Abstract
Using inverse polymerase chain reaction, we identified CD44, located on chromosome 11p13, as a novel translocation partner of IGH in 9 of 114 cases of gastric, nongastric extranodal, follicular, and nodal diffuse large B-cell lymphoma (DLBCL). Notably, these translocations involving IGHSμ were detected in follicular lymphomas and exclusively in germinal center B cell-ike (GCB)–DLBCLs. CD44 is not expressed in reactive GC B cells. The IGHSμ/CD44 translocations substitute Sμ for the CD44 promoter and remove exon 1 of CD44, resulting in the overexpression of Iμ-CD44 hybrid mRNA transcripts activated from derivative 11 that encode a new CD44 variant lacking the leader peptide and with a unique C-terminus (CD44ΔEx1). When overexpressed in vitro in the CD44− GCB-DLBCL cell line BJAB, CD44ΔEx1–green fluorescent protein localized to the cytoplasm and nucleus, whereas CD44s–green fluorescent protein (standard form) localized to the plasma membrane. The ectopic expression of CD44ΔEx1 in BJAB cells enhanced their proliferation rate and clonogenic ability, indicating a possible pathogenic role of the translocation.
Introduction
Mature B-cell non-Hodgkin lymphomas (NHLs) are often associated with chromosomal translocations involving IGH at chromosome band 14q32.1–5 V(D)J rearrangement and class switch recombination (CSR) are IG rearrangement processes that occur during B-cell development.6–9 In CSR, DNA breakage is mediated by activation-induced cytidine deaminase in the germinal centers (GCs) of secondary follicles.10,11 Any aberrant rearrangement during the DNA breakage and repair processes composed of V(D)J rearrangement and CSR may consequently generate a chromosomal translocation.12,13 IGH translocations contribute to the pathogenesis of B-cell lymphomas via deregulated expression of the genes located at the partner chromosome locus partly because of the presence of potent B cell–specific transcriptional enhancers within the IGH gene locus.1–5
Gastric B-cell lymphomas usually exhibit chromosomal translocations involving IGH.14–16 Molecular cloning of IGH translocation breakpoints has been successfully used to identify novel cancer-related genes in B-cell lymphomas.17,18 In this study, we used inverse polymerase chain reaction (PCR)18 to identify a novel translocation involving the 5′ Sμ region of the IGH gene (IGHSμ) and CD44 (located at chromosome 11p13) in gastric as well as other mature B-cell NHLs. Functional studies suggest a possible pathogenic role of this IGHSμ/CD44 translocation in mature B-cell malignancies.
Methods
Detection of translocations involving the IGHSμ by inverse PCR
Genomic DNA extracted from frozen sections of tumor specimens from gastric19 and other mature B-cell NHL cases (supplemental Table 1, available on the Blood website; see the Supplemental Materials link at the top of the online article) was digested with HindIII, and long-distance inverse PCR was performed to detect translocations involving the IGHSμ region using the primers SAE/JXE, followed by SAI/JXI18 (supplemental Table 2).
Overexpression of CD44ΔEx1-GFP tagged protein in transfected BJAB cells
CD44ΔEx1 (CD44 variant mRNA lacking exon 1) and CD44s (standard form; wild-type) cDNAs were amplified by reverse-transcribed (RT)–PCR (supplemental Table 2) from total RNA extracted from a case of gastric lymphoma with the IGHSμ/CD44 translocation (GL47) and from the peripheral blood lymphocytes of a healthy volunteer; they were then cloned in-frame with green fluorescent protein (GFP) at the C-terminus of the pmaxFP-Green-N vector (Amaxa) and transfected into the CD44− GC B cell–like (GCB)–diffuse large B-cell lymphoma (DLBCL) cell line BJAB by nucleofection (Amaxa). The subcellular localization of the GFP-tagged proteins was assessed by confocal laser microscopy. The effect of overexpressing CD44ΔEx1 on the growth rate of stably transfected BJAB cells in vitro was determined using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (Promega); clonogenic capacity was assessed with colony formation assays in methylcellulose.20
Results and discussion
Using an inverse PCR approach, we identified a novel translocation, t(11;14)(p13;q32), involving the 5′Sμ region of IGH and CD44 at 11p13 in 4 of 49 gastric lymphomas, 2 of 7 nongastric extranodal lymphomas, 2 of 30 nodal follicular lymphomas (FLs), and 1 of 26 nodal DLBCLs (Figure 1A; supplemental Table 1). This IGHSμ/CD44 translocation juxtaposes the IGHSμ enhancer to the 5′ regulatory region of CD44 in a tail-to-head orientation, leading to the removal of exon 1 of CD44 (Figure 1B). Functional switch recombination was also detected in the untranslocated allele in all translocation-positive cases. Further studies of all lymphoma cases by interphase fluorescence in situ hybridization (FISH) with CD44 break-apart probes (supplemental data) confirmed breakage at the CD44 locus in the 9 IGHSμ/CD44 translocation-positive cases identified by inverse PCR. No additional cases were identified with breakage at the CD44 locus among the inverse PCR− cases (Figure 1C).
CD44 was identified as a novel translocation partner of IGH in mature B-cell NHLs. (A) Ethidium bromide–stained gel showing the IGHSμ/CD44 translocation product resulting from illegitimate switch recombination (indicated by the arrow) and a self-rearrangement band (indicated by the arrowhead) resulting from the fusion of Sμ to Sγ3 of IGH (functional CSR) in a representative case (gastric GCB-type DLBCL; GL47) with an IGHSμ/CD44 translocation detected by inverse PCR. A vertical white line has been inserted to indicate a repositioned gel lane. (B) Ideogram showing that IGHSμ/CD44 translocations detected in mature B-cell lymphomas result in a classic promoter substitution. The positions and orientations of the primers (SAE/JXE followed by SAI/JXI) used for inverse PCR are indicated. The CD44ΔEx1 mRNA transcripts were activated from derivative 11. The intron/exon map of human CD44 is based on GenBank sequences (accession no. NT009237, DNA; and NM_000610, mRNA).21 Exons 1 to 5 and 15 to 18 (shown as black boxes) are the constitutive exons that code for CD44s (standard from). Exons 6 to 14 correspond to the alternatively spliced variant exons within the extracellular domain (shown as blue boxes). Exon 1 encodes the leader peptide (LP), exon 17 encodes the transmembrane domain (TM), and exon 18 encodes a short cytoplasmic domain. The open arrowhead indicates the coding initiation start site. (C) Interphase FISH confirmed breakage at the CD44 locus in IGHSμ/CD44 translocation-positive cases identified by inverse PCR. Break-apart FISH was performed using 2 BAC clones as probes: clone RP4-607I7 (151 kb) included the 5′ portion of intron 1 of CD44, whereas clone RP4-683L5 (135 kb) was approximately 64.5 kb away from the 3′ end of CD44 (top panel). Representative nuclei from cases with (GL47) or without (GL2) breakage of the CD44 locus are shown, exhibiting one orange, one green, and one yellow (colocalized orange and green) fusion signal pattern (1O1G1F). Vertical and horizontal white lines have been inserted to indicate different nuclei repositioned images. The images were captured by the Leica DMRBE (Leica Microsystem Wetzler GmbH) fluorescence microscope (100×/0.7 NA oil objective). The fluorescent images were imported and created by Cytovision software (Applied Imaging). (D) The IGHSμ/CD44 translocation breakpoints were located within small regions of IGHSμ and at the 5′ end of intron 1 of CD44 in mature B-cell lymphomas. The sequences shown are from GenBank (IGHSμ accession no. NG_001019.3; CD44: accession no. AL356215.11).21 The relative position of the 2 sets of seminested PCR primers used to confirm IGHSμ/CD44 translocations by direct PCR is shown. (E) Chromosomal locations of all IGH translocation partners (annotated genes only) reported in B-cell lymphomas. The CD44 gene (shown in bold) located at 11p13 was identified in this study as the first cell adhesion molecule22 involved in IGH chromosomal translocations in B-cell lymphomas. GL indicates gastric lymphoma; ENL, nongastric extranodal lymphoma; and NL, nodal DLBCL. *Gastric mucosa-associated lymphoid tissue (MALT) lymphoma. **Gastric composite DLBCL with residual MALT lymphoma (DLCLML).
CD44 was identified as a novel translocation partner of IGH in mature B-cell NHLs. (A) Ethidium bromide–stained gel showing the IGHSμ/CD44 translocation product resulting from illegitimate switch recombination (indicated by the arrow) and a self-rearrangement band (indicated by the arrowhead) resulting from the fusion of Sμ to Sγ3 of IGH (functional CSR) in a representative case (gastric GCB-type DLBCL; GL47) with an IGHSμ/CD44 translocation detected by inverse PCR. A vertical white line has been inserted to indicate a repositioned gel lane. (B) Ideogram showing that IGHSμ/CD44 translocations detected in mature B-cell lymphomas result in a classic promoter substitution. The positions and orientations of the primers (SAE/JXE followed by SAI/JXI) used for inverse PCR are indicated. The CD44ΔEx1 mRNA transcripts were activated from derivative 11. The intron/exon map of human CD44 is based on GenBank sequences (accession no. NT009237, DNA; and NM_000610, mRNA).21 Exons 1 to 5 and 15 to 18 (shown as black boxes) are the constitutive exons that code for CD44s (standard from). Exons 6 to 14 correspond to the alternatively spliced variant exons within the extracellular domain (shown as blue boxes). Exon 1 encodes the leader peptide (LP), exon 17 encodes the transmembrane domain (TM), and exon 18 encodes a short cytoplasmic domain. The open arrowhead indicates the coding initiation start site. (C) Interphase FISH confirmed breakage at the CD44 locus in IGHSμ/CD44 translocation-positive cases identified by inverse PCR. Break-apart FISH was performed using 2 BAC clones as probes: clone RP4-607I7 (151 kb) included the 5′ portion of intron 1 of CD44, whereas clone RP4-683L5 (135 kb) was approximately 64.5 kb away from the 3′ end of CD44 (top panel). Representative nuclei from cases with (GL47) or without (GL2) breakage of the CD44 locus are shown, exhibiting one orange, one green, and one yellow (colocalized orange and green) fusion signal pattern (1O1G1F). Vertical and horizontal white lines have been inserted to indicate different nuclei repositioned images. The images were captured by the Leica DMRBE (Leica Microsystem Wetzler GmbH) fluorescence microscope (100×/0.7 NA oil objective). The fluorescent images were imported and created by Cytovision software (Applied Imaging). (D) The IGHSμ/CD44 translocation breakpoints were located within small regions of IGHSμ and at the 5′ end of intron 1 of CD44 in mature B-cell lymphomas. The sequences shown are from GenBank (IGHSμ accession no. NG_001019.3; CD44: accession no. AL356215.11).21 The relative position of the 2 sets of seminested PCR primers used to confirm IGHSμ/CD44 translocations by direct PCR is shown. (E) Chromosomal locations of all IGH translocation partners (annotated genes only) reported in B-cell lymphomas. The CD44 gene (shown in bold) located at 11p13 was identified in this study as the first cell adhesion molecule22 involved in IGH chromosomal translocations in B-cell lymphomas. GL indicates gastric lymphoma; ENL, nongastric extranodal lymphoma; and NL, nodal DLBCL. *Gastric mucosa-associated lymphoid tissue (MALT) lymphoma. **Gastric composite DLBCL with residual MALT lymphoma (DLCLML).
The 9 different IGHSμ/CD44 translocation breakpoints were distributed within small regions of IGHSμ and at the 5′ end of intron 1 of CD44 (Figure 1D). Microhomology sequences found at the junctional breakpoints of all the IGHSμ/CD44 translocations suggest that these translocations were facilitated by homologous sequences present on both chromosomes (supplemental Figure 1).12,13 The CD44 gene at 11p13 is the first cell adhesion molecule22 known to be involved in an IGH translocation in B-cell lymphomas (Figure 1E). CSR occurs in antigen-stimulated B cells in the GC of secondary follicles.11 Notably, 2 FL cases were detected with IGHSμ/CD44 translocations, and all 4 translocation-positive DLBCL cases were GCB-type DLBCLs (supplemental Table 1), suggesting that these translocations resulted from illegitimate CSR occurring in GC B cells. This correlation is plausible, given that a high proportion of GCB-DLBCLs have undergone immunoglobulin class switching and that these tumors are trapped in the differentiation stage of the GC.23–25
As the principal molecular consequences of an IGH translocation is the deregulated expression of the partner gene,1–5 CD44 mRNA expression was then studied by semiquantitative RT-PCR. RT-PCR primers across exons 2 and 18 (supplemental Table 2) designed to amplify all CD44 mRNA variants26 were used to monitor CD44 expression in microdissected CD10+ GC B cells from reactive tonsils27 and the 9 translocation-positive cases (supplemental data). CD44 mRNA was either minimally or not expressed in CD10+ microdissected reactive GC B cells (Figure 2A), consistent with the results of Alizadeh et al who showed by gene expression profiling that CD44 mRNA expression is very low in reactive GC B cells.28 Our results suggest that this translocation caused overexpression of a new CD44 variant (CD44ΔEx1) activated from derivative 11 in all 6 IGHSμ/CD44 translocation-positive GC-derived lymphomas (Figure 2A).
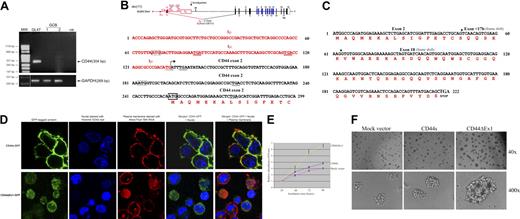
Subcellular localization and growth effects of the CD44ΔEx1 variant overexpressed in the CD44− GCB-DLBCL cell line BJAB. (A) No CD44 mRNA was detected by RT-PCR performed using primers CD44-F (exon 2) and CD44-R (exon 18) in total RNA extracted from the microdissected CD10+ GC B cells from reactive tonsils (2 preparations), whereas all the IGHSμ/CD44 translocation-positive cases showed the presence of CD44ΔEx1 mRNA; a representative translocation-positive case is shown (GL47). A vertical white line has been inserted in the bottom panel to indicate a repositioned gel lane. (B) 5′-RACE analysis detected the Iμ-CD44ΔEx1 hybrid transcripts in all 9 IGHSμ/CD44 translocation-positive cases; the 5′-RACE sequence (299 bp) from a representative translocation-positive case is shown (GL47). Stop codons (TGA) are underlined. The Iμ-CD44ΔEx1 ORF is not predicated to result in a fusion protein, as the CD44 exon 2 has an in-frame stop codon upstream of the predicted start codon ATG at nucleotide number 254 (boxed by solid line). The Iμ-CD44ΔEx1 ORF is predicted to encode for the CD44ΔEx1 protein starting from the ATG at nucleotide 254 (boxed by solid line) with a Kozak sequence29 ; the ATG codon at nucleotide 183 (boxed by dotted line) has a downstream in-frame stop codon (underlined by dotted line). (C) Nucleotide sequences of the CD44ΔEx1 ORF (219 bp) and predicted amino acid sequences of the protein (73 amino acids) are shown. Exon boundaries are marked with an arrow. Compared with CD44s (standard form; wild-type), the resultant CD44ΔEx1 protein lacks the first 62 amino acids at the N-terminus, including the 20 amino acids of the leader peptide. The CD44ΔEx1 protein also lacks the C-terminus of CD44s resulting from an altered reading frame starting from the junction of exons 2 and < 17b, which eliminates the transmembrane and intracellular domains. (D) When cloned in-frame with GFP at the C-terminus of the pmaxFP-Green-N vector and overexpressed in BJAB cells, CD44ΔEx1-GFP tagged protein was localized in the cytoplasm and nucleus instead of the plasma membrane, where CD44s-GFP was expressed. Representative images of cells taken by confocal laser microscopy are shown. The images were captured by the Zeiss LSM 510 (Carl Zeiss Microimaging GmbH) laser scanning microscope system (60× oil objective). The images in superimposed and split-image mode were displayed and exported as imaging files by LSM Image Browser software (Carl Zeiss Microimaging GmbH). Merged images were generated from the original confocal images using Adobe Photoshop software. (E-F) Overexpression of CD44ΔEx1 in stably transfected BJAB cells significantly enhanced the cell proliferation rate as recorded over 5 days of incubation (mean values of 3 separate experiments ± SD are shown; E) and clonogenic ability in methylcellulose as monitored after 6 days of incubation (F). The images in panel F were captured by the Nikon Eclipse TS100 inverted microscope (Nikon) and the Nikon DSU1 0507 11331 digital camera system and the images were acquired by ACT-2U software. The magnifications are written at the right side of each panel. Results similar to those in panels D through F were obtained when CD44ΔEx1 cloned in-frame with GFP at the N-terminus of the pmaxFP-Green-C vector was used for the aforementioned functional studies (results not shown).
Subcellular localization and growth effects of the CD44ΔEx1 variant overexpressed in the CD44− GCB-DLBCL cell line BJAB. (A) No CD44 mRNA was detected by RT-PCR performed using primers CD44-F (exon 2) and CD44-R (exon 18) in total RNA extracted from the microdissected CD10+ GC B cells from reactive tonsils (2 preparations), whereas all the IGHSμ/CD44 translocation-positive cases showed the presence of CD44ΔEx1 mRNA; a representative translocation-positive case is shown (GL47). A vertical white line has been inserted in the bottom panel to indicate a repositioned gel lane. (B) 5′-RACE analysis detected the Iμ-CD44ΔEx1 hybrid transcripts in all 9 IGHSμ/CD44 translocation-positive cases; the 5′-RACE sequence (299 bp) from a representative translocation-positive case is shown (GL47). Stop codons (TGA) are underlined. The Iμ-CD44ΔEx1 ORF is not predicated to result in a fusion protein, as the CD44 exon 2 has an in-frame stop codon upstream of the predicted start codon ATG at nucleotide number 254 (boxed by solid line). The Iμ-CD44ΔEx1 ORF is predicted to encode for the CD44ΔEx1 protein starting from the ATG at nucleotide 254 (boxed by solid line) with a Kozak sequence29 ; the ATG codon at nucleotide 183 (boxed by dotted line) has a downstream in-frame stop codon (underlined by dotted line). (C) Nucleotide sequences of the CD44ΔEx1 ORF (219 bp) and predicted amino acid sequences of the protein (73 amino acids) are shown. Exon boundaries are marked with an arrow. Compared with CD44s (standard form; wild-type), the resultant CD44ΔEx1 protein lacks the first 62 amino acids at the N-terminus, including the 20 amino acids of the leader peptide. The CD44ΔEx1 protein also lacks the C-terminus of CD44s resulting from an altered reading frame starting from the junction of exons 2 and < 17b, which eliminates the transmembrane and intracellular domains. (D) When cloned in-frame with GFP at the C-terminus of the pmaxFP-Green-N vector and overexpressed in BJAB cells, CD44ΔEx1-GFP tagged protein was localized in the cytoplasm and nucleus instead of the plasma membrane, where CD44s-GFP was expressed. Representative images of cells taken by confocal laser microscopy are shown. The images were captured by the Zeiss LSM 510 (Carl Zeiss Microimaging GmbH) laser scanning microscope system (60× oil objective). The images in superimposed and split-image mode were displayed and exported as imaging files by LSM Image Browser software (Carl Zeiss Microimaging GmbH). Merged images were generated from the original confocal images using Adobe Photoshop software. (E-F) Overexpression of CD44ΔEx1 in stably transfected BJAB cells significantly enhanced the cell proliferation rate as recorded over 5 days of incubation (mean values of 3 separate experiments ± SD are shown; E) and clonogenic ability in methylcellulose as monitored after 6 days of incubation (F). The images in panel F were captured by the Nikon Eclipse TS100 inverted microscope (Nikon) and the Nikon DSU1 0507 11331 digital camera system and the images were acquired by ACT-2U software. The magnifications are written at the right side of each panel. Results similar to those in panels D through F were obtained when CD44ΔEx1 cloned in-frame with GFP at the N-terminus of the pmaxFP-Green-C vector was used for the aforementioned functional studies (results not shown).
5′-Rapid amplification of cDNA ends (RACE) analysis (supplemental data) identified Iμ-CD44ΔEx1 hybrid mRNA transcripts in all 9 lymphomas with IGHSμ/CD44 translocation, resulting from splicing of the Iμ exon upstream of Sμ to exon 2 of CD44 (Figure 2B). Similar Iμ-BCL6 and Iμ-MMSET hybrid transcripts associated with t(3;14)23 and t(4;14)30,31 translocations have been previously described. Because CD44 exon 2 has an in-frame stop codon upstream of the predicted translational initiation codon ATG at nucleotide 254, the Iμ transcripts that splice to CD44 exon 2 would not result in a chimeric fusion protein, and the Iμ-CD44ΔEx1 ORF would encode for the CD44ΔEx1 protein starting from the ATG at nucleotide 254 with a strong Kozak sequence29 (Figure 2C). The new CD44ΔEx1 variant showed some similarity to CD44v532 ; but it lacked the leader peptide, and the 3′ end of exon 2 of CD44ΔEx1 was spliced into a new internal splice acceptor site present within exon 17b, altering the reading frame (Figure 2C).
Notably, the IGHSμ/CD44 translocation was detected in 4 patients with advanced (stages II-IV) gastric, follicular, and nodal DLBCL. CD44 is a cell adhesion molecule and signaling regulator.22 When overexpressed in vitro in the CD44− GCB-DLBCL cell line BJAB, CD44s-GFP was localized to the plasma membrane, whereas CD44ΔEx1-GFP was present in the cytoplasm and nucleus (Figure 2D). The ectopic expression of CD44ΔEx1 resulted in a significant increase in BJAB cell growth (Figure 2E) and clonogenicity in methylcellulose (Figure 2F).
Because of the unavailability of antibodies for the new CD44ΔEx1 isoform, immunohistochemical analysis to confirm the localization of CD44ΔEx1 in vivo could not be performed on the lymphoma biopsy specimens of IGHSμ/CD44 translocation-positive cases expressing the Iμ-CD44ΔEx1 hybrid mRNA. However, 7 of 9 translocation-positive lymphomas showed MIB1 (Ki-67 antibody; a proliferation marker) nuclear staining in the tumor cells (supplemental Figure 2), supporting the in vitro results showing that CD44ΔEx1 overexpression gives cells a proliferation advantage, and suggesting a possible pathogenic role of the IGHSμ/CD44 translocation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgment
This work was supported by the Research Grants Council of Hong Kong Special Administrative Region, China (grant HKU 7448/04M; G.S. and R.H.S.L.).
Authorship
Contribution: X.-T.H. and Y.-W.C. designed and performed experiments, interpreted data, and drafted the manuscript; A.C.T.L., K.-Y.W., T.S.K.W., M.L.Y.W., K.-K.C., and T.G. performed experiments; W.-Y.A., C.-S.C., Y.-L.K., and R.H.S.L. selected lymphoma cases; K.-M.C. provided gastric lymphoma surgical specimens; F.L., L.S., and W.W.L.C. selected tumor blocks and analyzed the immunohistochemistry slides; Q.T. and L.L. advised on the overall design of the study; C.-C.S. and L.C.C. analyzed interphase FISH data; and G.S. designed and supervised the study and finalized the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gopesh Srivastava, Department of Pathology, University of Hong Kong, Pokfulam, Hong Kong; e-mail: gopesh@pathology.hku.hk.
References
Author notes
X.-T.H. and Y.-W.C. contributed equally to this study.