Abstract
Glucocorticoids potently attenuate the production of inflammatory mediators by macrophages, a primary effector of innate immunity. Activation of different macrophage Toll-like receptors (TLRs) by their respective ligands presents a powerful system by which to evaluate stimulus-dependent glucocorticoid effects in the same cell type. Here, we test the hypothesis that glucocorticoids, acting through the glucocorticoid receptor, modulate macrophage activation preferentially depending upon the TLR-selective ligand and TLR adapters. We established that 2 adapters, Trif, MyD88, or both, determine the ability of glucocorticoids to suppress inhibitor of κB (IκB) degradation or Janus kinase (JNK) activation. Moreover, the sensitivity of transforming growth factor β–activated kinase 1 (TAK1) activation to glucocorticoids determines these effects. These findings identify TAK1 as a novel target for glucocorticoids that integrates their anti-inflammatory action in innate immunity signaling pathways.
Introduction
Glucocorticoids, produced endogenously by activation of the hypothalamic-pituitary-adrenal axis, or administered exogenously in immunosuppressive therapy, provide an essential restraint to limit the magnitude and duration of inflammatory responses.1 Recent studies in mice, with conditional inactivation of glucocorticoid receptors in immune cells, have demonstrated this function essential for organism survival and therapeutic efficacy.2-5 Understanding the mechanisms by which glucocorticoids exert their immunomodulatory functions holds promise for developing new therapies that may better limit inflammation and reduce detrimental sequelae these drugs are known to have. Important questions persist regarding mechanisms of glucocorticoid/glucocorticoid receptor action. What are the key signal transduction pathways acted upon? Do these differ between cell types? Do they differ within a given cell type depending upon the nature of the activating stimulus? The macrophage provides a useful model system to address these questions by virtue of the diverse repertoire of receptors they express that sense foreign or autoantigens. Among the most important of these are the Toll-like receptors.
Toll-like receptors (TLRs) are phylogenetically conserved molecular sensors that recognize pathogen-associated molecular patterns.6,7 Upon recognizing the molecular pattern present on invading pathogens, TLRs activate components of the innate and adaptive immune systems to restrict pathogen spread.8 In support of the notion that TLR initiation of innate immune responses is finely tuned to the activating stimulus, an increasing complexity of hierarchic regulation has been revealed. This complexity begins with the specific TLR isoform engaged, with at least 12 membrane-bound family members identified. These receptors then recruit the cytosolic adapter proteins Mal, MyD88, Trif, and TRAM9,10 to propagate their signals to intracellular effector molecules. Most TLRs use MyD88 with the exception of TLR3, which exclusively recruits Trif.11 TLR4 is the only member of the TLR family that exploits both MyD88 and Trif to induce the downstream targets of the signaling cascade,7,12 and notably produces levels of cytokines exceeding that caused by activation of the other TLRs.13,14 MyD88 and Trif next activate the nuclear factor κB (NFκB) signalosome and mitogen-activated protein kinases (MAPKs); both pathways facilitate transcription and stabilization of mRNAs for proinflammatory mediators including cyclooxygenase-2 (COX-2), interleukin-6 (IL-6), IL-1β, tumor necrosis factor-α (TNF-α), and IL-12.15-17 Direct activation of transforming growth factor β–activated kinase 1 (TAK1), a member of MAPK kinase kinase (MAP3K) family is required for TLR-mediated proinflammatory cytokine secretion in macrophages.18 TAK1 functions as an upstream signaling mediator for both NFκB and MAPK activation during TLR engagement in different immune cell types including macrophages.19,20
A number of mechanisms have been implicated for glucocorticoid receptor (GR)–mediated inhibition of TLR signals in macrophages, including induction of suppressor of cytokine signaling 1 (SOCS1),21,22 suppression of the transactivation potential of NFκB, and activator protein 1 via induction of glucocorticoid-inducible leucine zipper,23,24 and induction of MAP kinase phosphatases (MKPs).25,26 Whether these actions differ depending upon the context of macrophage activation and whether TAK1 activity is glucocorticoid regulated as a proximal signal in activation cascades have not been studied.
Our previous work, using GR-deficient macrophages, has demonstrated a key role for MKP-1 in down-regulating p38 MAPK activation after TLR4 ligation with lipopolysaccharide (LPS).2 Surprisingly, although robust increases in Janus kinase (JNK) and extracellular signal-related kinase (ERK) phosphorylation also occurred with LPS treatment, these MAPKs were not attenuated by glucocorticoids. Thus, p38 MAPK appears to be a relatively specific target for anti-inflammatory actions of glucocorticoids in TLR4-activated macrophages. Little information is available regarding GR effects on activation of MAPKs in macrophages by other members of the TLR superfamily. In this study, we test the hypothesis that glucocorticoids differentially inhibit NFκB and specific MAPKs depending upon which TLR isoform is activated and through which specific cytosolic adapter protein it interacts. Moreover, we identify TAK1 as one of the key targets of glucocorticoids for the differential regulation of NFκB- and MAPK-mediated inflammatory reactions depending on the nature of TLR/adapter protein recruited.
Methods
Animal handing
Mice were housed on a 12-hour light and 12-hour dark cycle. Blood was collected by retro-orbital phlebotomy into heparinized capillary tubes, with the time from first handling the animal to completion of the bleeding not exceeding 30 seconds. Mice used for the experimentation were 6 to 10 weeks old and were of C57BL/6×129/Sv background. The experimental protocols were approved by the Animal Care and Use Committee of Vanderbilt University. Trif−/− and Lps2 mutant (Trif mutant) mice were generated as described.27,28 Mice with conditional deletion of GR in macrophages (MGRKO) were generated using LysM promoter-driven, Cre recombinase–mediated excision of exons 1C and 2 of the GR gene.3,29 Sex-matched LysM-Cre–negative homozygous floxed GR littermates were used as controls for the MGRKO mice.
Isolation and culture of mouse peritoneal macrophages
Thioglycollate-elicited macrophages were isolated by peritoneal lavage 3 days after intraperitoneal injection of 2.0 mL of sterile 4% thioglycollate (Becton Dickinson). Cells were plated in complete medium containing Dulbecco modified Eagle medium (Cambrex BioSciences) with 10% fetal bovine serum (Invitrogen Corporation) and penicillin/streptomycin (Invitrogen Corporation). After 3 hours of attachment, medium was aspirated and the monolayer rinsed twice with serum-free Dulbecco modified Eagle medium. Fresh complete medium was then added to each well, and the cells were cultured under a humidified atmosphere of 5% CO2 in air at 37°C.
Reagents and antibodies
Poly (I:C) (Amersham Biosciences Corp) was solubilized at 2 mg/mL in sterile phosphate-buffered saline (PBS) by warming the mixture at 55°C. Once solubilized, the solution was slowly cooled to room temperature to allow for renaturation. Aliquots were stored at −20°C until use. CpG oligodeoxynucleotide (ODN) 1826 (5′-tcc atg acg ttc ctg acg tt-3′) and control ODN 1826 (5′-tcc atg acg ttc ctg acg tt-3′) were purchased from InvivoGen. Dexamethasone (Dex; Amtech), LPS (Escherichia coli 0111:B4; Sigma-Aldrich), and NFκB activation inhibitor II JSH-23 (Calbiochem, EMD Biosciences) were purchased and reconstituted according to the manufacturers' instructions. Antibodies used in this study were as follows: anti–COX-2 (160126; Cayman); anti-actin (A 5060; Sigma-Aldrich); anti-phospho–stress-activated protein kinase/JNK (9251), anti–stress-activated protein kinase/JNK (9252), anti–phospho-p38 MAPK (9211), anti–p38 MAPK (9212), anti–phospho-p44/42 MAPK (9101), anti–phospho-TAK1 (4508), anti-TAK1 (4505), anti–inhibitor of κBα (IκBα; 9242), anti–phospho-IκBα (2859), anti–phospho-p65 (3033; Cell Signaling Technology); anti-ERK2 (sc-154), anti-GR (sc-1004), and anti-p65 (sc-109; Santa Cruz Biotechnology).
Cytokine measurement
For in vivo cytokine studies, mice were injected intraperitoneally with PBS (Cambrex BioSciences) or Poly (I:C) in PBS at a dose of 10 mg/kg body weight. Plasma samples were collected after the indicated periods of time and stored at −80°C until assay. Serum proinflammatory cytokine concentrations were measured by enzyme-linked immunosorbent assay (ELISA) for TNF-α, IL-6, and IL-12 according to the manufacturer's instructions (BD Biosciences PharMingen).
For in vitro cytokine studies, cells were plated at 106 cells/mL in a 24-well plate (Becton Dickinson Labware) with 1.0 mL of complete medium. Cells were treated with Dex (Amtech) for 3 hours at a concentration of 100nM (except as indicated) followed by a variety of TLR ligands for 24 hours or as indicated. LPS (100 ng/mL), Poly (I:C) (50 μg/mL), CpG (2μM), and control oligonucleotide (ODNc, 2μM) were used to stimulate TLR-4, TLR-3, and TLR-9, respectively. For studies with MAPK inhibitors, cells were treated with 5 or 20μM SB 203580 (p38 MAPK inhibitor), SP 600125 (JNK inhibitor), and PD 98059 (ERK inhibitor; Calbiochem, EMD Biosciences) at indicated concentrations for 1 hour followed by stimulation with respective TLR ligand. Supernatants were collected and assessed for TNF-α, IL-6, and IL-12 as described in the preceding paragraph.
Real-time quantitative PCR
Cells were frozen in liquid nitrogen, and stored at −80°C until processing. Total RNA was extracted from tissue samples using TriZol reagent, and converted to cDNA using Quantitect Reverse Transcription Kit (QIAGEN) that includes the step for elimination of genomic DNA. Prevalidated primers for il6 (NM_031168), tnf alpha (NM_013693), and il12(p40) (NM_008352,) were used for the real-time quantitative polymerase chain reaction (qPCR) as described (OriGene Technologies). Glyceraldehyde-3-phosphate dehydrogenase was used for standardization in all experiments. Table 1 shows the primer sequences used for the real-time qPCR experiments.
Primers used for real-time qPCR
| Gene ID . | Gene name . | Sequence . |
|---|---|---|
| il6 | IL-6 | 5′-TACCACTTCACAAGTCGGAGGC-3′; 5′-CTGCAAGTGCATCATCGTTGTTC-3′ |
| tnf alpha | TNFa | 5′-GGTGCCTATGTCTCAGCCTCTT-3′; 5′-GCCATAGAACTGATGAGAGGGAG-3′ |
| il 12b | IL-12(p40) | 5′-TTGAACTGGCGTTGGAAGCACG-3′; 5′-CCACCTGTGAGTTCTTCAAAGGC-3′ |
| gapdh | GAPDH | 5′-ATT GTG GAA GGG CTC ATG AC-3′; 5′-AGT GGA TGC AGG GAT GAT GT-3′ |
| Gene ID . | Gene name . | Sequence . |
|---|---|---|
| il6 | IL-6 | 5′-TACCACTTCACAAGTCGGAGGC-3′; 5′-CTGCAAGTGCATCATCGTTGTTC-3′ |
| tnf alpha | TNFa | 5′-GGTGCCTATGTCTCAGCCTCTT-3′; 5′-GCCATAGAACTGATGAGAGGGAG-3′ |
| il 12b | IL-12(p40) | 5′-TTGAACTGGCGTTGGAAGCACG-3′; 5′-CCACCTGTGAGTTCTTCAAAGGC-3′ |
| gapdh | GAPDH | 5′-ATT GTG GAA GGG CTC ATG AC-3′; 5′-AGT GGA TGC AGG GAT GAT GT-3′ |
For each pair of primers, the forward sequence is listed first, followed by the reverse sequence.
qPCR indicates quantitative polymerase chain reaction; IL, interleukin; TNF, tumor necrosis factor, and GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Reactions were completed on the Applied Biosystems 7900HT Fast Real-Time PCR System. PCR conditions were as follows: 1 cycle at 50°C for 2 minutes, 1 cycle at 95°C for 5 minutes, and 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. A subsequent step to generate a dissociation curve was used to verify that a single amplicon was generated. qPCR reactions were performed with QuantiTect SYBR Green PCR Kit (QIAGEN). Sequence Detection Software (V2.3) was used to determine the threshold cycle (Ct) value for each reaction. The ΔΔCt method was used for quantification. Statistical analysis was by 1-way ANOVA, followed by Bonferroni posthoc test. Comparisons were considered statistically significant when P values were less than .05.
Western blot analysis
For Western blot analysis, whole-cell extracts were resolved through sodium dodecyl sulfate–polyacrylamide gel electrophoresis using 4% to 12% separating gel (Invitrogen). Proteins were transferred to Hybond enhanced chemiluminescence nitrocellulose membrane (Amersham Pharmacia Biotech) using a semidry transfer system (Bio-Rad) and blocked with 5% dried milk in PBS and 0.1% Tween-20 (Sigma-Aldrich). Blots were probed with primary antibody overnight at 4°C. Binding of horseradish peroxidase–labeled goat anti–rabbit antibody (sc-2004) or goat anti–mouse antibody (sc-2005; Santa Cruz Biotechnology) was determined using SuperSignalWest Chemiluminescent Substrate (Pierce). Blots were stripped with Restore Western Blot Stripping Buffer (Pierce) and reprobed with different antibodies.
EMSA
For electrophoretic mobility shift assay (EMSA), cytoplasmic and nuclear extracts were prepared, resolved, and analyzed as described.30 EMSA was performed using NFκB-binding, 32P-labeled double-stranded DNA probes from major histocompatibility complex class I H2K promoter, 5′-CAGGCTGGGGATTCCCATCTCCACAGTTTCACTTC-3′. Bands were visualized using a PhosphorImager (Molecular Dynamics).
Statistical analysis
Data are expressed as the mean (± SEM). Statistical significance was tested by unpaired 2-tailed Student t test. The differences were considered to be significant for P value less than .05.
Results
Dex suppresses TLR3- and TLR9-evoked signaling pathways
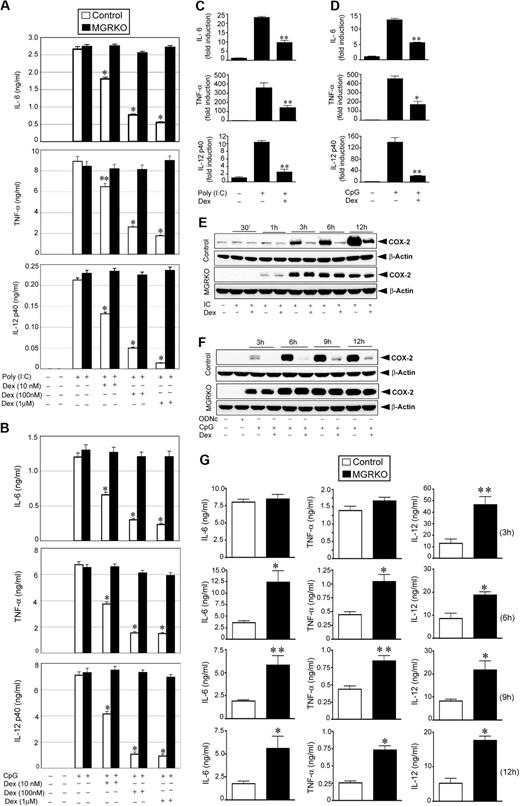
To determine the efficiency of ligand-activated GR to inhibit TLR signaling, we treated peritoneal macrophages from control and MGRKO mice with the TLR3 ligand, Poly (I:C), or the TLR9 ligand, CpG, in the presence and absence of Dex. Dex treatment resulted in dose-dependent inhibition of cytokine secretion in Poly (I:C)–treated control macrophages (Figure 1A), with 100 nM Dex, suppressing IL-6, TNF-α, and IL-12 by 72% to 77% in control macrophages compared with non–Dex-treated cells (P < .01). After the same treatment, cytokine secretion by MGRKO macrophages was not altered by Dex (Figure 1A). Activation of TLR9 by CpG showed a comparable profile for GR-mediated inhibition of cytokine secretion. At 100nM, Dex inhibited secretion of IL-6, TNF-α, and IL-12 by 75% to 85% in macrophages isolated from control mice compared with non–Dex-treated macrophages (P < .01; Figure 1B). MGRKO macrophages demonstrated no significant change in CpG-induced cytokine secretion with Dex (Figure 1B).
Dex suppresses TLR3- and TLR9-mediated inflammatory gene expression. Effect of Dexamethasone (Dex) on (A) Poly (I:C)–mediated (50 μg/mL) and (B) CpG-mediated (2μM) proinflammatory cytokine secretion in control and MGRKO macrophages. Cells were treated with the indicated doses of Dex for 3 hours followed by Poly (I:C) or CpG treatment for 24 hours. Concentrations of IL-6, TNF-α, and IL-12 in the culture media were analyzed by ELISA. Data are shown as mean ± SEM; n = 4; *P < .001 and **P < .005 for control macrophages compared with treatment group with Dex in MGRKO macrophages. Effect of Dex on (C) Poly (I:C)–mediated and (D) CpG-mediated proinflammatory cytokine mRNA transcript synthesis. Cells were pretreated with Dex followed by Poly (I:C) or CpG treatment for 4 hours. IL-6, TNF-α, and IL-12 mRNA was normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA, and expressed relative to that in untreated controls. Each bar in the figure represents the average across 3 independent experiments of 2 independently run experimental replicates. Data are shown as mean ± SEM; *P < .05 and **P < .01 for Dex-treated macrophages compared with treatment group treated with Poly (I:C) or CpG only. Effect of Dex on (E) Poly (I:C)–mediated and (F) CpG-mediated COX-2 expression. Cells were pretreated with Dex followed by Poly (I:C) or CpG treatment for the indicated periods of time. ODNc (2μM) was used as the oligonucleotide control for CpG. Cell lysates were analyzed by Western blot using anti–COX-2 and anti–β-actin antibodies. Representative of 4 to 5 independent experiments. (G) Deletion of GR in macrophages enhanced Poly (I:C)–induced proinflammatory responses in vivo. Control and MGRKO mice were treated with Poly (I:C) for the indicated periods of time (within parentheses). Plasma concentrations of IL-6, TNF-α, and IL-12 were analyzed by ELISA. Data presented as mean ± SEM; n = 8-10. *P < .001 and **P < .005 for MGRKO mice compared with treatment group with Poly (I:C) in control mice.
Dex suppresses TLR3- and TLR9-mediated inflammatory gene expression. Effect of Dexamethasone (Dex) on (A) Poly (I:C)–mediated (50 μg/mL) and (B) CpG-mediated (2μM) proinflammatory cytokine secretion in control and MGRKO macrophages. Cells were treated with the indicated doses of Dex for 3 hours followed by Poly (I:C) or CpG treatment for 24 hours. Concentrations of IL-6, TNF-α, and IL-12 in the culture media were analyzed by ELISA. Data are shown as mean ± SEM; n = 4; *P < .001 and **P < .005 for control macrophages compared with treatment group with Dex in MGRKO macrophages. Effect of Dex on (C) Poly (I:C)–mediated and (D) CpG-mediated proinflammatory cytokine mRNA transcript synthesis. Cells were pretreated with Dex followed by Poly (I:C) or CpG treatment for 4 hours. IL-6, TNF-α, and IL-12 mRNA was normalized to glyceraldehyde-3-phosphate dehydrogenase mRNA, and expressed relative to that in untreated controls. Each bar in the figure represents the average across 3 independent experiments of 2 independently run experimental replicates. Data are shown as mean ± SEM; *P < .05 and **P < .01 for Dex-treated macrophages compared with treatment group treated with Poly (I:C) or CpG only. Effect of Dex on (E) Poly (I:C)–mediated and (F) CpG-mediated COX-2 expression. Cells were pretreated with Dex followed by Poly (I:C) or CpG treatment for the indicated periods of time. ODNc (2μM) was used as the oligonucleotide control for CpG. Cell lysates were analyzed by Western blot using anti–COX-2 and anti–β-actin antibodies. Representative of 4 to 5 independent experiments. (G) Deletion of GR in macrophages enhanced Poly (I:C)–induced proinflammatory responses in vivo. Control and MGRKO mice were treated with Poly (I:C) for the indicated periods of time (within parentheses). Plasma concentrations of IL-6, TNF-α, and IL-12 were analyzed by ELISA. Data presented as mean ± SEM; n = 8-10. *P < .001 and **P < .005 for MGRKO mice compared with treatment group with Poly (I:C) in control mice.
Real-time qPCR experiments indicated 50% to 75% inhibition of Poly (I:C)–induced (4 hours of stimulation) synthesis of IL-6, TNF-α, and IL-12 mRNA transcript in Dex-treated control macrophages compared with non–Dex-treated macrophages (P < .01; Figure 1C). Dex caused 57% to 84% inhibition of CpG-mediated (4 hours of stimulation) IL-6, TNF-α, and IL-12 mRNA synthesis (Figure 1D). We observed a similar level of suppressive effect of Dex on mRNA synthesis after 2 hours of Poly (I:C) or CpG treatment (supplemental Figure 1A-B, available on the Blood website; see the Supplemental Materials link at the top of the online article). Similar pattern of inhibition on the synthesis of cyclooxygenase-2, a key enzyme for proinflammatory prostanoid production, by Dex after TLR3 or TLR9 engagement was also found (Figure 1E-F).
To evaluate the importance of GR engagement by endogenous glucocorticoid (GC) in macrophages in modulating effects of TLR3 activation in vivo, we measured the systemic release of proinflammatory cytokines after Poly (I:C) injection. After 6 to 12 hours of Poly (I:C) treatment, we observed 2- to 3.5-fold increases of plasma IL-6, TNF-α, and IL-12 concentrations in MGRKO mice in comparison with control mice (P < .01; Figure 1G). Previously, we reported a similar profile of systemic secretion of proinflammatory cytokine in control and MGRKO mice for in vivo TLR4 activation with LPS.2
Effect of NFκB inhibitor on TLR-mediated inflammatory cytokine secretion
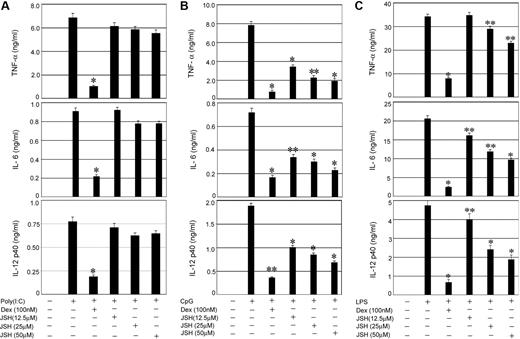
To establish the relative contribution of NFκB activity to the proinflammatory responses initiated by TLRs that signal through Trif, MyD88 plus Trif, or MyD88 alone, respectively, we used a specific inhibitor JSH-23 (JSH), which blocks nuclear translocation of NFκB.31-33 Whereas TLR3-Trif engagement with Poly (I:C) and subsequent induction of proinflammatory cytokines was Dex sensitive (Figure 2A), JSH-23 had little or no inhibitory effect on Poly (I:C)–mediated cytokine secretion. In contrast, 64% to 76% suppression of CpG-inducible IL-12, IL-6, and TNF-α secretion downstream of TLR9 was observed by treatment with 50μM JSH-23 (Figure 2B), implicating NFκB as a key component of MyD88-dependent inflammatory responses. Dex resulted in 77% to 90% inhibition of CpG-mediated cytokine secretion (Figure 2B). Engagement of TLR4 with LPS and coincident activation of Myd88-Trif–mediated cytokine secretion was also Dex sensitive, although only 15% to 50% inhibition of LPS-induced cytokine secretion was observed in JSH-23–treated cells (Figure 2C).
Effect of NFκB inhibition on TLR-mediated proinflammatory cytokine secretion. Control macrophages were pretreated with the indicated concentrations of the NFκB inhibitor (JSH-23) for 1 hour followed by stimulation with (A) Poly (I:C), (B) CpG, or (C) LPS for 24 hours. In other experimental sets, macrophages were pretreated with Dex (100nM) for 3 hours followed by TLR ligands as described. Concentrations of IL-6, TNF-α, and IL-12 in the culture media were measured. Data are shown as mean ± SEM; n = 3. *P < .01; and **P < .05 for Dex- or JSH-23–treated macrophages compared with treatment group treated with respective TLR ligand only.
Effect of NFκB inhibition on TLR-mediated proinflammatory cytokine secretion. Control macrophages were pretreated with the indicated concentrations of the NFκB inhibitor (JSH-23) for 1 hour followed by stimulation with (A) Poly (I:C), (B) CpG, or (C) LPS for 24 hours. In other experimental sets, macrophages were pretreated with Dex (100nM) for 3 hours followed by TLR ligands as described. Concentrations of IL-6, TNF-α, and IL-12 in the culture media were measured. Data are shown as mean ± SEM; n = 3. *P < .01; and **P < .05 for Dex- or JSH-23–treated macrophages compared with treatment group treated with respective TLR ligand only.
Effect of Dex on TLR-mediated activation of NFκB
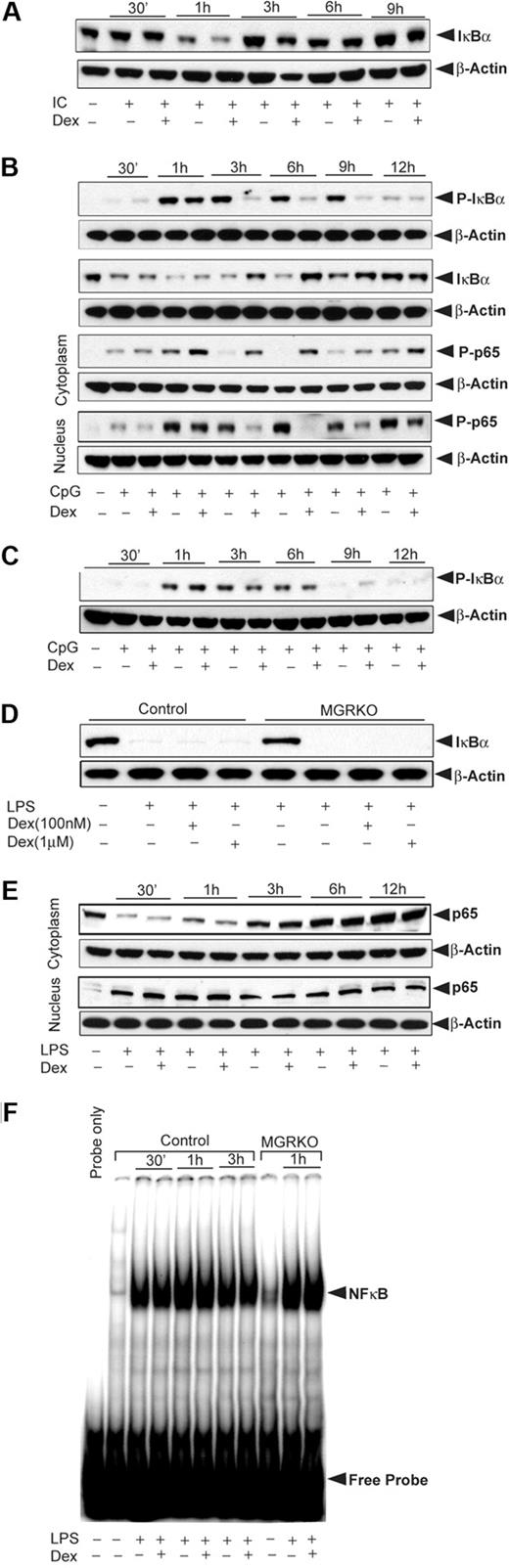
To explore the mechanisms by which GR regulates NFκB activity, macrophages from control and MGRKO mice were activated with TLR ligands in the presence and absence of Dex. Phosphorylation of IκBα is a prerequisite for IκBα degradation and NFκB activation.34 After TLR3 engagement, IκBα phosphorylation was not altered by Dex (data not shown), and Poly (I:C) treatment caused only transient, low-level IκBα degradation. Figure 3A shows 50% degradation of IκBα at 1 hour followed by rapid reaccumulation that was unchanged by Dex. This finding is consistent with the minimal effect of JSH-23 on TLR3-initiated macrophage responses.
Effect of Dex on TLR-3– and TLR-9–inducible NFκB activation. (A) Effect of Dex on Poly (I:C)–mediated IκBα degradation. Control macrophages were treated with Dex for 3 hours followed by Poly (I:C) for the indicated periods of time. Cytoplasmic extracts were analyzed by Western blotting using an anti-IκBα and anti–β-actin antibodies. (B) Effect of Dex on CpG-mediated NFκB activation in control macrophages. Cells were treated as described. Cytoplasmic and nuclear extracts were analyzed by Western blotting using anti–phospho-IκBα (P-IκBα), anti–total IκBα (IκBα), and anti–phospho-p65 (P-p65) antibodies. (C) Effect of Dex on CpG-mediated IκBα phosphorylation in MGRKO macrophages. Cytoplasmic extracts were analyzed by Western blotting using anti–phospho-IκBα and anti–β-actin antibodies. (D) Effect of Dex on LPS-mediated IκBα degradation. Macrophages from control and MGRKO mice were treated with the indicated doses of Dex for 3 hours followed by LPS for 30 minutes. Cytoplasmic extracts were analyzed by Western blotting using anti-IκBα and anti–β-actin antibodies. (E) Effect of Dex on LPS-mediated p65 redistribution. Cytoplasmic and nuclear extracts were analyzed by Western blotting using anti-p65 and anti–β-actin antibodies. (F) Effect of Dex on LPS-mediated NFκB DNA-binding activity. Nuclear extracts from control and MGRKO macrophages were prepared and DNA-binding activity to major histocompatibility complex class I H2K–specific oligonucleotide probe were analyzed using EMSA. Representative of 3 to 4 independent experiments.
Effect of Dex on TLR-3– and TLR-9–inducible NFκB activation. (A) Effect of Dex on Poly (I:C)–mediated IκBα degradation. Control macrophages were treated with Dex for 3 hours followed by Poly (I:C) for the indicated periods of time. Cytoplasmic extracts were analyzed by Western blotting using an anti-IκBα and anti–β-actin antibodies. (B) Effect of Dex on CpG-mediated NFκB activation in control macrophages. Cells were treated as described. Cytoplasmic and nuclear extracts were analyzed by Western blotting using anti–phospho-IκBα (P-IκBα), anti–total IκBα (IκBα), and anti–phospho-p65 (P-p65) antibodies. (C) Effect of Dex on CpG-mediated IκBα phosphorylation in MGRKO macrophages. Cytoplasmic extracts were analyzed by Western blotting using anti–phospho-IκBα and anti–β-actin antibodies. (D) Effect of Dex on LPS-mediated IκBα degradation. Macrophages from control and MGRKO mice were treated with the indicated doses of Dex for 3 hours followed by LPS for 30 minutes. Cytoplasmic extracts were analyzed by Western blotting using anti-IκBα and anti–β-actin antibodies. (E) Effect of Dex on LPS-mediated p65 redistribution. Cytoplasmic and nuclear extracts were analyzed by Western blotting using anti-p65 and anti–β-actin antibodies. (F) Effect of Dex on LPS-mediated NFκB DNA-binding activity. Nuclear extracts from control and MGRKO macrophages were prepared and DNA-binding activity to major histocompatibility complex class I H2K–specific oligonucleotide probe were analyzed using EMSA. Representative of 3 to 4 independent experiments.
In contrast to Poly (I:C) treatment, Dex pretreatment caused marked inhibition of CpG-induced IκBα phosphorylation in control macrophages (Figure 3B). This inhibitory effect was sustained for several hours. We observed comparable inhibition of IκBα degradation when macrophages were treated with Dex and CpG simultaneously (supplemental Figure 2). There was no effect of Dex on IκBα phosphorylation in MGRKO macrophages (Figure 3C). In accord with the attenuation of IκBα phosphorylation, CpG-mediated IκBα degradation in control macrophages was also inhibited by Dex (Figure 3B). In addition, Dex impaired translocation of phospho-p65 (Rel A) to the nucleus. Figure 3B shows cytoplasmic retention of phospho-p65 in Dex-treated macrophages. LPS induced IκBα degradation as early as 15 minutes after treatment, and this effect persisted at 1 hour after which IκBα reaccumulated in the cytoplasm (supplemental Figure 3). Irrespective of dose, Dex did not inhibit LPS-mediated IκBα degradation in either control or MGRKO macrophages (Figure 3D). Moreover, Dex had no effect on cytoplasmic or nuclear abundance of p65, indicating sustained nuclear localization of p65 after GR activation (Figure 3E). Because LPS-induced cytokine synthesis was GR suppressible (Figure 2C), we also examined LPS-mediated DNA-binding activity of NFκB. NFκB DNA-binding activity was found to be unaffected by Dex treatment for control or MGRKO macrophages (Figure 3F).
Effect of MAPK inhibitors on TLR3- or TLR9-mediated inflammatory cytokine secretion
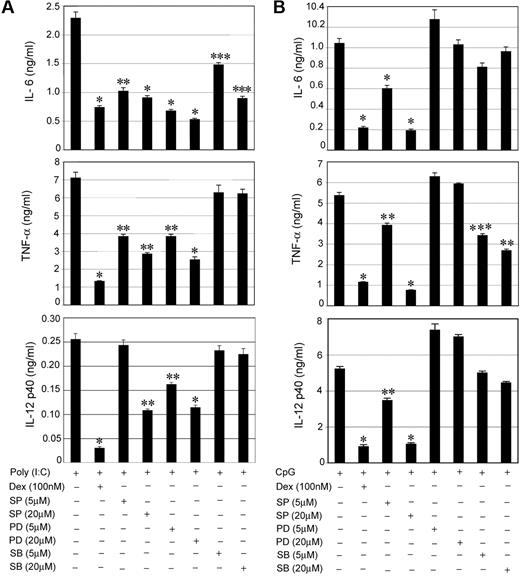
To explore the relative importance of individual MAPKs in TLR-mediated proinflammatory responses, specific pharmacologic inhibitors were used for members of the MAPK family. SP 600125 (SP), PD 98059 (PD), and SB 203580 (SB) were used to inhibit JNK, ERK, and p38 MAPK, respectively. For Poly (I:C), pretreatment with the individual MAPK inhibitors showed suppressive effects for IL-6, TNF-α, and IL-12 secretion (Figure 4A). SP and PD showed the most prominent inhibitory effects. We observed 73%, 65%, and 55% suppression of IL-6, TNF-α, and IL-12 in PD-pretreated cells compared with 68%, 81%, and 88% inhibition in Dex-treated cells (Figure 4A). Treatment with SP resulted in 60%, 60%, and 58% suppression of IL-6, TNF-α, and IL-12 compared with 60%, 12%, and 12% inhibition in case of SB treatment (Figure 4A). Our results define ERK and JNK as the major MAPKs that propagate TLR3-Trif activation into downstream proinflammatory responses. p38 MAPK contributed little to Poly (I:C)–mediated induction of TNF-α and IL-12.
Effect of MAPK inhibitors on TLR3- and TLR9-mediated proinflammatory cytokine secretion. Peritoneal macrophages were pretreated with the indicated concentrations of MAPK inhibitors SP 600125 (SP), PD 98059 (PD), and SB 203580 (SB) for 1 hour followed by stimulation with (A) Poly (I:C) or (B) CpG for 24 hours. The effect of MAPK inhibitors was compared with inhibitory effect of Dex on cytokine secretion. Macrophages pretreatment with Dex for 3 hours was followed by Poly (I:C) or CpG treatment for 24 hours. Concentrations of IL-6, TNF-α, and IL-12 in the culture media were measured. Data are shown as mean ± SEM; n = 4; *P < .001; **P < .01; and ***P < .05 for Dex or MAPK inhibitor(s)–treated macrophages compared with treatment group treated with Poly (I:C) or CpG only.
Effect of MAPK inhibitors on TLR3- and TLR9-mediated proinflammatory cytokine secretion. Peritoneal macrophages were pretreated with the indicated concentrations of MAPK inhibitors SP 600125 (SP), PD 98059 (PD), and SB 203580 (SB) for 1 hour followed by stimulation with (A) Poly (I:C) or (B) CpG for 24 hours. The effect of MAPK inhibitors was compared with inhibitory effect of Dex on cytokine secretion. Macrophages pretreatment with Dex for 3 hours was followed by Poly (I:C) or CpG treatment for 24 hours. Concentrations of IL-6, TNF-α, and IL-12 in the culture media were measured. Data are shown as mean ± SEM; n = 4; *P < .001; **P < .01; and ***P < .05 for Dex or MAPK inhibitor(s)–treated macrophages compared with treatment group treated with Poly (I:C) or CpG only.
We performed similar experiments to assess the contribution of individual MAPKs during TLR9-MyD88 engagement. Figure 4B shows 82%, 86%, and 80% suppression of IL-6, TNF-α, and IL-12, respectively, in SP-treated cells compared with 79%, 79%, and 83% inhibition in Dex-treated cells. Thus, JNK serves as an important target for the TLR9-MyD88 pathway for the induction of proinflammatory cytokines. SB had little or no effect on IL-6 and IL-12 and a moderate inhibitory effect on TNF-α secretion. Unexpectedly, all the cytokines exhibited resistance to PD treatment after TLR9 activation.
Effect of Dex on TLR3- and TLR9-mediated activation of MAPKs
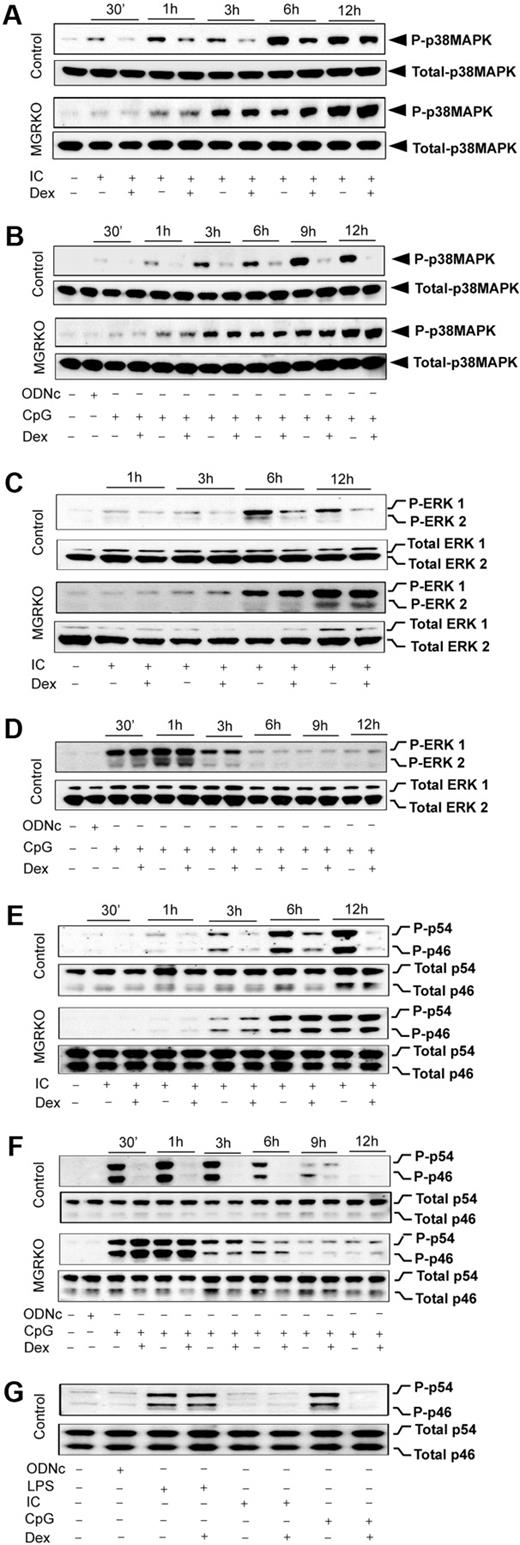
We reported glucocorticoid-mediated selective inhibition of p38 MAPK with no effect on ERK and JNK during TLR4 engagement with LPS.2 To examine the TLR3/9-specific suppression of MAPK by GR, we stimulated TLR3 and TLR9 either individually or together, and examined the effect of Dex on MAPK activation. Poly (I:C) resulted in induction of p38 MAPK phosphorylation at 30 minutes of stimulation, and treatment with Dex caused 2- to 2.5-fold suppression of p38 MAPK phosphorylation in control macrophages (Figure 5A). MGRKO macrophages displayed no suppression of p38 MAPK phosphorylation in the presence of Dex (Figure 5A). Figure 5B shows consistent suppression of CpG-mediated p38 MAPK phosphorylation by Dex. TLR9-mediated p38 MAPK phosphorylation was also found to be Dex resistant in MGRKO macrophages. Cotreatment of Dex and Poly (I:C) or CpG caused similar suppression of TLR3/9-mediated p38 MAPK phosphorylation (supplemental Figure 4A-B) as observed with Dex pretreatment (Figure 5A-B).
Dex suppresses TLR3- and TLR9-mediated activation of p38 MAPK. Peritoneal macrophages were administered with Dex 3 hours before Poly (I:C) or CpG treatment. Cells were treated with Poly (I:C) or CpG for the indicated periods of time. Cell lysates were analyzed by Western blot using (A-B) anti–phospho-p38 MAPK (P-p38MAPK) and total p38 MAPK, (C-D) anti–phospho-ERK1/2 (P-ERK1/2) and total ERK1/2, and (E-F) anti–phospho-JNK (P-p46/p54) and total JNK (Total p46/p54) antibodies. Representative of 3 to 4 independent experiments. (G) Comparative effect of Dex on JNK activation by TLR3 and TLR9 ligands. Macrophages stimulated with Poly (I:C), CpG, or LPS for 30 minutes with or without Dex pretreatment. Cell lysates were analyzed by Western blot using anti–phospho-JNK (P-p46/p54) and total JNK (p46/p54) antibodies. Representative of 3 independent experiments.
Dex suppresses TLR3- and TLR9-mediated activation of p38 MAPK. Peritoneal macrophages were administered with Dex 3 hours before Poly (I:C) or CpG treatment. Cells were treated with Poly (I:C) or CpG for the indicated periods of time. Cell lysates were analyzed by Western blot using (A-B) anti–phospho-p38 MAPK (P-p38MAPK) and total p38 MAPK, (C-D) anti–phospho-ERK1/2 (P-ERK1/2) and total ERK1/2, and (E-F) anti–phospho-JNK (P-p46/p54) and total JNK (Total p46/p54) antibodies. Representative of 3 to 4 independent experiments. (G) Comparative effect of Dex on JNK activation by TLR3 and TLR9 ligands. Macrophages stimulated with Poly (I:C), CpG, or LPS for 30 minutes with or without Dex pretreatment. Cell lysates were analyzed by Western blot using anti–phospho-JNK (P-p46/p54) and total JNK (p46/p54) antibodies. Representative of 3 independent experiments.
Dex has no effect on LPS-MyD88-Trif–mediated ERK phosphorylation in bone marrow–derived or peritoneal macrophages.2,25,35 To dissect the role of GR in regulating individual MyD88 or Trif pathways, we examined the effect of Dex on CpG- or Poly (I:C)–mediated activation of ERK. We found approximately 2-fold suppression of ERK phosphorylation by Dex 3 hours after Poly (I:C) treatment (Figure 5C). A more pronounced inhibitory effect of Dex was observed at later phases of ERK induction with no effect on ERK phosphorylation in MGRKO macrophages (Figure 5C). Simultaneous treatment with Dex and Poly (I:C) showed inhibitory effects of Dex on ERK phosphorylation (supplemental Figure 4C). In contrast, there was no effect of Dex on CpG-dependent ERK phosphorylation in either control or MGRKO macrophages (Figure 5D). We found that inhibition of ERK phosphorylation is indirect and results from inhibition of type 1 interferons, interferon-α (IFN-α), and IFN-β production and impairment of signaling through the IFN-α/β receptor (data not shown). To understand the role of type 1 interferons in regulating Poly (I:C)–induced MAPK activation, cells were pretreated with anti–IFN-α/β receptor 1 (IFNAR1) mouse monoclonal antibody (MARI) or isotype-matched control antibody (GIR) followed by stimulation with Poly (I:C). Supplemental Figure 5 shows that pretreatment with MARI abrogates Poly (I:C)–induced ERK phosphorylation.
Previously, we reported biphasic induction of JNK during TLR4 engagement with LPS, which was resistant to suppression by Dex in control macrophages.2 The kinetic profile of JNK induction after TLR3 or TLR9 engagement displayed monophasic activation (Figure 5E-F). Dex suppressed JNK phosphorylation (p44 and p54) in control macrophages as early as 1 hour after Poly (I:C) treatment, and more robustly at the later time points of treatment (Figure 5E). We observed robust phosphorylation of JNK after 30 minutes of CpG treatment, which was strongly suppressed by Dex pretreatment (Figure 5F). Cotreatment of Dex with Poly (I:C) or CpG resulted in equivalent inhibitory effect on ligand-induced JNK phosphorylation (supplemental Figure 4D-E). Figure 5G displays the relative effect of Dex on JNK activation after 30-minute stimulation of TLR4, TLR3, or TLR9 with their respective ligands. At this time point, induction of JNK by Poly (I:C) is minimal, as shown in Figure 5E. LPS-mediated JNK induction was completely Dex resistant, whereas CpG-dependent JNK phosphorylation was strongly Dex sensitive. Taken together, these results suggest a novel interaction between MyD88 and Trif pathways after TLR4 engagement that results in Dex-resistant JNK phosphorylation.
Effect of Dex on MAPK activation during TLR3/TLR9 coengagement and in Trif-deficient macrophages
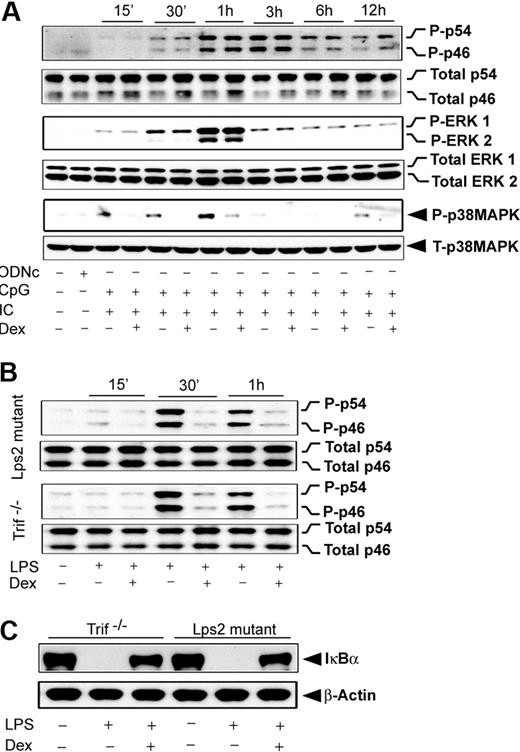
Our results revealed that induction of JNK by either MyD88- or Trif-selective mechanisms is Dex sensitive. In contrast, engagement of TLR4, which typically induces both of the signaling adapters, shows complete resistance to Dex during JNK activation (Figure 5G). To explore the possibility that coincident signals by MyD88 and Trif lead to Dex resistance, we evaluated the effect of Dex on MAPK activation during coengagement of TLR3 and TLR9 with their respective ligands. Figure 6A shows that JNK phosphorylation during cotreatment with Poly (I:C) and CpG was completely Dex resistant, which was similar to the case of TLR4 induction (Figure 5G). ERK was Dex resistant after costimulation (Figure 6B), in contrast to Dex sensitivity with Poly (I:C) alone (Figure 5C). Activation of p38 MAPK was Dex sensitive after costimulation (Figure 6A), as observed in the case of individually stimulated TLR4,2 TLR3, and TLR9. Interestingly, TLR4-dependent JNK activation was found to be strongly Dex sensitive in Trif-deficient macrophages stimulated with LPS. Robust suppression of LPS-mediated JNK phosphorylation was observed in Dex-treated macrophages isolated from Trif−/− or Lps2 mutant (Trif mutant) mice (Figure 6B).
Effect of Dex on MAPK activation during TLR3 and TLR9 coengagement. (A) Peritoneal macrophages were pretreated with Dex followed by Poly (I:C) and CpG cotreatment for the indicated periods of time. Cell lysates were analyzed by Western blot using anti–phospho-JNK (P-p46/p54) and total JNK (total p46/p54); anti–phospho-ERK1/2 (P-ERK1/2) and total ERK1/2; and anti–phospho-p38 MAPK (P-p38MAPK) and total p38 MAPK antibodies. (B) Effect of Dex on LPS-induced JNK activation in Lps2 mutant and Trif−/− macrophages. Peritoneal macrophages isolated from Lps2 mutant and Trif−/− mice treated with LPS for the indicated periods of time in the presence and absence of 100nM Dex. (C) Effect of Dex on LPS-induced IκBα degradation in Lps2 mutant and Trif−/− macrophages. Macrophages were treated with or without Dex for 3 hours followed by LPS for 30 minutes. Cytoplasmic extracts were analyzed by Western blotting using anti-IκBα and anti–β-actin antibodies. Representative of 2 to 3 independent experiments.
Effect of Dex on MAPK activation during TLR3 and TLR9 coengagement. (A) Peritoneal macrophages were pretreated with Dex followed by Poly (I:C) and CpG cotreatment for the indicated periods of time. Cell lysates were analyzed by Western blot using anti–phospho-JNK (P-p46/p54) and total JNK (total p46/p54); anti–phospho-ERK1/2 (P-ERK1/2) and total ERK1/2; and anti–phospho-p38 MAPK (P-p38MAPK) and total p38 MAPK antibodies. (B) Effect of Dex on LPS-induced JNK activation in Lps2 mutant and Trif−/− macrophages. Peritoneal macrophages isolated from Lps2 mutant and Trif−/− mice treated with LPS for the indicated periods of time in the presence and absence of 100nM Dex. (C) Effect of Dex on LPS-induced IκBα degradation in Lps2 mutant and Trif−/− macrophages. Macrophages were treated with or without Dex for 3 hours followed by LPS for 30 minutes. Cytoplasmic extracts were analyzed by Western blotting using anti-IκBα and anti–β-actin antibodies. Representative of 2 to 3 independent experiments.
Effect of Dex on NFκB activation TLR4 engagement and in Trif-deficient macrophages
After TLR4 engagement, we observed that NFκB activation is Dex resistant (Figure 3D-F), similar to the pattern observed for JNK. We explored whether MyD88-Trif coengagement would also be accountable for such NFκB resistance to Dex. Pretreatment with Dex markedly reduced the degradation of IκBα in Trif-deficient macrophages (Figure 6C). Densitometric analysis indicated 72% and 68% inhibition of IκBα degradation in Trif−/− and Lps2 mutant macrophages, respectively. In agreement with this result, we observed inhibition of IκBα phosphorylation by Dex pretreatment (data not shown).
Dex-mediated suppression of JNK activity during TLR3/TLR9 engagement is TAK1 dependent
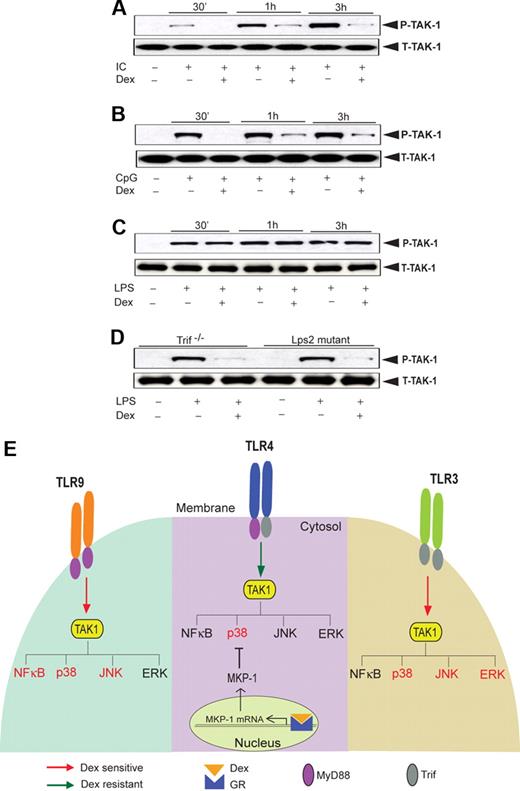
Next, we explored the activity of TAK1, an upstream kinase, which regulates JNK (and also NFκB) activity. TAK1 phosphorylation in Poly (I:C)–treated macrophages was significantly attenuated by Dex treatment (Figure 7A). Similarly, Dex inhibited CpG-MyD88–mediated TAK1 phosphorylation (Figure 7B). In contrast, we observed robust induction of TAK1 phosphorylation as early as 30 minutes after LPS treatment with no effect of Dex pretreatment (Figure 7C). Interestingly, we found marked suppression of LPS-mediated TAK1 phosphorylation in Dex-treated macrophages isolated from Trif−/− or Lps2 mutant mice (Figure 7D). These results are concordant with differential TAK1 regulation by Dex across different TLRs and implicate TAK1 as instrumental for glucocorticoid-mediated NFκB regulation (Figure 7E).
Effect of Dex on TLR-induced TAK1 activation. Macrophages were treated with Dex followed by (A) Poly (I:C), (B) CpG, and (C) LPS treatment for the indicated periods of time. Cell lysates were analyzed by Western blot using anti-TAK1 and total TAK1 antibodies. Representative of 2 to 3 independent experiments. (D) Effect of Dex on LPS-induced TAK1 phosphorylation in Lps2 mutant and Trif−/− macrophages. Peritoneal macrophages isolated from Lps2 mutant and Trif−/− mice treated with LPS for 30 minutes in the presence or absence of Dex. Cell lysates were analyzed by Western blot using anti-TAK1 and total TAK1 antibodies. Representative of 2 independent experiments. (E) Differential regulation of TLR-induced MAPK and NKκB activation by glucocorticoids. TAK1 induces activation of MAP kinases and NFκB irrespective of the nature of TLR ligation. However, activation of GR by Dex suppresses TAK1-induced MAPK and NFκB activation in a stimulus-specific manner. TLR4-MyD88-Trif–induced TAK1 activity is Dex resistant, and Dex attenuates p38 MAPK activity only via induction of MKP-1. TLR9-MyD88–induced TAK1 activity is Dex sensitive, and as such, NFκB, p38 MAPK, and JNK activity is as well. TLR3-Trif–induced TAK1 activity is also Dex sensitive, and Dex inhibits p38 MAPK, ERK, and JNK activity (NFκB is not significantly induced). Dex-sensitive TAK1 targets are designated in red font; Dex-resistant targets are in black font.
Effect of Dex on TLR-induced TAK1 activation. Macrophages were treated with Dex followed by (A) Poly (I:C), (B) CpG, and (C) LPS treatment for the indicated periods of time. Cell lysates were analyzed by Western blot using anti-TAK1 and total TAK1 antibodies. Representative of 2 to 3 independent experiments. (D) Effect of Dex on LPS-induced TAK1 phosphorylation in Lps2 mutant and Trif−/− macrophages. Peritoneal macrophages isolated from Lps2 mutant and Trif−/− mice treated with LPS for 30 minutes in the presence or absence of Dex. Cell lysates were analyzed by Western blot using anti-TAK1 and total TAK1 antibodies. Representative of 2 independent experiments. (E) Differential regulation of TLR-induced MAPK and NKκB activation by glucocorticoids. TAK1 induces activation of MAP kinases and NFκB irrespective of the nature of TLR ligation. However, activation of GR by Dex suppresses TAK1-induced MAPK and NFκB activation in a stimulus-specific manner. TLR4-MyD88-Trif–induced TAK1 activity is Dex resistant, and Dex attenuates p38 MAPK activity only via induction of MKP-1. TLR9-MyD88–induced TAK1 activity is Dex sensitive, and as such, NFκB, p38 MAPK, and JNK activity is as well. TLR3-Trif–induced TAK1 activity is also Dex sensitive, and Dex inhibits p38 MAPK, ERK, and JNK activity (NFκB is not significantly induced). Dex-sensitive TAK1 targets are designated in red font; Dex-resistant targets are in black font.
Discussion
In the present study, we evaluated the regulation of different members of the TLR family that preferentially use MyD88 or Trif to transmit activation signals to changes in cellular function. We hypothesized the glucocorticoid regulation of specific signaling pathways induced after TLR engagement would show different sensitivity to GR-mediated inhibition important for both actions of endogenous glucocorticoid and physiologic effects, and anti-inflammatory actions of pharmacologic doses. We found that both TLR3-Trif– and TLR9-MyD88–induced proinflammatory gene expression were sensitive to GR-mediated suppression. We reported a similar pattern of regulation of inflammatory genes by GR for TLR4, which uses both Trif and MyD88. Our in vivo data similarly show a significantly higher level of systemic cytokine in Poly (I:C)–injected (Figure 1E) or LPS-injected2 MGRKO mice, implicating an essential role for endogenous glucocorticoids in controlling TLR-mediated inflammatory responses.
We found that glucocorticoids selectively impair NFκB activation in primary macrophages for isolated MyD88 engagement but they have little effect on NFκB during Trif or MyD88-Trif coincident engagement. After TLR9-MyD88 activation, glucocorticoids interfere with the phosphorylation of IκBα, which in turn inhibits degradation of IκBα, and phosphorylation and nuclear translocation of p65. The inhibitory effect of Dex on TLR9-MyD88–induced NFκB activation is later than anticipated. Earlier studies indicated that glucocorticoids enhance synthesis of IκBα depending on the cell type and target gene.36,37 In our case, instead of direct inhibition of IκBα degradation, Dex may enhance the synthesis of IκBα and thereby dampen oscillation of NFκB activity. TLR4-MyD88-Trif engagement strongly induces NFκB activity but resists GR-mediated suppression of IκBα degradation or NFκB DNA-binding activity. These findings are in agreement with a previous report that demonstrated GR disruption of p65–interferon regulatory factor 3 interaction, but not p65 DNA binding itself.38 Thus, the majority of Dex modulation of TLR4 signaling can be ascribed to a moderate contribution of NFκB inhibition along with induction of MKP-1 and inhibition of p38 MAPK, as we and others have previously reported.2,25,26 Our results indicate proinflammatory processes associated primarily with TLR3-Trif activation would likely not benefit from treatment by NFκB inhibitors that are currently in clinical trials, as NFκB plays little role downstream of this receptor. Inflammatory disorders that involve TLR9-MyD88, in contrast, would be candidates for intervention with these drugs.
Engagement of TLR3, TLR9, or TLR4 results in augmented phosphorylation of p38 MAPK, JNK, and ERK (Figure 5 and Bhattacharyya et al2 ). For TLR3 and TLR9, phosphorylation of p38 MAPK and JNK was effectively inhibited by Dex, independent of whether their peak induction occurred within minutes of cell activation or after hours of stimulation (Table 2). Taken in the context of our previous results with MAPK activation after TLR4 ligation, p38 MAPK appears to be glucocorticoid suppressible2 downstream of MyD88, Trif, or combined MyD88-Trif activation (Figure 7E). Our new findings demonstrate a novel pattern of regulation of JNK, however, depending upon the nature of TLR activation. Individual activation of either MyD88 or Trif confers robust JNK induction that is glucocorticoid suppressible (Figure 7E). In contrast, coincident activation of MyD88 and Trif by either TLR4 or combined treatment with ligands that bind TLR3 and TLR9 confers a state of glucocorticoid resistance to JNK activation. Moreover, results from Trif-deficient macrophages show ablation of Trif signaling during TLR4 engagement restores glucocorticoid sensitivity to JNK, indicating MyD88-Trif coincident signaling mediates glucocorticoid resistance to JNK. JNK activation is responsible for a substantial contribution of cytokine production from primary macrophages in vitro, as shown by our experiments with MAPK isoform–specific inhibitors. The pattern of glucocorticoid resistance of ERK activation depending upon TLR isoform activated is unique, with TLR3 showing Dex-suppressible ERK phosphorylation, whereas TLR3 and TLR9 exhibit Dex-resistant ERK activation. We found that the ERK sensitivity to glucocorticoids associated with TLR3-Trif was an indirect effect of GR suppression of IFN-α and IFN-β (S.B. and L.J.M., unpublished results, December 2007). Neutralization of IFNAR1 with anti-IFNAR1 mouse monoclonal antibody indicated selective inhibitory effect on TLR3-Trif–induced ERK activation but no effect on other members of the MAPK family.
Temporal sensitivity of MAPKs to glucocorticoids
| Ligand . | p38 MAPK . | JNK . | ERK . | |||
|---|---|---|---|---|---|---|
| Early . | Late . | Early . | Late . | Early . | Late . | |
| LPS | ++ | +++ | R | R | R | R |
| Poly (I:C) | ++ | + | + | +++ | + | +++ |
| CpG | ++ | +++ | R | R | +++ | + |
| Ligand . | p38 MAPK . | JNK . | ERK . | |||
|---|---|---|---|---|---|---|
| Early . | Late . | Early . | Late . | Early . | Late . | |
| LPS | ++ | +++ | R | R | R | R |
| Poly (I:C) | ++ | + | + | +++ | + | +++ |
| CpG | ++ | +++ | R | R | +++ | + |
Degree of GC sensitivity: high (+++), moderate (++), low (+), GR resistant (R).
MAPK indicates mitogen-activated protein kinase; JNK, Janus kinase; ERK, extracellular signal-related kinase; LPS, lipopolysaccharide; and GC, glucocorticoid.
The physiologic ramifications of the effects of coincident MyD88-Trif signaling are substantial. First, as has been noted in several experiments, TLR4 activation by LPS produces very high levels of cytokines in vivo and in vitro2 compared with the other individually activated TLRs. Although glucocorticoids are capable for suppressing approximately 70% to 80% of LPS-stimulated cytokine production from macrophages, the absolute suppressed levels still exceed those associated with stimulation of the other TLRs that individually signal through MyD88 or Trif. Induction of both MyD88- and Trif-dependent signals is evident in several pathologic states, for example, both TLR3 and TLR9 are activated during herpes simplex virus and murine cytomegalovirus infection. Moreover, involvement of TLR3 and TLR9 has been implicated in diabetes,39,40 cardiomyopathy,41 asthma, and chronic obstructive pulmonary disease.42,43
Our data indicate JNK and NFκB resistance to glucocorticoids during coincident MyD88-Trif engagement for LPS or Poly (I:C)–CpG cotreatment. Such a selective resistance of the 2 most important members of TLR inflammosome led us to hypothesize differential sensitivity of an upstream regulator of MAPK and NFκB pathways to glucocorticoids. A recent report demonstrated that TAK1 regulates JNK and p38MAPK activity in a direct manner and ERK activity via IKK-β.44 However, TAK1 is dispensable for LPS-induced activation of ERK,45 and its role in regulating p38 MAPK is yet to be fully delineated. Several groups have found that JNK and NFκB are the major targets of TAK1.46-48 We found TAK1 phosphorylation is glucocorticoid resistant after TLR4 engagement with LPS (Figure 7G). Intriguingly, TAK1 activation is strongly glucocorticoid sensitive for TLR4 engagement in Trif-deficient macrophages and also for individual TLR3 or TLR9 engagement, and parallels the downstream effects on JNK and IκB (Figure 7G). Taken together, our findings suggest that TAK1 resistance to glucocorticoid during MyD88-Trif coincident signaling may be responsible for downstream resistance of both JNK and NFκB and pathways. The activation of TAK1 follows a complex cascade of signaling events involving interleukin-1 receptor–associated kinase M (IRAK-M), IRAK-2, IRAK-4, TNF receptor–associated factor 6 (TRAF-6), and TAB 1/2/3. Sequential phosphorylation of IRAK4 and IRAK1 activates TRAF6, which promotes the synthesis of lysine 63–linked polyubiquitin chains. TAB2 contains a zinc finger domain that binds to polyubiquitin chains and thereby facilitates TAK1 activation.20 A recent report, however, indicated that lysine 63–linked polyubiquitin chains, which are not conjugated to an intermediate protein, can directly activate TAK1.49 The biochemical pathway(s) accountable for the differential suppressive effect of glucocorticoid on TLR-induced TAK1 activation promises to be an important avenue for future investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Jacek Hawiger for review of the paper.
This work was supported by National Institutes of Health grant AI50653 and Pfizer Inc.
National Institutes of Health
Authorship
Contribution: S.B. designed and performed experiments and participated in writing the paper; C.K.R., S.K.V., and C.K. performed experiments; M.C. and R.D.S. provided essential reagents and designed experiments; and L.J.M. designed experiments, generated essential reagents, and participated in writing the paper.
Conflict-of-interest disclosure: L.J.M. received a grant from Pfizer Inc. The remaining authors declare no competing financial interests.
Correspondence: Sandip Bhattacharyya, Vanderbilt University Medical Center, 2215 Garland Ave, MRBIV, Rm 1115, Nashville, TN 37232; e-mail: sandip.bhattacharyya@vanderbilt.edu.