The mechanism by which chronic thromboembolic pulmonary hypertension (CTEPH) develops after acute pulmonary thromboembolism is unknown. We previously reported that fibrin from CTEPH patients is relatively resistant to fibrinolysis in vitro. In the present study, we performed proteomic, genomic, and functional studies on fibrin(ogen) to investigate whether abnormal fibrin(ogen) might contribute to the pathogenesis of CTEPH. Reduced and denatured fibrinogen from 33 CTEPH patients was subjected to liquid chromatography–mass spectrometry analysis. Fibrinogen from 21 healthy controls was used to distinguish atypical from commonly occurring mass peaks. Atypical peaks were further investigated by targeted genomic DNA sequencing. Five fibrinogen variants with corresponding heterozygous gene mutations (dysfibrinogenemias) were observed in 5 of 33 CTEPH patients: Bβ P235L/γ R375W, Bβ P235L/γ Y114H, Bβ P235L, Aα L69H, and Aα R554H (fibrinogensSan Diego I-V). Bβ P235L was found in 3 unrelated CTEPH patients. Functional analysis disclosed abnormalities in fibrin polymer structure and/or lysis with all CTEPH-associated mutations. These results suggest that, in some patients, differences in the molecular structure of fibrin may be implicated in the development of CTEPH after acute thromboembolism.
Introduction
Chronic thromboembolic pulmonary hypertension (CTEPH) is a complication of acute thrombotic pulmonary embolism (PE) characterized by persistent obstruction of the proximal pulmonary arteries with fibrotic material, elevated pulmonary vascular resistance, and life-threatening right ventricular failure.1 Definitive treatment requires surgical resection of the obstruction by a highly invasive procedure known as pulmonary thromboendarterectomy.2 Because the mechanism of CTEPH development after acute PE is poorly understood, it is currently difficult to prevent CTEPH or predict its development after acute PE.
We recently reported that fibrinogen purified from patients with CTEPH and clotted in vitro is partially resistant to plasmin-mediated lysis, relative to healthy controls.3 This observation raises the possibility that structural or functional abnormalities of fibrinogen in CTEPH patients may confer resistance to fibrinolysis in vivo and constitute an important etiologic step in the development of CTEPH. Structurally abnormal fibrin might persist within thromboemboli and stimulate remodeling of the thrombotic material into vascular scars.
We undertook this study to search for fibrinogen structural variants and underlying genetic mutations (dysfibrinogenemias) that might be present in patients with CTEPH. Patient and control fibrinogen was analyzed for the presence of structural variants by mass spectroscopic analysis, and the presence of variants suggested by this analysis was further investigated by targeted fibrinogen gene sequencing. We observed 5 dysfibrinogenemias in our group of 33 CTEPH patients: 2 with genetic mutations reported in neither the open literature nor the single nucleotide polymorphism (SNP) databases (Seattle SNPs: National Heart, Lung, and Blood Institute [NHLBI] Program for Genomic Applications, http://pga.gs.washington.edu4 ; National Center for Biotechnology Information [NCBI] Single Nucleotide Polymorphism, http://www.ncbi.nlm.nih.gov/projects/SNP5 ) at the time of this writing.
Methods
Materials
Human α thrombin and human glu-plasminogen were purchased from Enzyme Research Laboratories. Recombinant human tissue-type plasminogen activator (t-PA), 2-chain form, was obtained from Burroughs Wellcome Company. A synthetic peptide corresponding to the first 18 residues of the β chain of human fibrin (designated B knob standard) was obtained from Bachem Americas. A murine monoclonal antibody (isotype IgG1) raised against the first 7 residues of the β chain of human fibrin (designated B knob antibody) was obtained from Hybritech Inc. A peroxidase-conjugated goat anti–mouse IgG (Fc fragment specific) was purchased from Jackson ImmunoResearch Laboratories. All other chemicals were reagent grade or better.
Patient enrollment
Patients who were being evaluated at the University of California, San Diego (UCSD) Medical Center for the diagnosis and treatment of CTEPH were considered for recruitment into the study. Patients with CTEPH were defined as those with cardiopulmonary symptoms lasting more than one month, accompanied by the objective findings of elevated right ventricular or pulmonary artery pressures as well as pulmonary angiography confirming the characteristic obstruction of the pulmonary arteries at the segmental or more proximal levels.6 The diagnosis was confirmed by histologic examination of the obstructive tissue removed during pulmonary thromboendarterectomy or during autopsy.7 This material distinguishes patients with CTEPH from those with more unusual causes of chronic proximal pulmonary arterial obstruction, such as pulmonary artery sarcoma and pulmonary arteritis.
Forty consecutive patients being evaluated for CTEPH between October 2003 and February 2004 were enrolled in the study. Eight patients did not have hemodynamically significant CTEPH. Thirty-two remaining patients had CTEPH confirmed by the removal of significant amounts of intra-arterial organized thrombotic material during pulmonary thromboendarterectomy and were included in the analysis. One additional histologically confirmed CTEPH patient, referred because of postoperative thrombosis despite therapeutic-range INRs during warfarin therapy, was also analyzed, for a total of 33 CTEPH patients (mean age, 54 ± 13 years; 9 males and 24 females). One of these patients has been reported previously.8 Twenty-one healthy volunteers with no history of thrombotic diseases or events were enrolled to represent “typical mass spectrometry data” so that abnormalities could be visually identified.
After written informed consent was obtained in accordance with the Declaration of Helsinki, blood samples were collected from patients (before surgery) and controls. The patients' clinical courses were followed until discharge after pulmonary thromboendarterectomy. The UCSD Institutional Review Board approved this study.
Blood sample collection and processing
Blood (20 mL) was collected from each subject into a one-tenth volume of buffered sodium citrate (3.8%) and plasma was obtained by centrifugation at 2000g for 10 minutes at 4°C. The white blood cell layer was collected as a source of genomic DNA. Functional fibrinogen plasma levels were determined by the Clauss method and antigenic levels by a commercially available kit (Enzyme Research Laboratories) using the Factor Assay Control plasma from George King Bio-Medical as standard for both assays.
Fibrinogen was purified from plasma as previously described.3 The fibrinogen preparations were more than 90% pure, as judged by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) analysis under reducing conditions on 10% NuPAGE gels (Invitrogen), and contained more than 90% clottable protein. Genomic DNA was isolated using a QIAamp DNA Blood Kit (QIAGEN) according to the manufacturer's instructions. All samples were stored at −70°C until used.
Liquid chromatography–mass spectrometry
Fibrinogen was suspended in 100 mM Tris buffer (pH 8.0) containing 8 M urea and 20 mM dithiothreitol, and incubated for 4 hours at 37°C to denature the subunits and reduce disulfide bonds.8 The reduced protein (10 μg) was then acidified with 0.05% trifluoroacetic acid and injected onto a 1 mm × 50 mm, 300 Å C-4 reversed phase column (Phenomenex). Individual fibrinogen Aα, Bβ, and γ chains were eluted in that order with a linear acetonitrile gradient containing 0.05% trifluoroacetic acid and directed into an electrospray Micromass Q-tof mass spectrometer (Waters Corporation) with continuous acquisition of MS1 spectra (150-2000 m/z).8 Data were acquired and processed using MassLynx 4.5 software (Waters Corporation). The mass to charge (M/Z) range 1000 to 2000 was used to transform the data onto a true molecular mass scale using the maximum entropy (MaxEnt 1) component of the software. Peak areas were determined using the peak integration component of the MassLynx software.8
Screening of liquid chromatography–mass spectrometry spectra for uncommon fibrinogen variants
Normal liquid chromatography–mass spectrometry (LC-MS) fibrinogen component peaks (narrow, bilaterally symmetrical peaks that were observed in both patient and healthy volunteers and that corresponded to the predicted masses of common fibrinogen isotypes) were identified (Table 1), including the common fibrinogen polymorphisms Aα T312A and Bβ R448K.9 Variant LC-MS peaks (peaks with masses not found in Table 1, or wide asymmetrical peaks that suggested the superimposition of 2 distinct isotypes with mass differences too small to be resolved by LC-MS) were also identified.
Predicted and observed fibrinogen chain masses in 21 healthy subjects
| Fibrinogen chain/phosphorylation state* . | Sialylation state† . | Common polymorphisms . | Predicted mass, Da‡ . | Observed mass, mean (SD) . |
|---|---|---|---|---|
| Aα | ||||
| 312A | 66 102 | 66 101 (4) | ||
| 312T | 66 132 | 66 134 (4) | ||
| 1 | 312A | 66 182 | 66 180 (5) | |
| 1 | 312T | 66 212 | 66 214 (4) | |
| 2 | 312A | 66 262 | 66 259 (4) | |
| 2 | 312T | 66 292 | 66 293 (4) | |
| Bβ | ||||
| 1 | 448K | 54 185 | 54 182 (2) | |
| 1 | 448R | 54 213 | 54 213 (3) | |
| 2 | 448K | 54 476 | 54 473 (2) | |
| 2 | 448R | 54 504 | 54 504 (3) | |
| γ | ||||
| 1 | 48 368 | 48 381 (3) | ||
| 2 | 48 659 | 48 672 (3) |
| Fibrinogen chain/phosphorylation state* . | Sialylation state† . | Common polymorphisms . | Predicted mass, Da‡ . | Observed mass, mean (SD) . |
|---|---|---|---|---|
| Aα | ||||
| 312A | 66 102 | 66 101 (4) | ||
| 312T | 66 132 | 66 134 (4) | ||
| 1 | 312A | 66 182 | 66 180 (5) | |
| 1 | 312T | 66 212 | 66 214 (4) | |
| 2 | 312A | 66 262 | 66 259 (4) | |
| 2 | 312T | 66 292 | 66 293 (4) | |
| Bβ | ||||
| 1 | 448K | 54 185 | 54 182 (2) | |
| 1 | 448R | 54 213 | 54 213 (3) | |
| 2 | 448K | 54 476 | 54 473 (2) | |
| 2 | 448R | 54 504 | 54 504 (3) | |
| γ | ||||
| 1 | 48 368 | 48 381 (3) | ||
| 2 | 48 659 | 48 672 (3) |
Denotes the number of phosphate groups (80 Da each) attached to the chain.
Denotes the number of sialic acid groups (291 Da each) attached to the chain.
Predicted masses of the fibrinogen chains16 were used to identify predominant LC-MS peaks that would be expected in a normal population. These include nonphosphorylation, monophosphorylation, and diphosphorylation of the Aα chain; monosialylation and disialylation of the Bβ and γ chains; and the common polymorphisms Aα T312A and Bβ R448K, which have allele frequencies of 24% and 15%, respectively, in healthy subjects.9
Gene sequencing
Exons contained within the fibrinogen genes FGA, FGB, and FGG (encoding the Aα, Bβ, and γ fibrinogen chains, respectively) were amplified from 50 ng genomic DNA using 10 pmol each of forward and reverse primers in a 50-μL amplification mixture containing AmpliTaq Gold PCR Master Mix (Applied Biosystems). The samples were amplified for 35 cycles of denaturation (94°C, 30 seconds), annealing (58°C, 30 seconds), and extension (72°C, 30-90 seconds depending on fragment size), preceded by a 12-minute denaturation step at 94°C, and followed by a 7-minute extension step at 72°C. After confirmation of the presence of polymerase chain reaction (PCR) fragments of the expected size by electrophoresis in 2% agarose gels, PCR products were purified using a QIAquick PCR Purification Kit (QIAGEN). DNA sequencing of the PCR fragments was performed at the UCSD DNA Sequencing Facility according to standard protocols. All missense mutations were confirmed by sequencing of the complimentary strand. Primer sequences are available from the authors on request.
Frequency of CTEPH-associated mutations among historical controls
We performed a comprehensive literature search to determine whether the fibrinogen gene variations (other than the common polymorphisms described in Table 1) we observed in CTEPH patients had been reported among patients with acute venous thromboembolism (VTE). We searched the MEDLINE database10 using the following search terms: (“Fibrinogen/genetics”[Mesh] AND [“Venous Thromboembolism”[Mesh] OR “Pulmonary Embolism”[Mesh] OR “Venous Thrombosis”[Mesh]]) OR (“Venous Thrombosis/genetics”[Mesh] AND fibrinogen). The bibliographies from review articles were also searched for relevant publications. Identified articles were included in the analysis if they met the inclusion criteria recommended by Friedenreich,11 modified as follows: (1) the study sampled healthy subjects as well as patients with acute deep vein thrombosis or PE12 ; (2) subjects were selected for enrollment consecutively, randomly, or by a method unlikely to bias the genetic analysis; (3) the study was judged to be of “high quality” by 2 independent readers (T.A.M. and N.-C.L.); and (4) all exons from all 3 fibrinogen genes were sequenced and the results were reported in all enrolled subjects.
To estimate further the prevalence of the CTEPH-associated dysfibrinogenemias among the general population, we searched the NCBI and Seattle SNPs databases for the corresponding allele frequencies.
Molecular modeling of variant fibrinogens
The locations of each amino acid substitution were modeled with the “O” software package (Uppsala University, Uppsala, Sweden),13 using x-ray crystallographic data derived from fibrinogen.14,15 The most likely orientations of the amino acid side chains were estimated by the “O” package, in relation to the existing crystallographic data.
Turbidimetric fibrin clot lysis assay
Fibrin clots were simultaneously formed and lysed by combining fibrinogen (patient or control; 1.4 mg/mL), plasminogen (10 μg/mL), thrombin (0.2 NIH units/mL), t-PA (0.1 μg/mL), and CaCl2 (10 mM) in 200 μL TN buffer (50 mM Tris, pH 7.0 containing 150 mM NaCl). The mixture was immediately added to a microtiter plate well and the turbidity of the solution (absorbance at 405 nm) was monitored in a plate reader (Molecular Devices) at 1-minute intervals for a period of 1 hour at room temperature. Each curve was characterized by an increase in absorbance (clot formation), followed by brief plateau and then a decrease in absorbance (clot lysis) to baseline. Net turbidity was defined as the difference between the maximum and baseline absorbance values. The lysis rate was determined from the slope of the lysis portion of the curve at 50% clot lysis. A typical experiment included analysis of fibrinogen from 1 patient and 2 controls, and each experiment was repeated 2 to 3 times. The 2 controls used in each experiment were randomly selected from a set of fibrinogen preparations from 5 healthy controls. The relative turbidity (or lysis rate) was calculated as the net turbidity (or lysis rate) of the patient's fibrin relative to the average net turbidity (or lysis rate) of fibrin from 2 controls run in parallel.
Fibrin B knob release assay
As an independent method for evaluating fibrin lysis, an assay for measuring the release of B knob–containing fibrin fragments during lysis was developed. Fibrin clots were prepared in microcentrifuge tubes by combining fibrinogen (1.4 mg/mL), plasminogen (20 μg/mL), thrombin (0.2 NIH units/mL), and CaCl2 (10 mM) in 50 μL TN buffer. After clotting for 1 hour at room temperature, fibrinolysis was initiated by adding 50 μL t-PA (0.1 μg/mL) to the surface of the clot. After 6 hours of incubation at 37°C, d-Val-Phe-Lys-chloromethyl ketone was added (10 μM final concentration) to arrest plasmin activity, and the residual fibrin clot (about 25% of the initial amount) was removed by centrifugation. The supernatant was applied to a Microcon YM-30 Centrifugal Device (Millipore) and centrifuged (12 minutes, 10 000g) to remove intact fibrin, plasmin(ogen), and t-PA. The filtrate (containing fibrinolytic fragments) was then assayed for B knob–containing peptides using a competition enzyme-linked immunosorbent assay.
Enzyme-linked immunosorbent assay plate wells (Nunc Maxisorp) were coated with fibrinogen solution (2 μg/mL in TN buffer; 50 μL/well) for 3 hours at room temperature. After washing with PBS containing 0.05% Tween-20, wells were treated with thrombin solution (0.2 IU/mL in PBS containing 1 mg/mL BSA; 100 μL/well) for 1 hour, which simultaneously exposes the fibrin B knob on surface-bound fibrinogen and blocks residual protein binding sites on the wells. Meanwhile, preincubation mixtures composed of B knob antibody (0.5 μg/mL) and B knob standard (0-5 μM) or B knob antibody and subject-derived fibrinolytic fragments were each prepared in 220 μL PBS containing 1 mg/mL BSA and 0.05% Tween 20 (dilution buffer). After 3 to 4 hours of incubation, 100 μL of each preincubation mixture was applied to duplicate fibrinogen-coated/thrombin-treated wells that had been previously washed. After a 1-hour incubation, the wells were washed again and peroxidase-conjugated anti–mouse IgG antibody (1:5000 in dilution buffer; 100 μL/well) was added. After a 1-hour incubation, the wells were washed a final time and chromogenic peroxidase substrate (o-phenylene diamine; 100μL/well) was added. The reaction was stopped after 5 minutes by adding 3N H2SO4 (100 μL/well) and the absorbance at 490 nm of each well was read in a microplate reader. A standard curve was generated from a 4-parameter fit of concentration versus absorbance values of the standards. The concentration of B knob peptide in each sample was interpolated from the standard curve. Results of each patient sample are expressed as a percentage of the mean B knob peptide concentration of 14 different control samples included in the assay. The assay was validated by comparing the competition curve obtained with the synthetic B knob peptide with that of a crude peptide mixture derived from a complete plasmin digest of normal fibrin of known concentration. As shown in supplemental Figure 1 (available on the Blood website; see the Supplemental Materials link at the top of the online article), the 2 curves were essentially superimposable, demonstrating the suitability of the synthetic peptide as a standard in this assay.
Results
LC-MS of fibrinogen from healthy subjects
LC-MS data from 21 healthy subjects (mean age, 45.2 ± 10.6 years; 11 males, 10 females) were used to validate the precision and accuracy of the technique for measuring the masses of fibrinogen Aα, Bβ, and γ chains and to identify peaks corresponding to commonly occurring polymorphisms.16 Observed masses in the healthy subjects that corresponded to the calculated masses of the common fibrinogen isotypes16 are presented in Table 1.
LC-MS of fibrinogen from CTEPH patients
LC-MS data from 33 CTEPH patients and the 21 healthy controls were examined for the presence of fibrinogen chain peaks that are not found in the general population. Five CTEPH patients were observed to have abnormal peak patterns (Table 2). Patients nos. 1, 2, and 3 all exhibited a similar abnormal peak pattern in their Bβ chain spectra. An abnormal peak pattern was also observed in the γ chain spectrum of patient no. 2, and 2 distinct abnormal peak patterns were observed in the Aα chain spectra of patients nos. 4 and 5. No abnormal peak patterns were found among the control subjects.
Fibrinogen variants in CTEPH patients
| LC-MS findings . | Patients with the finding . | Gene . | Exon . | Nucleotide variation* (rs #)† . | Amino acid variation‡ . |
|---|---|---|---|---|---|
| Increase in the size of disialylated Bβ and γ peaks | 1 | FGG | 9 | 9163C>T | γ R375W |
| Additional γ peak pair 28 Da lighter than typical peak pair | 2 | FGG | 5 | 3977T>C (rs2066870) | γ Y114H |
| Atypical Bβ peak pair 14 Da heavier than typical peak pair | 1, 2, 3 | FGB | 5 | 7337C>T (rs6054) | Bβ P235L |
| Atypical Aα peak pattern 24 Da heavier than typical peak pattern | 4 | FGA | 3 | 4075T>A | Aα L69H |
| Additional Aα peak pattern 19 Da lighter than typical peak pattern | 5 | FGA | 5 | 7261G>A | Aα R554H |
| LC-MS findings . | Patients with the finding . | Gene . | Exon . | Nucleotide variation* (rs #)† . | Amino acid variation‡ . |
|---|---|---|---|---|---|
| Increase in the size of disialylated Bβ and γ peaks | 1 | FGG | 9 | 9163C>T | γ R375W |
| Additional γ peak pair 28 Da lighter than typical peak pair | 2 | FGG | 5 | 3977T>C (rs2066870) | γ Y114H |
| Atypical Bβ peak pair 14 Da heavier than typical peak pair | 1, 2, 3 | FGB | 5 | 7337C>T (rs6054) | Bβ P235L |
| Atypical Aα peak pattern 24 Da heavier than typical peak pattern | 4 | FGA | 3 | 4075T>A | Aα L69H |
| Additional Aα peak pattern 19 Da lighter than typical peak pattern | 5 | FGA | 5 | 7261G>A | Aα R554H |
Nucleotide numbering based on GenBank DNA accession numbers AF361104(FGA), AF388026(FGB), and AF350254(FGG).17 Mutations reported herein have been submitted to the NCBI Single Nucleotide Polymorphism database, and have been assigned the following identification (ss) numbers: 120032890 (FGG 9163C>T), 120032889 (FGG 3977T>C), 120032888 (FGB 7337C>T), 120032886 (FGA 4075T>A), and 120032887 (FGA 7261G>A).
Reference cluster identification numbers assigned in the SNP database, if previously reported.
Codon numbering based on the mature polypeptide chains.
Gene sequencing
Fibrinogen gene sequencing disclosed missense mutations corresponding to all the abnormal LC-MS peaks identified in the 5 CTEPH patients (Table 2). Patients nos. 1 and 2 were found to have 2 mutations each, one of which they had in common with each other and with patient no. 3. Targeted sequencing of genomic DNA from each of the 21 healthy controls was performed to search for the missense mutations that had been identified in the CTEPH patients. The search disclosed none of these mutations.
FibrinogenSan Diego I: compound of heterozygous γ R375W (associated with excessive Bβ and γ chain disialylation) and heterozygous Bβ P235L
CTEPH patient no. 1 exhibited a much higher percentage of disialylated Bβ chains (77.3%) and disialylated γ chains (56.0%) than the controls (40.8% ± 8.3% and 25.3% ± 6.6%, respectively) on mass spectrometry (supplemental Figure 2). DNA sequencing disclosed a heterozygous C>T substitution in exon 9 of the patient's FGG gene, resulting in a R375W amino acid substitution in the γ chain (corresponding to an R401W substitution in the precursor polypeptide). The mass difference affected by this substitution (−30 Da) was not reflected in the γ chain mass spectrum. However, fibrinogen molecules with this type of variant γ chain never reach the circulation,18,–20 but contribute to excessive disialylation of otherwise normal Bβ and γ chains (Figure 1). The Bβ P235L variant also found in this patient is described in “FibrinogenSan Diego III: heterozygous Bβ P235L.”
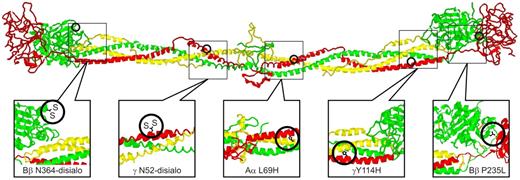
Structural locations of fibrinogen variants in patients with CTEPH. The positions of the alterations expected from the various mutations are illustrated on a ribbon model of chicken fibrinogen.21 In fibrinogenSan Diego I, the oligosaccharides attached to Bβ Asn-364 and γ Asn-52 are excessively disialylated, which imparts negative charges to the regions, thereby promoting the formation of “thin” fibrin fibers.22,23 A similar situation exists for fibrinogenSan Diego V (Aα R554H), in which the arginine→histidine substitution would also reduce the net charge in that region (not shown because it occurs in a region too flexible for crystallographic resolution). In fibrinogenSan Diego II (γ Y114H) and fibrinogenSan Diego IV (Aα L69H) the substitutions result in the insertion of a polar imidazole side chain within the “helix-permissive” hydrophobic center of the coiled coil,24 which could disrupt the molecular structure. In fibrinogensSan Diego I-III, the Bβ P235L substitution occurs at an articulation site in the βC region that ordinarily allows exposure of a t-PA binding site within the coiled coil. Substitution of leucine for the conformationally rigid proline in this area could affect the flexibility between the βC region and the coiled coil,14 which normally allows exposure of a region (circa Aα-157) that has been implicated in the activation of t-PA by fibrin.25
Structural locations of fibrinogen variants in patients with CTEPH. The positions of the alterations expected from the various mutations are illustrated on a ribbon model of chicken fibrinogen.21 In fibrinogenSan Diego I, the oligosaccharides attached to Bβ Asn-364 and γ Asn-52 are excessively disialylated, which imparts negative charges to the regions, thereby promoting the formation of “thin” fibrin fibers.22,23 A similar situation exists for fibrinogenSan Diego V (Aα R554H), in which the arginine→histidine substitution would also reduce the net charge in that region (not shown because it occurs in a region too flexible for crystallographic resolution). In fibrinogenSan Diego II (γ Y114H) and fibrinogenSan Diego IV (Aα L69H) the substitutions result in the insertion of a polar imidazole side chain within the “helix-permissive” hydrophobic center of the coiled coil,24 which could disrupt the molecular structure. In fibrinogensSan Diego I-III, the Bβ P235L substitution occurs at an articulation site in the βC region that ordinarily allows exposure of a t-PA binding site within the coiled coil. Substitution of leucine for the conformationally rigid proline in this area could affect the flexibility between the βC region and the coiled coil,14 which normally allows exposure of a region (circa Aα-157) that has been implicated in the activation of t-PA by fibrin.25
FibrinogenSan Diego II: compound of heterozygous γ Y114H and heterozygous Bβ P235L
The γ chain of patient no. 2 disclosed a variant peak pattern that was 28 Da lighter than expected (supplemental Figure 3). DNA sequence analysis revealed a heterozygous T>C substitution in exon 5 of the patient's FGG gene, resulting in a Y114H amino acid substitution in the fibrinogen γ chain (corresponding to a Y140H substitution in the precursor polypeptide). This mutation localizes to the coiled coil region of fibrinogen (Figure 1). The predicted mass difference affected by this substitution is −26 Da. The Bβ P235L variant also found in this patient is described in “FibrinogenSan Diego III: heterozygous Bβ P235L.”
FibrinogenSan Diego III: heterozygous Bβ P235L
The Bβ chain from patients nos. 1, 2, and 3 exhibited a variant peak pattern that was 15 Da heavier than expected (supplemental Figure 4). DNA sequencing disclosed that all 3 CTEPH patients had the same heterozygous C>T substitution in exon 5 of the FGB gene, resulting in a P235L amino acid substitution in the Bβ chain (a P265L substitution in the precursor polypeptide). This mutation localizes to the globular “D” region of fibrin(ogen) (Figure 1). The predicted mass difference affected by this substitution is +16 Da.
FibrinogenSan Diego IV: heterozygous Aα L69H
The Aα chain of patient no. 4 disclosed a variant peak pattern that was 24 Da heavier than expected (supplemental Figure 5). DNA sequence analysis revealed a heterozygous T>A substitution in exon 3 of the FGA gene, resulting in an L69H amino acid substitution in the Aα chain (an L88H substitution in the precursor polypeptide). This mutation localizes to the coiled coil region of fibrinogen (Figure 1) and the predicted mass difference affected by this substitution is +24 Da.
FibrinogenSan Diego V: heterozygous Aα R554H
The Aα chain of patient no. 5 disclosed a variant peak pattern that was 17 Da lighter than that expected for the common Aα T312A polymorphism26 (supplemental Figure 6). In addition to the T312A polymorphism, DNA sequence analysis disclosed a heterozygous G>A substitution in exon 5 of the FGA gene, resulting in a R554H amino acid substitution in the Aα chain (an R573H substitution in the precursor polypeptide). This mutation localizes to a region near the carboxyl terminus of the Aα chain outside of the globular D region. The observed mass difference is consistent with both genetic variants residing on the same allele in this patient.
Frequency of CTEPH-associated mutations among patients with VTE and in the general population
The MEDLINE search and subsequent bibliography review disclosed 86 candidate references, only one of which met the predetermined criteria for subject similarity, design, enrollment methodology, and quality.27 This publication enrolled 474 consecutively diagnosed patients with objectively confirmed first episodes of deep vein thrombosis (57.4% women) and 474 controls frequency matched for sex and age. However, the selected study did not report complete sequencing of all exons from all 3 fibrinogen genes. The study included only haplotype tagging of single nucleotide polymorphisms (htSNPs) that were common among the healthy subjects of European descent (n = 23) reported in the Seattle SNPs database. These polymorphisms did not include the mutations responsible for the 5 CTEPH-associated dysfibrinogenemias identified in the current study. In fact, the common Aα T312A variant (observed with an allele frequency of 20% in the referenced Seattle SNPs sample) was the only single nucleotide polymorphism studied that occurred in a coding region of the fibrinogen genes.
Clinical characteristics of CTEPH patients with variant fibrinogens
The clinical characteristics of CTEPH patients with variant fibrinogens are presented in Table 3. Three of the 5 patients had documented histories of previous venous thromboembolism, but no other systemic thrombotic disorders. None of the 5 patients had clinical, radiographic, or laboratory evidence of cirrhosis, liver dysfunction, or amyloidosis. Although severe hepatic dysfunction has been present in patients previously reported with heterozygous γ R375W,18,–20 our patient had no clinical manifestations of liver dysfunction and only a slight (< 10%) increase in alanine amino transferase upon presentation. Serum albumin, aspartate aminotransferase, alkaline phosphatase, and bilirubin levels were normal.
Clinical characteristics of CTEPH patients with fibrinogen variants
| Patient ID . | Fibrinogen variant . | Age, y/sex . | History* . | Severity† . | Duration of symptoms . | PVR, dyn•s/cm5 . | Plasma fibrinogen, mg/dL . | ||
|---|---|---|---|---|---|---|---|---|---|
| Preop . | Postop . | Clauss . | Antigen . | ||||||
| 1 | San Diego I | 23/M | No | Severe | 7 y | 311 | 157 | 94 | 96 |
| 2 | San Diego II | 45/F | No | Moderate | 9 mo | 1175 | 378 | 319 | 317 |
| 3 | San Diego III | 54/F | Yes | Mild | 18 mo | 1570 | 172 | 260 | 244 |
| 4 | San Diego IV | 49/M | Yes | Mild | 20 mo | 679 | 166 | NA | NA |
| 5 | San Diego V | 33/F | No | Moderate | 8 mo | 772 | 242 | 367 | 451 |
| Patient ID . | Fibrinogen variant . | Age, y/sex . | History* . | Severity† . | Duration of symptoms . | PVR, dyn•s/cm5 . | Plasma fibrinogen, mg/dL . | ||
|---|---|---|---|---|---|---|---|---|---|
| Preop . | Postop . | Clauss . | Antigen . | ||||||
| 1 | San Diego I | 23/M | No | Severe | 7 y | 311 | 157 | 94 | 96 |
| 2 | San Diego II | 45/F | No | Moderate | 9 mo | 1175 | 378 | 319 | 317 |
| 3 | San Diego III | 54/F | Yes | Mild | 18 mo | 1570 | 172 | 260 | 244 |
| 4 | San Diego IV | 49/M | Yes | Mild | 20 mo | 679 | 166 | NA | NA |
| 5 | San Diego V | 33/F | No | Moderate | 8 mo | 772 | 242 | 367 | 451 |
PVR indicates pulmonary vascular resistance; and NA, not available.
Family history of acute thrombosis.
Severity of initial acute PE: severe indicates unstable in ICU; moderate, hospitalized; and mild, outpatient.
Plasma fibrinogen levels were measured in 4 of the 5 patients with mutations (Table 3). Patient no. 1 had plasma fibrinogen levels that were just under half the lower limit of normal. Patients nos. 2, 3, and 5 had normal fibrinogen levels. There was insufficient plasma available to assay the fibrinogen levels of patient no. 4, who had been lost to follow up before further blood samples could be obtained; however, based on the yield of purified fibrinogen, hypofibrinogenemia in this patient seems unlikely.
Fibrin clot turbidity and susceptibility to lysis
Analysis of the purified variant fibrinogens by SDS PAGE under reducing conditions revealed typical heterogeneity of the Aα chains but no evidence of degradation of the fibrinogen chains (Figure 2). Turbidimetric clot lysis assays revealed abnormalities in fibrin clot turbidity, susceptibility to lysis, or both (Figure 3). Relative clot turbidities and lysis rates determined from the lysis curves as described in “Turbidimetric fibrin clot lysis assay” for each of the variant fibrinogens are presented in Table 4.
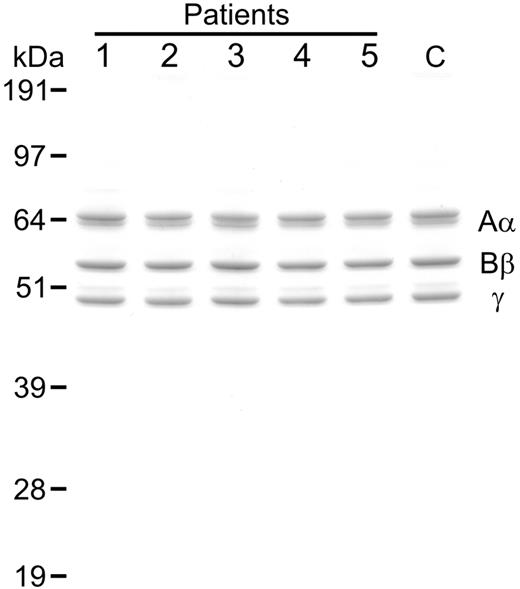
Analysis of purified fibrinogen. Samples of variant fibrinogen (1 μg) from CTEPH patients (patients nos. 1-5) as well as normal fibrinogen from a representative healthy control (C) were subjected to SDS PAGE (10% reducing gel) followed by staining with colloidal blue. Mobilities of the fibrinogen chains (Aα, Bβ, γ) and molecular mass markers (kilodaltons) are shown.
Analysis of purified fibrinogen. Samples of variant fibrinogen (1 μg) from CTEPH patients (patients nos. 1-5) as well as normal fibrinogen from a representative healthy control (C) were subjected to SDS PAGE (10% reducing gel) followed by staining with colloidal blue. Mobilities of the fibrinogen chains (Aα, Bβ, γ) and molecular mass markers (kilodaltons) are shown.
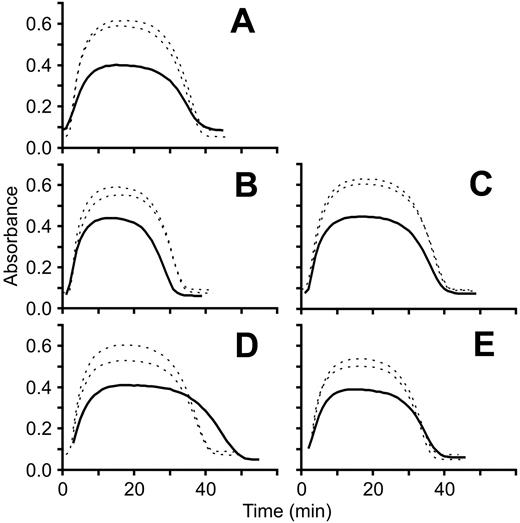
Turbidimetric clot lysis curves. Fibrin clot polymerization/lysis curves for each patient (solid line) were generated in parallel with 2 different healthy controls (dotted lines) by measuring absorbance at 405 nm at 1-minute intervals for 1 hour at room temperature. Each panel represents a separate experiment and each experiment was repeated 2 to 3 times. A representative experiment for each fibrinogen variant is shown. (A) FibrinogenSan Diego I, Bβ P235L/γ R375W; (B) fibrinogenSan Diego II, Bβ P235L/γ Y114H; (C) fibrinogenSan Diego III, Bβ P235L; (D) fibrinogenSan Diego IV, Aα L69H; (E) fibrinogenSan Diego V, Aα R554H.
Turbidimetric clot lysis curves. Fibrin clot polymerization/lysis curves for each patient (solid line) were generated in parallel with 2 different healthy controls (dotted lines) by measuring absorbance at 405 nm at 1-minute intervals for 1 hour at room temperature. Each panel represents a separate experiment and each experiment was repeated 2 to 3 times. A representative experiment for each fibrinogen variant is shown. (A) FibrinogenSan Diego I, Bβ P235L/γ R375W; (B) fibrinogenSan Diego II, Bβ P235L/γ Y114H; (C) fibrinogenSan Diego III, Bβ P235L; (D) fibrinogenSan Diego IV, Aα L69H; (E) fibrinogenSan Diego V, Aα R554H.
Fibrin clot turbidity and lysis
| Fibrinogen variant . | Amino acid substitution(s) . | Turbidity, % of control . | Lysis rate, % of control . | B knob release, % of control . |
|---|---|---|---|---|
| San Diego I | Bβ P235L/γ R375W | 59.3 | 56.9 | 66.3 |
| San Diego II | Bβ P235L/γ Y114H | 77.1 | 78.1 | 64.8 |
| San Diego III | Bβ P235L | 71.3 | 70.7 | 73.0 |
| San Diego IV | Aα L69H | 74.2 | 55.2 | 49.0 |
| San Diego V | Aα R554H | 71.9 | 64.6 | 80.2 |
| Fibrinogen variant . | Amino acid substitution(s) . | Turbidity, % of control . | Lysis rate, % of control . | B knob release, % of control . |
|---|---|---|---|---|
| San Diego I | Bβ P235L/γ R375W | 59.3 | 56.9 | 66.3 |
| San Diego II | Bβ P235L/γ Y114H | 77.1 | 78.1 | 64.8 |
| San Diego III | Bβ P235L | 71.3 | 70.7 | 73.0 |
| San Diego IV | Aα L69H | 74.2 | 55.2 | 49.0 |
| San Diego V | Aα R554H | 71.9 | 64.6 | 80.2 |
As an independent measure of clot lysis, t-PA–mediated release of B knob–containing peptide from preformed fibrin clots was also evaluated. In general, there was a reasonably good concordance between the lysis rate and B knob release for each patient (Table 4).
Discussion
CTEPH is an uncommon disease caused by the persistence of pulmonary thromboemboli and eventual organization into intra-arterial scars. We observed 5 dysfibrinogenemias in our group of 33 CTEPH patients. The prevalence of dysfibrinogenemia in our sample was 15% (95% confidence interval, 3%-27%). Although the prevalence of fibrinogen mutations in the general population has not been determined, 454 variants (of any type) have been reported in the Groupe d'Etudes sur l'Hemostase et la Thrombose (GEHT: Study Group on Hemostasis and Thrombosis) international database28,29 by the time of this writing.
The γ R375W mutation had been previously reported in a family without thrombotic disease.18,19 An additional 2 mutations that we found have been previously reported in the NCBI SNP database. The Bβ P235L mutation (rs6054) occurred with a frequency of 9.1% in our cohort of CTEPH patients, which is 11 times the frequency of heterozygotes reported in the database (0.8%). A previously published series, which had not been included in our structured literature review, found the Bβ P235L mutation with an allele frequency of 5% among 31 healthy subjects enrolled in New Zealand.30 The differences in the frequencies of the Bβ P235L mutation between that sample and the population samples included in the NCBI database, our own healthy subjects, and the reports included in our literature review are as yet unexplained. The γ Y114H polymorphism (rs2066870) occurred with a frequency of 3.0% in our cohort of CTEPH patients. The overall average frequency for heterozygous rs2066870 was not computed in the SNP database, but in a sample composed of approximately 50% persons of African descent, the frequency was 4.8%. Of note, our CTEPH patient with this polymorphism is of African descent. No mutations were found in our healthy control group aside from the common polymorphisms listed in Table 1.
Unfortunately, our structured literature review did not disclose sufficient evidence to determine whether the CTEPH-associated dysfibrinogenemias reported here are associated with VTE itself. The one study that met our criteria for clinical trial methodology did not search for the dysfibrinogenemias that we now report. An additional trial, which enrolled perimenopausal and postmenopausal women with first-time VTE (n = 349) and healthy women controls (n = 1680),31 was also a haplotype-tagged search for common polymorphisms, which did not include the 5 CTEPH-associated dysfibrinogenemias. In fact, none of the citations produced by our MEDLINE search (regardless of inclusion criteria) reported an association between the CTEPH-associated mutations we discovered and acute VTE itself.
The unusually high prevalence of these 5 dysfibrinogenemias in CTEPH patients raises the possibility that they have in common the capacity to impart resistance to the fibrinolytic mechanisms that normally lead to resolution of pulmonary thromboemboli. Modeling13 of the regions affected by fibrinogensSan Diego I-V within the structure of fibrinogen determined by x-ray crystallography discloses potential mechanisms by which each mutant could influence the structure of the fibrin clot (Figure 1). Our turbidity data suggest that the mutations may lead to “thin” fibrin fibers,22,23 which contain relatively few lateral protofibril associations (Table 4, Figure 3). Such fibers form clots that are transparent and more resistant to lysis than opaque clots composed of “thick” fibers.32 We speculate that alterations in net charge caused by oversialylation in fibrinogenSan Diego I or the arginine to histidine substitution in fibrinogenSan Diego V might cause abnormalities in fibrin polymerization. In fibrinogenSan Diego II and fibrinogenSan Diego IV, perturbations of the basic structure of the fibrin coiled coils, the regions targeted by fibrinolytic enzymes, might lead to fibrinolytic defects. Finally, the conformational changes resulting from fibrinogenSan Diego III could interfere with the binding of lytic agents to fibrin.
Our findings are consistent with a previous report of another heterozygous fibrinogen mutation associated with CTEPH, fibrinogen Bellingham (γ R275C).33 This variant is associated with a relative resistance to t-PA–mediated fibrinolysis, presumably related to steric interference with the normal polymerization/cross-linking structure due to the cysteine residue or a structure bound to it. Interestingly, relatives and other individuals with the γ R275C polymorphism alone did not develop CTEPH. The mutation was not sufficient to cause CTEPH; rather, CTEPH required the combination of acute PE (in the reported case, due to knee replacement surgery as well as an additive thrombogenic genetic risk factor) along with the resistance to fibrinolysis.
Although the molecular alterations we describe for the mutations are speculative, it seems unlikely that they occurred by coincidence in our small sample of patients with an extreme manifestation of unresolved pulmonary thromboembolism. It is more likely that the rare mutations we report are implicated in the development of CTEPH after acute pulmonary embolism. The fact that fibrinogen variants were not observed in all of the patients who we studied makes it likely that other clinical factors also contribute to this disease progression. Nevertheless, our findings support the conclusion that incomplete lysis by any number of means may predispose to CTEPH.
Our findings may also be relevant to PE patients who have not developed CTEPH, per se. After a PE, thrombotic material in the pulmonary arteries either resolves by fibrinolysis or is remodeled into organized scars. The extent to which remodeling occurs, and the severity of the resulting vascular obstruction, varies among patients. In some, lung perfusion is rapidly restored,34 although the resolution in the first week is typically incomplete. Recovery continues at a slower pace for the next 1 to 2 months,35,36 during which clot remodeling may be occurring. Residual defects persist commonly beyond this period, suggesting that the clots have been remodeled into permanent vascular scars.37 Long-term follow-up studies have consistently demonstrated that incomplete perfusion recovery, even years after acute PE, occurs in one-third to more than two-thirds of patients.38,,–41 Up to 15% of acute PE patients remain symptomatically compromised 2 years after treatment42 and may have abnormal pulmonary gas exchange (O2 gradients, dead space, etc) as well.43 These findings suggest that incomplete clot resolution has clinical manifestations in a significant proportion of acute PE patients, the most severe of which is CTEPH. We are currently organizing clinical studies to determine whether the fibrinogen variants we discovered would predict incomplete clot resolution in patients presenting with acute PE.
The online version of this article contains a data supplement.
Portions of this data have been presented in abstract form at the International Conference of the American Thoracic Society44,,–47 (2007 Conference, San Francisco, CA, May 23, 2007; 2008 Conference, Toronto, Canada, May 18, 2008) and the XXIst Congress of the International Society on Thrombosis and Haemostasis, Geneva, Switzerland, July 11, 2007.48,49
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr R. Doolittle for assistance with the x-ray crystallography ribbon diagrams and for critical reading of the paper; Dr M. Diccianni for assistance with development of the PCR and DNA sequencing strategies; and V. A. Woods for assistance with mass spectrometry data reduction. We also thank Dr N. Smith for his helpful comments regarding fibrinogen genetics and epidemiology. Finally, we thank the Pulmonary Thromboendarterectomy Service in the Division of Pulmonary and Critical Care Medicine at the University of California, San Diego (UCSD), which allowed its CTEPH patients to participate in this study.
This work was supported by National Institutes of Health grants HL-080302 and HL-095089 (T.A.M., J.J.M., and P.G.C.), CA099835, CA118595, and AI076961 (V.L.W.) as well as a Discovery Grant (bio06-10591) from the University of California Industry–University Cooperative Research Program, Biogen Idec, corporate sponsor (V.L.W.). DNA sequencing was performed by the UCSD DNA Sequencing Shared Resource, which is funded in part by a grant from the National Cancer Institute (2 P30 CA023100-23).
National Institutes of Health
Authorship
Contribution: T.A.M. designed experiments, analyzed data, performed statistical analysis, and wrote the paper; J.J.M. designed and performed experiments, analyzed data, and edited the paper; P.G.C. designed and performed experiments and prepared figures; M.M.M. designed and performed experiments and analyzed data; N.-C.L. performed experiments and statistical analysis; X.S. and D.J.D. analyzed data; D.N. compiled and analyzed clinical data; V.L.W. designed and performed experiments and analyzed data; and all authors reviewed the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Timothy Morris, UCSD Medical Center, 200 West Arbor Dr, San Diego, CA 92103-8378; e-mail: t1morris@ucsd.edu.