells/0.1 mL human CD34 cell Nucleofector solution (Amaxa Biosystems). Cells were nucleofected with transposon and/or transposase-expressing plasmids as indicated using program U-08 on the Nucleofector (Amaxa Biosystems) device. Transfected cells were immediately transferred to 24-well plates containing 37°C prewarmed medium described in in vitro differentiation assays.
In vitro T-cell differentiation assays were carried out as previously described.39 Briefly, transfected CD34+ cells (104) were plated in 24-well plates containing subconfluent layers of murine Delta-like 1-expressing OP9 stromal cells (OP9-DL1) or OP9-GFP stromal cells (kindly provided by Dr Juan Carlos Zúňiga-Pflücker, University of Toronto, Toronto, ON) with α-minimal essential medium (Invitrogen) supplemented with 20% fetal bovine serum (FBS; HyClone), 50 U/mL penicillin (Invitrogen), and 50 μg/mL streptomycin (Invitrogen). Recombinant human Fms-like tyrosine kinase 3 ligand (Flt-3L; 5 ng/mL) and interleukin-7 (IL-7; 5 ng/mL; PeproTech) were added to the cultures first on day 0 and then every subsequent 3 to 4 days during media changes. NK-cell developmental potential was assayed by coculture of transfected CD34+ cells (1000 cells per well) onto irradiated (3000 cGy) murine AFT024 stromal cells (3 × 104 cells per well) in 24-well plates in Ham12 plus Dulbecco modified Eagle medium (1:2 ratio) supplemented with 20% human male AB serum (SeraCare Life Sciences), ethanolamine (50 μM), ascorbic acid (20 mg/L), 5 μg/L sodium selenite (NaSeO3), β-mercaptoethanol (24 μM; Sigma-Aldrich), and penicillin (100 U/mL) and streptomycin (100 U/mL). At the start of cultures, IL-3 (5 ng/mL), IL-7 (20 ng/mL), IL-15 (10 ng/mL), stem cell factor (SCF; 20 ng/mL), and Flt-3L (10 ng/mL) were added. Weekly thereafter, cultures were refed by semidepletion (50% volume change) supplemented with IL-7, IL-15, SCF, and Flt-3L as previously described.40
B-cell developmental potential was assayed using the UCB CD34+ cells (1000 cells per well)/murine MS-5 stromal cell (2 × 103 per well) culture in 96-well plates with Dulbecco modified Eagle medium-10% FBS supplemented with human G-CSF (10 ng/mL) and SCF (10 ng/mL) as described previously.41 To assess myelopoietic potential, transfected CD34+ cells were again cultured on MS-5 stromal cells in Dulbecco modified Eagle medium/20% FBS supplemented with IL-3 (10 ng/mL), SCF (50 ng/mL), IL-6 (10 ng/mL), and thrombopoietin (10 ng/mL). After overnight culture, the medium was replaced with Dulbecco modified Eagle medium/20% FBS supplemented with SCF, IL-3, and IL-6 (10 ng/mL each), and the cells were cultured for 28 days for flow cytometric analysis.42
Colony-forming unit assay
Colony-forming cell assays were performed using human methylcellulose complete media (catalog no. HSC003, R&D Systems), containing 25% FBS and erythropoietin (3 IU/mL), granulocyte macrophage-colony stimulating factor (GM-CSF; 10 ng/mL), interleukin-3 (IL-3; 10 ng/mL), and SCF (50 ng/mL). Transfected CD34+ cells were seeded in triplicate and cultured for 14 days. Colonies were scored in a blind manner using an inverted fluorescence light microscope (Leica DMIL S90). Images of colonies in methylcellulose complete media were taken at room temperature using the Magnafire 2.0 software (Optronics; original magnification, ×100).
Flow cytometric analysis
Single-cell suspensions were analyzed by staining with antibodies specific for CD34 (allophycocyanin [APC]/clone 581: phycoerythrin/clone 563), CD45 (APC/clone HI30; fluorescein isothiocyanate [FITC]/clone HI30), CD4 (APC/clone L200), CD8 (FITC/clone G428), CD7 (FITC/clone MT-701), CD1a (APC/clone HI149), CD56 (APC/clone B159), CD33 (APC/clone WM53), CD14 (APC/clone MϕP9), CD15 (APC/clone HI198), CD19 (FITC/clone 1D3), and CD16 (FITC/clone 3G8; BD Biosciences PharMingen). Data were collected on a FACSCalibur flow cytometer (BD Biosciences) and analyzed using FlowJo software (TreeStar).
Mapping transposition sites by linker-mediated PCR
Genomic DNA was isolated from differentiated T, B, NK, and myeloid cells 4 weeks after nucleofection and differentiation in in vitro culture and from bone marrow and spleen 12 weeks after transplantation using the Purgen DNA purification kit (QIAGEN). The transposition sites were cloned based on linker-mediated polymerase chain reaction (PCR) as described previously.26 Genomic DNA samples (1 μg) were digested with BfaI and ApoI and ligated to the linker flanking corresponding enzyme compatible sequence. Nested PCR products were purified, concentrated with PCR purification kit (QIAGEN), cloned into pCR2.1-TOPO vector (Invitrogen), and transformed into TOP10 cells (Invitrogen). Plasmid DNA was purified, and transposon/chromosome junctions were confirmed by EcoRI digests and sequencing. DNA sequencing was performed at the BioMedical Genomics Center at the University of Minnesota. The sequence results were subjected to BlastN analysis against the human genome using the University of California–Santa Cruz (UCSC) database. Bioinformatic analysis of insertion sites was performed at the Center for Functional Genomics, Northwestern University. Cancer gene data were obtained from CancerGenes at Memorial Sloan-Kettering Cancer Center and from the UCSC OMIM database. Transcriptional start site (TSS) data were extracted from the UCSC database originally from SwitchGear Genomics. The closest TSS on the same stand with the mapped region was used.
NOD-scid IL2γcnull engraftment assay
Six-week-old female NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (stock number 005557, abbreviated as NOD-scid IL2γcnull or NOG) mice were purchased from the Jackson Laboratory and housed in specific pathogen-free conditions at the University of Minnesota Animal Facility in accordance with institutional guidelines and approval. Nucleofected CD34+ cells were cultured overnight in 24-well plates in X-VIVO15 medium (Cambrex) supplemented with SCF (50 ng/mL), IL-3 (10 ng/mL), and IL-6 (20 ng/mL), washed, and resuspended in 250 μL phosphate-buffered saline. Mice were irradiated (2.5 Gy, cesium-137) in a J. L. Shepherd Mark 1 Model 30 Irradiator and intravenously injected with nucleofected CD34+ cells (8 × 104-2 × 105 live cells per mouse) within 6 hours of irradiation. Mice were killed 12 to 14 weeks after transplantation, and spleen and bone marrow tissue was harvested. Engraftment of human hematopoietic cells was assessed by immunophenotyping.
Statistical analyses
A two-way analysis of variance analysis with factors of treatment and cell lines was used to analyze the data in Table 1. To make the outcome closer to normal distribution and homogeneous variance, we chose the natural log transformation on the outcome. Two main factors and their interaction terms were tested. The overall mean of outcome for treatments was the main interested effect. The Tukey adjustment method was used to adjust the multiple comparison P values on the treatment effect. The Wilcoxon two-sample rank-sum test was use to analyze the data in Figure 1. Bonferroni P value adjustment was used for pair-wise comparison. Statistical analyses were conducted using SAS (Version 9.1) software. P values less than .05 were considered statistically significant.
Summary of DsRed+ cells after differentiation of SB100X-transfected CD34+ HPCs into lymphoid and myeloid lineages in vitro
| . | % of DsRed+ cells (> 3 weeks after transfection) . | ||
|---|---|---|---|
| Mock . | Transposon alone . | Transposon + SB100X . | |
| T cells | |||
| 1* | 0.80 | 0.37 | 19.37 |
| 2 | ND | ND | 6.23 |
| 3 | 0.12 | 0.26 | 3.17 |
| 4 | ND | ND | 21.84 |
| 5 | 0.28 | 0.40 | 5.67 |
| 6 | ND | ND | 7.97 |
| 7 | ND | ND | 13.80 |
| Median | 0.28 | 0.37 | 7.97 |
| B cells | |||
| 1 | 0.22 | 0.12 | 5.80 |
| 2 | ND | ND | 9.88 |
| 3 | 0.10 | 0.03 | 3.83 |
| 4 | ND | ND | 18.66 |
| Median | 0.22 | 0.12 | 7.84 |
| NK cells | |||
| 1 | 0.21 | 0.27 | 13.11 |
| 2 | ND | ND | 3.53 |
| 3 | 0.15 | 0.17 | 5.72 |
| 4 | ND | ND | 79.98 |
| 5 | 0.08 | 0.01 | 7.88 |
| Median | 0.15 | 0.17 | 7.88 |
| Myeloid cells | |||
| 1 | 0.14 | 0.61 | 11.37 |
| 2 | ND | ND | 6.11 |
| 3 | 0.39 | 0.19 | 15.63 |
| 4 | ND | ND | 7.59 |
| Median | 0.27 | 0.4 | 9.48 |
| . | % of DsRed+ cells (> 3 weeks after transfection) . | ||
|---|---|---|---|
| Mock . | Transposon alone . | Transposon + SB100X . | |
| T cells | |||
| 1* | 0.80 | 0.37 | 19.37 |
| 2 | ND | ND | 6.23 |
| 3 | 0.12 | 0.26 | 3.17 |
| 4 | ND | ND | 21.84 |
| 5 | 0.28 | 0.40 | 5.67 |
| 6 | ND | ND | 7.97 |
| 7 | ND | ND | 13.80 |
| Median | 0.28 | 0.37 | 7.97 |
| B cells | |||
| 1 | 0.22 | 0.12 | 5.80 |
| 2 | ND | ND | 9.88 |
| 3 | 0.10 | 0.03 | 3.83 |
| 4 | ND | ND | 18.66 |
| Median | 0.22 | 0.12 | 7.84 |
| NK cells | |||
| 1 | 0.21 | 0.27 | 13.11 |
| 2 | ND | ND | 3.53 |
| 3 | 0.15 | 0.17 | 5.72 |
| 4 | ND | ND | 79.98 |
| 5 | 0.08 | 0.01 | 7.88 |
| Median | 0.15 | 0.17 | 7.88 |
| Myeloid cells | |||
| 1 | 0.14 | 0.61 | 11.37 |
| 2 | ND | ND | 6.11 |
| 3 | 0.39 | 0.19 | 15.63 |
| 4 | ND | ND | 7.59 |
| Median | 0.27 | 0.4 | 9.48 |
ND indicates not done.
Number of experiments.
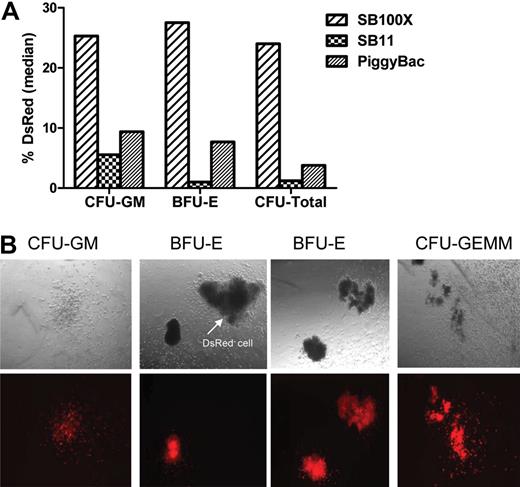
Transposon-mediated stable reporter gene expression in CD34+ HPCs. (A) Bar graph represents the percentage of DsRed+ colonies generated by CD34+ cells after nucleofection using 15 μg SB transposon with 5 μg SB100X or SB11, or using 15 μg piggyBac transposon plus 5 μg piggyBac transposase. Median of 5 replicates is shown. There were no DsRed+ colonies derived from CD34+ cells transfected with SB transposon plasmid without transposase (data not shown). (B) Morphology of DsRed+ progenitor colonies. (Top panel) Dark field microscopic view (original magnification, ×100) of human hematopoietic committed progenitors. (Bottom panel) DsRed fluorescence of the same colonies. BFU-E indicates burst-forming unit-erythrocyte; CFU-GM, colony-forming unit-granulocyte/macrophage; CFU-GEMM, colony-forming unit- granulocyte/erythrocyte/monocyte/macrophage.
Transposon-mediated stable reporter gene expression in CD34+ HPCs. (A) Bar graph represents the percentage of DsRed+ colonies generated by CD34+ cells after nucleofection using 15 μg SB transposon with 5 μg SB100X or SB11, or using 15 μg piggyBac transposon plus 5 μg piggyBac transposase. Median of 5 replicates is shown. There were no DsRed+ colonies derived from CD34+ cells transfected with SB transposon plasmid without transposase (data not shown). (B) Morphology of DsRed+ progenitor colonies. (Top panel) Dark field microscopic view (original magnification, ×100) of human hematopoietic committed progenitors. (Bottom panel) DsRed fluorescence of the same colonies. BFU-E indicates burst-forming unit-erythrocyte; CFU-GM, colony-forming unit-granulocyte/macrophage; CFU-GEMM, colony-forming unit- granulocyte/erythrocyte/monocyte/macrophage.
Results
A hyperactive SB100X transposase mediates high efficiency gene transfer and stable transgene expression in hematopoietic progenitor cells
To determine whether a hyperactive transposase, SB100X, can mediate stable gene transfer and expression in CD34+ hematopoietic progenitor cells (HPCs), we first optimized the amount of transposon (containing a DsRed reporter gene) and transposase-expressing plasmids required for efficient nucleofection of CD34+ HPCs. Colony-forming cell assays were used to quantify gene transfer efficiency. Supplemental Figure 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) shows that the greatest number of DsRed+ colonies (∼ 22%) was achieved when 15 μg transposon and 5 μg SB100X transposase (3:1 ratio) plasmid were used. After nucleofection with 15 μg SB transposon and 5 μg SB100X transposase-expressing plasmids and overnight culture, the viability of transfected CD34+ cells was 46.9% plus or minus 11.7% (n = 5) as determined by trypan blue exclusion. These results contrast with our previously published data showing that the highest level of long-term transgene expression was obtained in human primary T cells when 5 μg transposon and 10 to 15 μg SB10 or SB11 transposase-expressing plasmids (1:2-3 ratio) were conucleofected.26,27 All subsequent in vitro and in vivo nucelofection experiments (except Supplemental Figure 2) described herein were carried out using either 15 μg SB transposon with 5 μg SB100X transposase plasmids, or using 5 μg SB or piggyBac transposon with 10 to 15 μg SB11 or piggyBac transposase plasmids, respectively, to compare the ability of each transposase to mediate stable gene transfer and expression in CD34+ HPCs.
Figures 1A and 1B illustrate that the highest efficiency of stable gene transfer occurred in erythroid (22.41%-29.16%; median, 25.31% DsRed+ burst-forming unit-erythroid [BFU-E]), granulocyte and macrophage (19.48%-35.11%; median, 27.54% DsRed+ colony-forming unit-granulocyte/macrophage [CFU-GM]), and total lineage (21.23%-31.64%; median, 26% DsRed+) committed progenitor cells derived from CD34+ cells after transfection with SB transposon and SB100X transposase (SB100X vs SB11 and SB100X vs piggyBac, P = .024). The piggyBac transposase appeared as efficient as SB11 transposase in mediating piggyBac transposon gene transfer in BFU-E (2.18-17.64%; median, 9.38% vs 0%-7.69%; median, 5.56% DsRed+ colonies, P = 1), CFU-GM (2.4%-14.28%; median, 7.69% vs 0%-2.45%; median, 1% DsRed+ colonies, P = .09), and CFU-total (2.19%-6.09%; median, 3.8% vs 0%-4.88%; median, 1.23% DsRed+ colonies, P = .45; Figure 1A-B). As shown in Figure S2, the total number and number of BFU-E and CFU-GM colonies generated from SB transposon + SB100X transposase-transfected CD34+ cells was approximately 50% to 60% fewer than the number generated by mock-transfected CD34+ cells because of DNA toxicity. However, SB transposon + SB100X-transfected CD34+ cells generated approximately 50% more CFU-GM colonies compared with SB transposon + SB11 transposase and piggyBac transposon + transposase-transfected CD34+ cells. As expected, CD34+ cells transfected with either SB or piggyBac transposon alone failed to establish stable DsRed+ colonies (data not shown). These results demonstrate that the hyperactive SB100X transposase mediates higher efficiency stable gene transfer in CD34+ HPCs than SB11 or piggyBac transposase. We conclude that common myeloid progenitor, megakaryocyte/erythroid progenitor, and granulocyte/macrophage progenitor cells and/or primitive HSCs may have been transfected by both transposon and transposase, giving rise to red blood cells, megakaryocytes, and monocytes/macrophages stably expressing DsRed.
SB100X-mediated stable gene transfer in differentiated T, B, NK, and myeloid cells in vitro
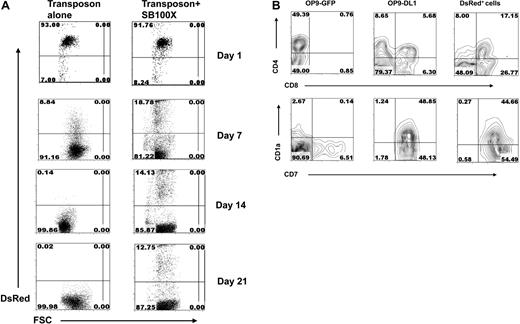
Next, we tested the capacity of transfected CD34+ HPCs to differentiate into lymphoid cells stably expressing the DsRed transgene. Transfected CD34+ HPCs were cultured on stromal cells in the presence of cytokines that promote differentiation to the T, B, and NK lineages. Figure 2A shows the kinetics of DsRed expression in CD34+ HPCs cultured on OP9-DL1 after transfection with either the SB transposon alone or the SB transposon with SB100X transposase. The percentage of DsRed+ cells was similar after one day regardless of whether cells were transfected with the transposon alone (∼ 93% DsRed+) or with both transposon and SB100X transposase (∼ 92% DsRed+). However DsRed expression in the transposon-only–transfected cells declined over time and after 14 days of culture was undetectable. Importantly, under the same culture conditions, DsRed expression was maintained (14% on day 14 and 13% on day 21) when cells were transfected with both SB transposon and SB100X transposase. At day 28, cells that were phenotypically early T cells were observed in cells cultured on OP9-DL1 but not on OP9-GFP control cells. DsRed+ early T cells were only observed in cultures initiated with SB transposon + SB100X transposase-transfected CD34+-transfected cells (Figure 2B).
T-cell development and transgene expression in SB100X-transfected CD34+ HPCs. (A) The percentage of DsRed+ cells after transfection of CD34+ HPCs with SB transposon alone or both transposon and SB100X transposase. Nucleofected CD34+ cells were cocultured onto OP9-DL1 stromal cells + Flt-3L and IL-7 and analyzed for DsRed expression by flow cytometric analysis on days 1, 7, 14, and 21. (B) T-cell development and phenotyping. Transfected CD34+ HPCs were cocultured with OP9-DL1 or OP9-GFP stromal cells for 28 days and assayed by flow cytometry for the expression of CD4, CD8, CD1a, and CD7. In CD34+ SB transposon + SB100X transposase-transfected HSCs, the DsRed+ cells were gated and analyzed for the expression of CD4/CD8 and CD1a/CD7.
T-cell development and transgene expression in SB100X-transfected CD34+ HPCs. (A) The percentage of DsRed+ cells after transfection of CD34+ HPCs with SB transposon alone or both transposon and SB100X transposase. Nucleofected CD34+ cells were cocultured onto OP9-DL1 stromal cells + Flt-3L and IL-7 and analyzed for DsRed expression by flow cytometric analysis on days 1, 7, 14, and 21. (B) T-cell development and phenotyping. Transfected CD34+ HPCs were cocultured with OP9-DL1 or OP9-GFP stromal cells for 28 days and assayed by flow cytometry for the expression of CD4, CD8, CD1a, and CD7. In CD34+ SB transposon + SB100X transposase-transfected HSCs, the DsRed+ cells were gated and analyzed for the expression of CD4/CD8 and CD1a/CD7.
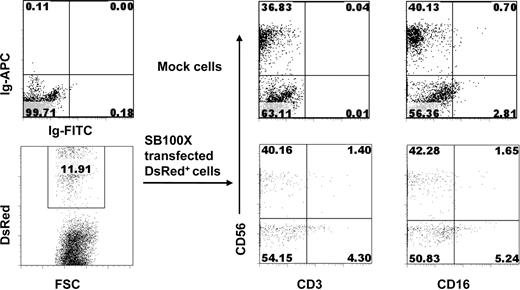
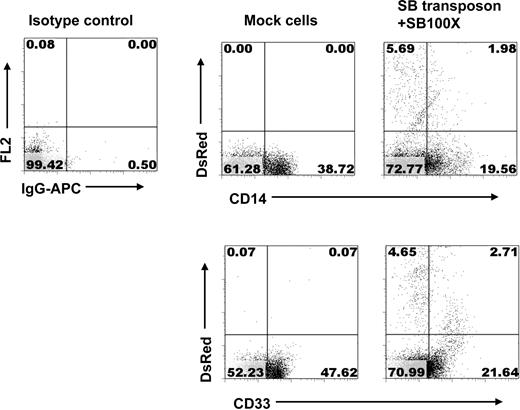
We next evaluated whether stable gene transfer by SB100X transposase can also occur in differentiated B, NK, and myeloid cells. The murine MS-5 stromal cell line was used to support development of CD19+ B-lineage cells from CD34+ HPCs after nucleofection. Figure 3 shows that after 28 days a greater percentage of DsRed+/CD19+ cells were derived from SB transposon and SB100X transposase-transfected CD34+ HPCs than from piggyBac transposon and piggyBac transposase-transfected CD34+ HPCs (∼ 18.66% vs 1.78%, respectively). As expected, untransfected, mock-transfected, or transposon-only-transfected CD34+ HPCs cultured in the same manner as controls all gave rise to CD19+ B-lineage cells, but no DsRed+ cells were observed. Figures 4 and 5 show that CD34+ cells transfected with SB transposon and SB100X transposase cultured on AFT024 murine fetal liver stromal cells in the presence of cytokines that promote NK cell development or on MS5 with cytokines that promote myeloid cell development gave rise to transgene-expressing NK cells (DsRed+/CD56+) and myeloid cells (DsRed+/CD33+).
Development of B cells stably expressing transgene from SB100X-transfected CD34+ HPCs. CD34+ cells were nucleofected with SB transposon alone or with SB transposon + SB100X, or with piggyBac transposon + piggyBac, transposase or without DNA, respectively. Transfected cells were cocultured with MS-5 in medium supplemented with SCF and G-CSF. At day 28, cells were collected and analyzed for expression of CD19 and DsRed by flow cytometry.
Development of B cells stably expressing transgene from SB100X-transfected CD34+ HPCs. CD34+ cells were nucleofected with SB transposon alone or with SB transposon + SB100X, or with piggyBac transposon + piggyBac, transposase or without DNA, respectively. Transfected cells were cocultured with MS-5 in medium supplemented with SCF and G-CSF. At day 28, cells were collected and analyzed for expression of CD19 and DsRed by flow cytometry.
Stable transgene expression in NK cells differentiated from SB100X-transfected CD34+ HPCs. Transfected CD34+ cells were cocultured with irradiated AFT024 cells and the indicated cytokines. On day 28, cells were harvested and analyzed by flow cytometry. DsRed+ cells were derived from SB transposon + SB100X-transfected CD34+ HPCs after coculture on AFT024 stromal cells.
Stable transgene expression in NK cells differentiated from SB100X-transfected CD34+ HPCs. Transfected CD34+ cells were cocultured with irradiated AFT024 cells and the indicated cytokines. On day 28, cells were harvested and analyzed by flow cytometry. DsRed+ cells were derived from SB transposon + SB100X-transfected CD34+ HPCs after coculture on AFT024 stromal cells.
Stable transgene expression in myeloid cells derived from SB100X-transfected CD34+ HPCs. After nucleofection, CD34+ cells were cocultured with MS-5 cells + SCF, IL-6, and IL-3. On day 28, the cells were analyzed with flow cytometry for expression of transgene and markers of myeloid cells.
Stable transgene expression in myeloid cells derived from SB100X-transfected CD34+ HPCs. After nucleofection, CD34+ cells were cocultured with MS-5 cells + SCF, IL-6, and IL-3. On day 28, the cells were analyzed with flow cytometry for expression of transgene and markers of myeloid cells.
Table 1 summarizes the efficiency of gene transfer in 4 to 7 independent experiments where differentiated T, B, NK, and myeloid cells were derived from SB transposon + SB100X transposase-transfected CD34+ HPCs. The overall frequency of cells stably expressing DsRed derived from CD34+ HPCs transfected with both SB transposon and SB100X transposase was significantly higher than that from CD34+ HPCs transfected with SB transposon alone (P < .001). No difference was observed between mock- and transposon-only–transfected CD34+ HPCs (P = .83).
To confirm that stable DsRed expression in T, B, NK, and myeloid cells was the result of transposition and not unintegrated episomal DNA, a linker-mediated PCR technique was used to recover sequences flanking transposon inserts on the 5′ end. SB transposase-mediated transposition requires a TA dinucleotide for integration. As summarized in Table 2, 31 representative integration sequences from T, B, NK, and myeloid cells were recovered at TA sites, the hallmark of transposition. These junction sequences were mapped to their intronic or intergenic location, and their proximity to cancer-related genes and TSSs on human chromosomes was noted. Taken together, these results demonstrate that the hyperactive SB100X transposase can mediate genomic integration and stable transgene expression in the lymphoid and myeloid progeny of CD34+ HPCs with high efficiency.
Molecular evidence of SB100X-mediated transposition in multilineage cells derived from human HPCs
| Cells/transposition site sequences . | Chromosome location:hit from . | Located gene . | Gene symbol . | Cancer-related gene . | Proximal TSS . | Distance to TSS, bp . |
|---|---|---|---|---|---|---|
| T | ||||||
| CAACTGTACATCCTTCATTCTAACTACTGAGTTAACTATCCA | chr12:32279251 | Intronic | BICD1 | BICD1 | chr12:32151168 | 128630 |
| CAACTGTACAGTATGGATGGCTCTCATAAATAGAATGTTGAG | chrX:40617753 | Intergenic | NA | NA | NA | NA |
| CAACTGTACAGTATGATTTCGTTTGGGTAAAAACAATGACAG | chr16:25261378 | Intergenic | NA | NA | NA | NA |
| CAACTGTATTATATGGAAATTATTATGCTAGTCCCTTAAGCG | chr12:27063331 | Intergenic | NA | NA | NA | NA |
| CAACTGTATTATTAAGTGCTAGTCCCTTAAGCGGAGCCCTAA | chrX:71598419 | Intronic | HDAC8 | HDAC8 | chrX:71731790 | 133350 |
| CAACTGTAAAATCTGCCCTTACTTACCTGCCCGCATCCTCGT | chr11:43002377 | Intergenic | NA | NA | NA | NA |
| CAACTGTACATTCCGACAGCCTGGGGAAATGGATCTTTGAGA | chr3:58431902 | Intergenic | NA | NA | NA | NA |
| CAACTGTATTTCAAACACTGAAGATCTGACTCAGGAAGTGCT | chr10:59705796 | Intronic | CISD1 | CISD1, MITONEET | chr10:59698939 | 6857 |
| B | ||||||
| CAACTGTACTAAGTATAGGCATCCTTAATTGGTGCAATTCTA | chr7:147222151 | Intronic | CNTNAP2 | CNTNAP2, CASPR2, NRXN4, CDFE, AUTS15 | chr7:146999704 | 222447 |
| CAACTGTATGTTGGAATGCCCCAGAATTTGGAGTTTATCTCT | chr13:45671894 | Intergenic | NA | NA | NA | NA |
| CAACTGTAGGCCTGAAAGCGCTCCAAATGTCCACTTCCAGA | chr2:91688345 | Intergenic | NA | NA | NA | NA |
| CAACTGTACATTCCGACAGCCTGGGGAAATGGATCTTTGAG | chr3:58431902 | Intergenic | NA | NA | NA | NA |
| CAACTGTATGTTATATATATATGCAAATATAAACACAGAAAA | chr11:72120697 | Intronic | CENTD2 | CENTD2, ARAP1, KIAA0782 | chr11:72141107 | 19946 |
| CAACTGTAAATCAGGTGAAGCCCTATTAAAGATGTCCTGAAA | chr18:35338281 | Intronic | AK090603 | (PIK3C3) | chr18:35634274 | 295573 |
| CAACTGTATTCTCAGAATATTTGCAACAATCACTCAAAAGGT | chr2:182456397 | Intergenic | NA | NA | NA | NA |
| CAACTGTACAAATCTGGAGTCCTTCCAAAACAGGACAAGTAA | chr12:3737128 | Intergenic | NA | NA | NA | NA |
| NK | ||||||
| CAACTGTAAGTTCCTTCCACAAAAATTGGGCAGCTTCTAGAAT | chr8:102548406 | Intergenic | NA | NA | NA | NA |
| CAACTGTACATATATAGTCTATTAATTGAGATAATATCTGTAA | chr2:192099107 | Intergenic | NA | NA | NA | NA |
| CAACTGTAGGTGTTTAGAGGGAAAGAAGAAAGGACATTCTGT | chr17:41583312 | Intronic | KIAA1267 | KIAA1267 | chr17:41605366 | 21942 |
| CAACTGTATAATTTTAGGTTACCATCTTCCATGGGGGAAATAT | chr12:69015183 | Intronic | CNOT2 | CNOT2, NOT2 | chr12:68923454 | 91729 |
| CAACTGTATATGGCACATGGGCTTTTGCAGGTGTGATGAAACT | chr16:30803923 | Intronic | BCL7C | BCL7C | chr16:30812887 | 8898 |
| CAACTGTATTCTCAGAATATTTGCAACAATCACTCAAAAGGTT | chr2:182456397 | Intergenic | NA | NA | NA | NA |
| CAACTGTAATATCCCAAGACTCTTTAAAGGTGGCAATGGCCG | chr7:294398 | Intronic | FAM20C | FAM20C, DMP4 | chr7:291895 | 2503 |
| M | ||||||
| CAACTGTACATACTTTCTTTCTTAAGGTAGTGTTTTGACAGAG | chr8:108578342 | Intronic | ANGPT1 | ANGPT1, ANG1 | chr8:108579262 | 920 |
| CAACTGTAGTTGAGGTCACACAAGACCTAAGTAGGGGAAACT | chr5:172192814 | Intergenic | NA | NA | NA | NA |
| CAACTGTACAATCATGTCGTCTGCGAACAGGGACAATTTGACT | chr5:38207284 | Intergenic | NA | NA | NA | NA |
| CAACTGTATATGTAAAGGTTTTTTTAAGTGGGTATATTGCGTGA | chr4:126348665 | Intergenic | NA | NA | NA | NA |
| CAACTGTAGATGTTGTGAGCATAATGAGTTAGGTGTTCCAAAG | chr3:178255770 | Intronic | TBL1XR1; TBLR1 | TBL1XR1 | chr3:178397855 | 141978 |
| CAACTGTAGCAACATGTTTAAGAGATTATACACCATGACCCAC | chr2:115950838 | Intronic | DPP10 | DPP10, DPRP3, KIAA1492 | chr2:115635383 | 315455 |
| CAACTGTATATACAGACTCTAAGTATGCTTACCTAGTCCCTTA | chr6:13990710 | Intergenic | NA | NA | NA | NA |
| CAACTGTATGTCCATCTATTGAGGCCCTAAATTAAGTCTACAG | chr5:131806233 | Intronic | LOC441108 | (IRF1, MAR) (SLC22A5, OCTN2, CDSP, SCD) | chr5:131774556 | 31677 |
| Cells/transposition site sequences . | Chromosome location:hit from . | Located gene . | Gene symbol . | Cancer-related gene . | Proximal TSS . | Distance to TSS, bp . |
|---|---|---|---|---|---|---|
| T | ||||||
| CAACTGTACATCCTTCATTCTAACTACTGAGTTAACTATCCA | chr12:32279251 | Intronic | BICD1 | BICD1 | chr12:32151168 | 128630 |
| CAACTGTACAGTATGGATGGCTCTCATAAATAGAATGTTGAG | chrX:40617753 | Intergenic | NA | NA | NA | NA |
| CAACTGTACAGTATGATTTCGTTTGGGTAAAAACAATGACAG | chr16:25261378 | Intergenic | NA | NA | NA | NA |
| CAACTGTATTATATGGAAATTATTATGCTAGTCCCTTAAGCG | chr12:27063331 | Intergenic | NA | NA | NA | NA |
| CAACTGTATTATTAAGTGCTAGTCCCTTAAGCGGAGCCCTAA | chrX:71598419 | Intronic | HDAC8 | HDAC8 | chrX:71731790 | 133350 |
| CAACTGTAAAATCTGCCCTTACTTACCTGCCCGCATCCTCGT | chr11:43002377 | Intergenic | NA | NA | NA | NA |
| CAACTGTACATTCCGACAGCCTGGGGAAATGGATCTTTGAGA | chr3:58431902 | Intergenic | NA | NA | NA | NA |
| CAACTGTATTTCAAACACTGAAGATCTGACTCAGGAAGTGCT | chr10:59705796 | Intronic | CISD1 | CISD1, MITONEET | chr10:59698939 | 6857 |
| B | ||||||
| CAACTGTACTAAGTATAGGCATCCTTAATTGGTGCAATTCTA | chr7:147222151 | Intronic | CNTNAP2 | CNTNAP2, CASPR2, NRXN4, CDFE, AUTS15 | chr7:146999704 | 222447 |
| CAACTGTATGTTGGAATGCCCCAGAATTTGGAGTTTATCTCT | chr13:45671894 | Intergenic | NA | NA | NA | NA |
| CAACTGTAGGCCTGAAAGCGCTCCAAATGTCCACTTCCAGA | chr2:91688345 | Intergenic | NA | NA | NA | NA |
| CAACTGTACATTCCGACAGCCTGGGGAAATGGATCTTTGAG | chr3:58431902 | Intergenic | NA | NA | NA | NA |
| CAACTGTATGTTATATATATATGCAAATATAAACACAGAAAA | chr11:72120697 | Intronic | CENTD2 | CENTD2, ARAP1, KIAA0782 | chr11:72141107 | 19946 |
| CAACTGTAAATCAGGTGAAGCCCTATTAAAGATGTCCTGAAA | chr18:35338281 | Intronic | AK090603 | (PIK3C3) | chr18:35634274 | 295573 |
| CAACTGTATTCTCAGAATATTTGCAACAATCACTCAAAAGGT | chr2:182456397 | Intergenic | NA | NA | NA | NA |
| CAACTGTACAAATCTGGAGTCCTTCCAAAACAGGACAAGTAA | chr12:3737128 | Intergenic | NA | NA | NA | NA |
| NK | ||||||
| CAACTGTAAGTTCCTTCCACAAAAATTGGGCAGCTTCTAGAAT | chr8:102548406 | Intergenic | NA | NA | NA | NA |
| CAACTGTACATATATAGTCTATTAATTGAGATAATATCTGTAA | chr2:192099107 | Intergenic | NA | NA | NA | NA |
| CAACTGTAGGTGTTTAGAGGGAAAGAAGAAAGGACATTCTGT | chr17:41583312 | Intronic | KIAA1267 | KIAA1267 | chr17:41605366 | 21942 |
| CAACTGTATAATTTTAGGTTACCATCTTCCATGGGGGAAATAT | chr12:69015183 | Intronic | CNOT2 | CNOT2, NOT2 | chr12:68923454 | 91729 |
| CAACTGTATATGGCACATGGGCTTTTGCAGGTGTGATGAAACT | chr16:30803923 | Intronic | BCL7C | BCL7C | chr16:30812887 | 8898 |
| CAACTGTATTCTCAGAATATTTGCAACAATCACTCAAAAGGTT | chr2:182456397 | Intergenic | NA | NA | NA | NA |
| CAACTGTAATATCCCAAGACTCTTTAAAGGTGGCAATGGCCG | chr7:294398 | Intronic | FAM20C | FAM20C, DMP4 | chr7:291895 | 2503 |
| M | ||||||
| CAACTGTACATACTTTCTTTCTTAAGGTAGTGTTTTGACAGAG | chr8:108578342 | Intronic | ANGPT1 | ANGPT1, ANG1 | chr8:108579262 | 920 |
| CAACTGTAGTTGAGGTCACACAAGACCTAAGTAGGGGAAACT | chr5:172192814 | Intergenic | NA | NA | NA | NA |
| CAACTGTACAATCATGTCGTCTGCGAACAGGGACAATTTGACT | chr5:38207284 | Intergenic | NA | NA | NA | NA |
| CAACTGTATATGTAAAGGTTTTTTTAAGTGGGTATATTGCGTGA | chr4:126348665 | Intergenic | NA | NA | NA | NA |
| CAACTGTAGATGTTGTGAGCATAATGAGTTAGGTGTTCCAAAG | chr3:178255770 | Intronic | TBL1XR1; TBLR1 | TBL1XR1 | chr3:178397855 | 141978 |
| CAACTGTAGCAACATGTTTAAGAGATTATACACCATGACCCAC | chr2:115950838 | Intronic | DPP10 | DPP10, DPRP3, KIAA1492 | chr2:115635383 | 315455 |
| CAACTGTATATACAGACTCTAAGTATGCTTACCTAGTCCCTTA | chr6:13990710 | Intergenic | NA | NA | NA | NA |
| CAACTGTATGTCCATCTATTGAGGCCCTAAATTAAGTCTACAG | chr5:131806233 | Intronic | LOC441108 | (IRF1, MAR) (SLC22A5, OCTN2, CDSP, SCD) | chr5:131774556 | 31677 |
Bold letters represent transposon sequence. Terms in parentheses indicate the closest neighboring cancer-related gene.
M indicates myeloid cells; NA, not applicable; and TSS, transcriptional start site.
In vivo engraftment and stable transgene expression in multilineage cells
To determine whether the SB-transfected CD34+ HPCs retain their ability to engraft and differentiate into multilineage cells with stable transgene expression, sublethally irradiated NOG mice were transplanted with cord blood CD34+ HPCs transfected either with transposon-only or both SB transposon and SB100X transposase. Twelve to 14 weeks after transplantation, the level of human CD34+ HPC engraftment and transgene expression from both bone marrow and spleen was evaluated by flow cytometry. As shown in Figure 6A and B, similar levels of engraftment in bone marrow and spleen were seen when the mice were transplanted with mock-, transposon-only-, or SB transposon + SB100X transposase-tranfected CD34+ HPCs. Importantly, a high percentage of DsRed+ cells was detected in both marrow and spleen after transplantation with SB transposon + SB100X transposase-transfected CD34+ HPCs, whereas no DsRed+ cells were observed in mice transplanted with transposon-only-transfected CD34+ HPCs. Mock-, SB transposon-only-, and SB transposon + SB100X transposase-transfected CD34+ HPCs were all capable of differentiating into CD19+ B, CD33+ myeloid, and CD14+ monocyte lineages in vivo (Figure 6C). Few CD3+ T cells were detected in any of the mice, confirming the lack of generation of human CD3+ T cells in NOG mice because of lack of cytokines, eg, tumor necrosis factor-α.43
Engraftment and stable expression of transgene in human CD45+ cells in NOG mice after transplantation of transfected CD34+ HPCs. (A) Engraftment and transgene expression in bone marrow cells at 12 weeks after transplantation. (B) Engraftment and transgene expression in spleen at 12 weeks after transplantation. (C) Phenotyping of bone marrow cells 12 weeks after transplantation. The percentage of DsRed+ and human CD19+, CD3+, CD33+, CD14+, and CD34+ cells is shown.
Engraftment and stable expression of transgene in human CD45+ cells in NOG mice after transplantation of transfected CD34+ HPCs. (A) Engraftment and transgene expression in bone marrow cells at 12 weeks after transplantation. (B) Engraftment and transgene expression in spleen at 12 weeks after transplantation. (C) Phenotyping of bone marrow cells 12 weeks after transplantation. The percentage of DsRed+ and human CD19+, CD3+, CD33+, CD14+, and CD34+ cells is shown.
Table 3 summarizes in vivo engraftment and gene transfer efficiency in 21 mice 12 to 14 weeks after transplantation. All mice receiving SB transposon + SB100X transposase-tranfected CD34+ HPCs had high levels of engraftment and stable transgene expression in B, T, myeloid, and monocytoid cells. Transgene transposition in engrafted human CD45+ cells was confirmed at the molecular level. Transgene integration sites were mapped to specific locations, and the proximity of the integration to cancer-related genes and TSSs was noted (Table 4). We conclude that SB100X transposase-transfected CD34+ HPCs retain their ability to engraft and differentiate into multiple lineages of cells, a high percentage of which exhibit sustained transgene expression.
In vivo engraftment of SB100X-transfected CD34+ HPCs
| CD34 cells/mouse ID . | BM . | Spleen . | ||
|---|---|---|---|---|
| hCD45+, % . | DsRed+hCD45+, % . | hCD45+, % . | DsRed+hCD45+, % . | |
| Mock | ||||
| 1 | 45.92 | 0.58 | 83.62 | 1.64 |
| 2 | 18.71 | 0.05 | 14.25 | 0.10 |
| 3 | 60.41 | 0.25 | 81.49 | 5.81 |
| Median | 45.92 | 0.25 | 81.49 | 1.64 |
| Transposon alone | ||||
| 4 | 63.73 | 0.60 | 48.00 | 0.36 |
| 5 | 27.17 | 0.34 | 47.24 | 3.22 |
| 6 | 42.87 | 0.00 | 48.01 | 0.15 |
| 7 | 55.02 | 0.63 | 39.41 | 0.66 |
| Median | 48.95 | 0.47 | 47.60 | 0.51 |
| Transposon + SB100X | ||||
| 8 | 64.61 | 1.32 | 70.81 | 2.59 |
| 9 | 23.29 | 2.92 | 23.69 | 13.43 |
| 10 | 8.68 | 1.61 | 33.08 | 0.49 |
| 11 | 45.95 | 7.32 | 26.85 | 7.11 |
| 12 | 29.39 | 7.62 | 18.94 | 5.89 |
| 13 | 22.74 | 4.02 | 47.99 | 3.32 |
| 14 | 70.73 | 27.14 | 69.99 | 18.11 |
| 15 | 76.82 | 1.56 | 25.39 | 7.37 |
| 16 | 47.42 | 8.25 | 52.95 | 6.22 |
| 17 | 39.42 | 2.65 | 57.30 | 3.99 |
| 18 | 16.37 | 28.96 | 18.07 | 34.50 |
| 19 | 68.73 | 0.57 | 43.29 | 4.28 |
| 20 | 78.67 | 0.69 | 69.83 | 5.29 |
| 21 | 55.57 | 1.06 | 72.16 | 2.12 |
| Median | 46.69 | 2.79 | 45.64 | 5.59 |
| CD34 cells/mouse ID . | BM . | Spleen . | ||
|---|---|---|---|---|
| hCD45+, % . | DsRed+hCD45+, % . | hCD45+, % . | DsRed+hCD45+, % . | |
| Mock | ||||
| 1 | 45.92 | 0.58 | 83.62 | 1.64 |
| 2 | 18.71 | 0.05 | 14.25 | 0.10 |
| 3 | 60.41 | 0.25 | 81.49 | 5.81 |
| Median | 45.92 | 0.25 | 81.49 | 1.64 |
| Transposon alone | ||||
| 4 | 63.73 | 0.60 | 48.00 | 0.36 |
| 5 | 27.17 | 0.34 | 47.24 | 3.22 |
| 6 | 42.87 | 0.00 | 48.01 | 0.15 |
| 7 | 55.02 | 0.63 | 39.41 | 0.66 |
| Median | 48.95 | 0.47 | 47.60 | 0.51 |
| Transposon + SB100X | ||||
| 8 | 64.61 | 1.32 | 70.81 | 2.59 |
| 9 | 23.29 | 2.92 | 23.69 | 13.43 |
| 10 | 8.68 | 1.61 | 33.08 | 0.49 |
| 11 | 45.95 | 7.32 | 26.85 | 7.11 |
| 12 | 29.39 | 7.62 | 18.94 | 5.89 |
| 13 | 22.74 | 4.02 | 47.99 | 3.32 |
| 14 | 70.73 | 27.14 | 69.99 | 18.11 |
| 15 | 76.82 | 1.56 | 25.39 | 7.37 |
| 16 | 47.42 | 8.25 | 52.95 | 6.22 |
| 17 | 39.42 | 2.65 | 57.30 | 3.99 |
| 18 | 16.37 | 28.96 | 18.07 | 34.50 |
| 19 | 68.73 | 0.57 | 43.29 | 4.28 |
| 20 | 78.67 | 0.69 | 69.83 | 5.29 |
| 21 | 55.57 | 1.06 | 72.16 | 2.12 |
| Median | 46.69 | 2.79 | 45.64 | 5.59 |
hCD45 indicates human CD45.
Molecular evidence of SB100X-mediated transposition in vivo
| Tissue/transposition site sequences . | Chromosome location:hit from . | Sequence frequency . | Located gene . | Gene symbol . | Cancer-related gene . | Proximal TSS . | Distance to TSS, bp . |
|---|---|---|---|---|---|---|---|
| BM-12 | |||||||
| CAACTGTATATTGTCTAGTCCCTTAAGCGGAGCCCT | short sequence | 2 | NA | NA | NA | NA | NA |
| Spleen-12 | |||||||
| CAACTGTATAATTTTAGGTTACCATCTTCCATGGGGGAA | chr12:69,015,183 | 1 | Intronic | CNOT2 | CNOT2, NOT2 | chr12:68923454 | 91729 |
| CAACTGTAGCTTATTCAGATTTTGTATATACATAGAAAAT | chr1:225568155 | 1 | Intronic | CDC42BPA | CDC42BPA, PK428, MRCKA | chr1:225571573 | 3359 |
| CAACTGTACATTCCGACAGCCTGGGGAAATGGATCTTTG | chr3:58,431,902 | 1 | Intergenic | NA | NA | NA | NA |
| BM-18 | |||||||
| CAACTGTATGTCAAAATGCCCCTGTAGGCAGAACCTACA | multi | 3 | NA | NA | NA | NA | NA |
| CAACTGTACCAGGTACCTTCTCTGTGCCAGCCTCTTCCC | chr6:144,514,825 | 1 | Intronic | STX11 | STX11, FHL4, HPLH4, HLH4 | chr6:144513369 | 1456 |
| CAACTGTATGTCACAATGATCCCTGTAGGCAAAGCCTAG | multi | 2 | NA | NA | NA | NA | NA |
| Spleen-18 | |||||||
| CAACTGTATGTGTGGGTGAACCAGGTAGGAAGGTATGTG | chr12:62,625,888 | 1 | Intronic | SRGAP1 | SRGAP1, KIAA1304 | chr12:62524327 | 101561 |
| CAACTGTATATAGTATCTGGAGTTTCCTAGTCCCTTAAGC | multi | 1 | NA | NA | NA | NA | NA |
| CAACTGTATATAGTATCTGAAGTTTCCTAGTCCCTTAAGC | chr4:154,192,090 | 3 | Intergenic | NA | NA | NA | NA |
| CAACTGTATCAATGATAATGAAGAAGCTACAACTACATAT | chr9:93,833,074 | 3 | Intergenic | NA | NA | NA | NA |
| CAACTGTATCAAATGTAAGATACTAGTCCCTTAAGCGGAG | chr17:59,141,036 | 1 | Intronic | LYK5; GH1 | LYK5, PMSE; GH1, GHN | not available | NA |
| BM-14 | |||||||
| CAACTGTACTTGACCCCATTAAAATGTCAGTAAGTTGAATT | chr3:103,263,186 | 2 | Intergenic | NA | NA | NA | NA |
| CAACTGTACCAGGTACCTTCTCTGTGCCAGCCTCTTCCCT | multi | 1 | NA | NA | NA | NA | NA |
| CAACTGTAGCTTATTCAGATTTTGTATATACATAGAAAATA | chr1:225568155 | 1 | Intronic | CDC42BPA | CDC42BPA, PK428, MRCKA | chr1:225571573 | 3359 |
| Tissue/transposition site sequences . | Chromosome location:hit from . | Sequence frequency . | Located gene . | Gene symbol . | Cancer-related gene . | Proximal TSS . | Distance to TSS, bp . |
|---|---|---|---|---|---|---|---|
| BM-12 | |||||||
| CAACTGTATATTGTCTAGTCCCTTAAGCGGAGCCCT | short sequence | 2 | NA | NA | NA | NA | NA |
| Spleen-12 | |||||||
| CAACTGTATAATTTTAGGTTACCATCTTCCATGGGGGAA | chr12:69,015,183 | 1 | Intronic | CNOT2 | CNOT2, NOT2 | chr12:68923454 | 91729 |
| CAACTGTAGCTTATTCAGATTTTGTATATACATAGAAAAT | chr1:225568155 | 1 | Intronic | CDC42BPA | CDC42BPA, PK428, MRCKA | chr1:225571573 | 3359 |
| CAACTGTACATTCCGACAGCCTGGGGAAATGGATCTTTG | chr3:58,431,902 | 1 | Intergenic | NA | NA | NA | NA |
| BM-18 | |||||||
| CAACTGTATGTCAAAATGCCCCTGTAGGCAGAACCTACA | multi | 3 | NA | NA | NA | NA | NA |
| CAACTGTACCAGGTACCTTCTCTGTGCCAGCCTCTTCCC | chr6:144,514,825 | 1 | Intronic | STX11 | STX11, FHL4, HPLH4, HLH4 | chr6:144513369 | 1456 |
| CAACTGTATGTCACAATGATCCCTGTAGGCAAAGCCTAG | multi | 2 | NA | NA | NA | NA | NA |
| Spleen-18 | |||||||
| CAACTGTATGTGTGGGTGAACCAGGTAGGAAGGTATGTG | chr12:62,625,888 | 1 | Intronic | SRGAP1 | SRGAP1, KIAA1304 | chr12:62524327 | 101561 |
| CAACTGTATATAGTATCTGGAGTTTCCTAGTCCCTTAAGC | multi | 1 | NA | NA | NA | NA | NA |
| CAACTGTATATAGTATCTGAAGTTTCCTAGTCCCTTAAGC | chr4:154,192,090 | 3 | Intergenic | NA | NA | NA | NA |
| CAACTGTATCAATGATAATGAAGAAGCTACAACTACATAT | chr9:93,833,074 | 3 | Intergenic | NA | NA | NA | NA |
| CAACTGTATCAAATGTAAGATACTAGTCCCTTAAGCGGAG | chr17:59,141,036 | 1 | Intronic | LYK5; GH1 | LYK5, PMSE; GH1, GHN | not available | NA |
| BM-14 | |||||||
| CAACTGTACTTGACCCCATTAAAATGTCAGTAAGTTGAATT | chr3:103,263,186 | 2 | Intergenic | NA | NA | NA | NA |
| CAACTGTACCAGGTACCTTCTCTGTGCCAGCCTCTTCCCT | multi | 1 | NA | NA | NA | NA | NA |
| CAACTGTAGCTTATTCAGATTTTGTATATACATAGAAAATA | chr1:225568155 | 1 | Intronic | CDC42BPA | CDC42BPA, PK428, MRCKA | chr1:225571573 | 3359 |
Genomic DNA was extracted from total mouse bone marrow cells and spleen cells at 12–14 weeks after transplantation with SB transposon + SB100X transfected CD34+ HPCs.
Bold letters indicate transposon sequence; BM-12, bone marrow cells from the mouse 12; multi, multi chromosme locations due to the sequence similarity; NA, not applicable; and TSS, transcriptional start site.
Discussion
Several new observations with relevance to gene therapy in hematopoietic cells emerged from our study. First, a hyperactive SB transposase, SB100X, can achieve an approximately 19-fold and approximately 6.3-fold higher efficiency of stable gene transfer than the SB11 transposase and the piggyBac system, respectively, in committed HPCs. Second, SB transposon + SB100X transposase-transfected CD34+ HPCs are capable of differentiating into cells in the T, B, NK, and myeloid lineages while maintaining stable transgene expression in vitro. Third, SB transposon + SB100X transposase-transfected CD34+ HPCs can efficiently engraft and differentiate into CD19+ B, CD33+ myeloid, and CD14+ monocytoid lineage cells stably expressing a DsRed transgene in NOG mice. Molecular analyses confirmed that stable transgene expression in differentiated lymphoid and myeloid lineage cells in vitro and in vivo resulted from transposition and not expression of episomal DNA.
DNA transposons are the most frequently used mobile element for manipulating and transforming the genomes of prokaryotic and eukaryotic organisms. Recently, they have been harnessed for nonviral gene delivery and show promise for gene therapy applications in humans. Currently, the most widely used transposon system for preclinical gene therapy studies is SB, the first approved transposon vector for use in clinical trials in the United States.44 SB is a synthetic DNA transposon of the Tc1/mariner superfamily that was resurrected from the fish genome and uses a “cut-and-paste” mechanism of transposition.16 The piggyBac system, derived from the cabbage looper moth, represents the most active alternative transposon for gene delivery into mammalian cells.17,20-22
Both the SB and piggyBac systems have been shown to mediate efficient transposition and long-term expression in a wide range of vertebrate cells and tissues.15-36 However, the low efficiency of transposition in primary cell types, including human HSCs, is limiting for the development of certain applications. Our study demonstrates successful engineering of CD34+ HPCs to stably express a reporter gene as a consequence of transposon-mediated chromosomal integration while retaining the capability for multilineage differentiation using the recently developed SB100X hyperactive system.36 Importantly, SB transposon + SB100X transposase-transfected CD34+ HPCs can successfully engraft in NOG mice and differentiate into DsRed+ B, myeloid, and monocyte lineage cells. Although Hollis et al35 were able to demonstrate 1% to 6% of human CD34+ HPCs expressed an eGFP (enhanced green fluorescence protein) reporter gene regulated by the retroviral MNDU3 promoter after nucleofection using the SB system, stable reporter gene expression was absent in the cells that engrafted in immune-deficient mice. In that report, the original SB10 transposase, also regulated by the MNDU3 promoter, was used for transposition. In contrast, our use of the newest generation of SB transposase, hyperactive SB100X, regulated by the CMV promoter, has allowed us to achieve engraftment of human hematopoietic cells in NOG mice that retain expression of a DsRed reporter gene during differentiation into B, myeloid, and monocytoid lineage cells. Although it is known that the CMV promoter is a weaker promoter than MNDU3 in human hematopoietic cells,35 transient expression of SB100X in CD34+ cells (in this report) or in human primary T cells regulated by the CMV promoter (data not shown) is apparently sufficient to achieve high-level transposition. It is probable that the low level of stable gene transfer in CD34+ cells reported by Hollis et al35 results from the use of the lower efficiency SB10 transposase.
SB100X was created by in vitro evolution of transposase gene variants each containing amino acid replacements in the encoded transposase resulting in hyperactivity.36 SB100X contains a particular combination of hyperactive mutations that results in approximately 100-fold higher transposition activity compared side-by-side with the original SB10 transposase in HeLa cells, especially under experimental conditions where the availability of transposon DNA is limited in the cell.36 Thus, SB100X will be a superior reagent for stable gene transfer in hard-to-transfect cell lines as well as in primary cell types, such as HPCs. At present, it is not known why the SB100X transposase is so much more efficient than SB11 or piggyBac in mediating transposition in HPCs. One notable difference is that SB100X transposase appears to be more thermo-stable than SB10,36 but whether this difference may contribute to different efficiency of gene transfer in HPCs is not clear. Other possible explanations include enhanced binding affinity to a cofactor involved in transposition or reduced binding affinity to an inhibitor protein.45-47
One of the major advantages offered by the SB transposon system over γ-retroviruses and lentiviruses is random integration of the transgene without preferential targeting of actively transcribed genes.19 The regional preferences associated with SB integrations (39% RefSeq genes) are much less pronounced than with γ-retroviral (51%) and lentiviral vectors (83%).15,19 Importantly, microarray analysis revealed no correlation between the integration profile of SB and the transcriptional status of targeted genes,19 suggesting that SB might be a safer vector for gene therapy. In support of this view, we have demonstrated by genome-wide analysis that SB integrants in primary human T cells are randomly distributed with respect to TSSs, CpG island regions, and DNase hypersensitive sites (X.H. et al, unpublished data). It has been well documented that MLV-derived γ-retroviral vectors favor integration within transcribed genes and around promoters and CpG islands.9 In contrast, lentiviral vectors strongly favor integration within active transcriptional units while showing no particular preference for promoter regions.10 In addition, γ-retroviral but not lentiviral integration hot spots in human CD34+ HSCs are highly enriched in proto-oncogenes, cancer-associated common insertion sites, and growth-controlling genes.48 Indeed, insertional activation of proto-oncogenes (eg, LMO2, BMI1, CCND2) in T cells has been correlated with the occurrence of acute T-cell lymphoblastic leukemia in 4 patients after retrovirus-mediated gene therapy for X-SCID.11-13 However, it remains to be determined whether the SB system can be used safely for HSC gene therapy at a genome-wide level because the insertion site analysis presented in this report is not sufficient to assess the issue of insertional mutagenesis.
Another advantage offered by DNA transposons over viral vectors in gene transfer for some applications is that individual transposon insertions can be removed from transposed cells. Indeed, 3 recent papers29-31 have highlighted this unique advantage by showing that the piggyBac transposon/tranposase system can efficiently generate induced pluripotent stem cells. More importantly, piggyBac transposons are completely removable from their integration site without any residual change using subsequent transposase transfection. Although we did not observe a superior activity of piggyBac in CD34+ hematopoietic cells compared with SB100X, it is possible that the piggyBac system could be further improved.
The overall efficiency of reporter gene transfer observed in human cells isolated from the bone marrow and spleen of animals injected with cord blood-derived CD34+ cells (median, 2.79% and 5.59%, or mean, 6.84% and 8.19%, respectively, Table 3) is generally lower than that observed when lentiviral vectors are used (mean, 10%-12% of human cells in the BM and spleen of nonobese diabetic/severe combined immunodeficiency mice injected with cord blood-derived CD34+ cells transduced with lentivirus at an multiplicity of infection of 60).49 This reduced level of gene transfer might be the result of nucleofection toxicity (∼ 50% viability after nucleofection with 20 μg total DNA). However, it may be possible to improve SB100X-mediated in vivo transposition efficiency by cotransfection of SB100X mRNA and SB transposon DNA or using other means of transfection to reduce toxicity and increase transfection efficiency.
In conclusion, we have presented evidence that the nonviral SB transposon/SB100X transposase system can efficiently mediate gene transfer and stable transgene expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells and in multiple cell lineages when differentiated both in vitro and in vivo. Our results have the potential to facilitate the development of a SB transposon-based HSC therapy for patients with inherited and acquired immunodeficiencies, metabolic diseases, and hematologic malignancies.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr San Ming Wang and Dr Yeong C. Kim (Northwestern University, Evanston, IL) and Dr Zheng Jin Tu (University of Minnesota Supercomputing Institute, Minneapolis, MN) for help in integration site analysis, Dr Malcolm J. Fraser (University of Notre Dame, Notre Dame, IN) for providing piggyBac vectors, Dr Juan Carlos Zúňiga-Pflücker (University of Toronto, Toronto, ON) for OP9-DL1 and OP9-GFP cells, Dr Jeffery S. Miller (University of Minnesota, Minneapolis, MN) for AFT024 cells, and Ms Marianna Wong for technical assistance.
This work was supported by grants from the Children's Cancer Research Fund in Minneapolis, Alliance for Cancer Gene Therapy, the Gabrielle's Angel (formerly G&P) Foundation for Cancer Research, the Sidney Kimmel Foundation for Cancer Research Kimmel Scholar Program, the University of Minnesota Translational Research Grant, the University Minnesota Medical School Dean's Commitment, and Leukemia Research Fund at the University of Minnesota (X.Z.).
Authorship
Contribution: X.X. and X.H. designed and performed the research, analyzed the data, and wrote the paper; S.E.N. performed the research and the major editing of the paper; L. Ma performed the statistical analyses; L. Mátés, Z. Izsvák, and Z. Ivics provided critical reagents and edited the paper; T.W.L. and R.S.M. discussed the work and edited the paper; J.E.W. provided critical reagents, discussed the work, and edited the paper; X.Z. designed the research and wrote the paper.
Conflict-of-interest disclosure: R.S.M. has a financial interest in Discovery Genomics Inc. The remaining authors declare no competing financial interests.
Correspondence: Xianzheng Zhou, University of Minnesota Masonic Cancer Center, MMC 366, 420 Delaware St, Minneapolis, MN 55455; e-mail: zhoux058@umn.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal